Abstract
Nisin is a class I bacteriocin (lantibiotic), which is employed by the food and veterinary industries and exhibits potent activity against numerous pathogens. However, this activity could be further improved through the targeting and inhibition of factors that contribute to innate nisin resistance. Here we describe a novel locus, lmo1967, which is required for optimal nisin resistance in Listeria monocytogenes. The importance of this locus, which is a homologue of the tellurite resistance gene telA, was revealed after the screening of a mariner random mutant bank of L. monocytogenes for nisin-susceptible mutants. The involvement of telA in nisin resistance was confirmed through an analysis of a nonpolar deletion mutant. In addition to being 4-fold-more susceptible to nisin, the ΔtelA strain was also 8-fold-more susceptible to gallidermin and 2-fold-more susceptible to cefuroxime, cefotaxime, bacitracin, and tellurite. This is the first occasion upon which telA has been investigated in a Gram-positive organism and also represents the first example of a link being established between a telA gene and resistance to cell envelope-acting antimicrobials.
Nisin is a ribosomally synthesized cationic antimicrobial peptide that has been employed commercially for over 50 years in the preservation of foodstuffs (22) and, more recently, as an antimastitis agent (15). It also exhibits great potency against a number of human clinical pathogens, including many multidrug-resistant strains (33), and as a consequence of the continually diminishing options available to clinicians when targeting such microorganisms, the application of nisin has been the subject of renewed attention. Nisin is the prototypical example of the class I group of bacteriocins, which are also known as lantibiotics by virtue of the presence of unusual posttranslationally introduced structures known as lanthionines (12). In addition to being the most thoroughly characterized lantibiotic, nisin is also an example of a cell envelope-acting antimicrobial, acting through a combination of inhibiting peptidoglycan synthesis and forming pores in the cell membrane of target cells (3, 4, 18, 45).
Despite the potency of nisin, there is evidence that suggests that its activity would be even greater were it not for factors that contribute to the innate resistance of some target microorganisms; the deletion of virR and mprF is known to result in 32- and 16-fold reductions in resistance to nisin in Listeria monocytogenes (9). For clarity, we discriminate between the mechanisms underpinning acquired resistance (resistance occurring in a formerly susceptible strain) and innate resistance (resistance intrinsically associated with particular genera or species). One example of a system contributing to innate resistance is DltA, which is required for the d-alanyl decoration of teichoic acid in the cell wall of many Gram-positive microorganisms. Its role in antimicrobial resistance was first noted when a disruption of dltA resulted in the sensitization of Staphylococcus aureus to the lantibiotic gallidermin as a consequence of a reduced capacity to repulse positively charged compounds (30). This susceptibility and, indeed, a susceptibility to a wider range of cationic antimicrobial peptides (CAMPs), including nisin, defensins, vancomycin, polymyxin B, and colistin, are also apparent in dltA mutants of Streptococcus pneumoniae, Streptococcus agalactiae, Enterococcus faecalis, and Listeria monocytogenes (14, 20, 25, 30, 35). An altered cell envelope charge, in this case due to the nonlysinylation of membrane phospholipids, is also the basis for the enhanced susceptibility of mprF mutants of S. aureus, L. monocytogenes, and Bacillus anthracis to nisin and other CAMPs (29, 36, 42). Unsurprisingly, eliminating the VirR regulator component of the two-component signal transduction system (VirRS) that regulates the expression of both dltA and mprF in L. monocytogenes also impacts susceptibility to CAMPs (23, 42). Other loci that play a role in the innate resistance of Gram-positive bacteria to nisin include lisRK, lmo0327, pbp2229, sigB, graS, nsr, and abcAB (1, 11, 16, 24, 34, 37, 40). There have also been a number of loci linked with acquired resistance to nisin through gene expression-based studies. An analysis of nisin-resistant mutants of Lactococcus lactis Il1403 revealed the increased expression of a number of different genes, including cell wall-related loci, operons involved in metabolism, as well as a number of genes involved in transport and stress responses (21). The contribution of some of these genes, i.e., dltD, galKMT, and ahrC, to nisin resistance was subsequently confirmed by using genetic knockouts. The involvement of another set of genes, ysaBC, was confirmed when overexpression was found to lead to increased nisin resistance (21). Expression studies have also established that the histidine kinase-encoding gene (lmo1021), a penicillin binding protein determinant (lmo2229), and a gene encoding a protein of unknown function (lmo2487) are all upregulated in spontaneously nisin-resistant L. monocytogenes strains (16).
Here the screening of a mariner transposon bank of L. monocytogenes EGDe has resulted in the identification of a mutant that is susceptible to nisin as a consequence of the disruption of lmo1967, a gene homologous to tellurite resistance loci, designated telA, found in many members of the Firmicutes and Proteobacteria. This is the first occasion in which a telA gene has been associated with resistance to a cell envelope-acting antimicrobial and the first time that it has been studied in a Gram-positive bacterium. The study of this transposon mutant, and of another mutant in which the gene was removed in a nonpolar manner, revealed that telA also contributes to the pathogen's natural resistance to tellurite and to many cell envelope-acting antimicrobials, including gallidermin, bacitracin, cefuroxime, and cefotaxime.
MATERIALS AND METHODS
Strains, plasmids, and media.
The bacterial strains, plasmids, and culture conditions used in this study are listed in Table 1. Strains were grown at 37°C with shaking unless otherwise stated. L. monocytogenes was grown aerobically in tryptic soy broth with yeast extract (TSB-YE). Escherichia coli cells were cultured in Luria-Bertani medium. Antibiotics were used at the following concentrations: erythromycin at 5 μg/ml, kanamycin at 50 μg/ml, and chloramphenicol (Cm) at 10 μg/ml.
TABLE 1.
Strains, plasmids, and PCR primers used
| Strain, primer, or plasmid | Relevant feature(s)f | Source or reference |
|---|---|---|
| Strains | ||
| DH10B | Cloning host strain | Invitrogen |
| L. monocytogenes EGDe | Serotype 1/2a strain; genome sequenced | 15a |
| Primers | ||
| Lmo1967_PstI_SOEA | CTCTGCAGTGGAGCTAACTGa | This study |
| Lmo1967_SOEB | CACTTGGCTTGTTCTCGGTC | This study |
| Lmo1967_SOEC | GACCGAGAACAAGCCAAGTGACAAAACCCCACTCTTACGb | This study |
| Lmo1967_XbaI_SOED | CCTCTAGAATGCTTGCGACc | This study |
| Lmo1967_OutFor | CCAAGAGCGTAACAAAACGC | This study |
| Lmo1967_OutRev | GGAATGATTCATGCCTGTG | This study |
| Lmo1967_comp_NCO_F | CAAACCATGGCCGAGAACAAGCCd | This study |
| Lmo1967_comp_PST_R | GGGCTGCAGTTATTTCATTTCe | This study |
| Marq207 | GGCCACGCGTCGACTAGTACNNNNNNNNNNGTAAT | 6 |
| Marq208 | GGCCACGCGTCGACTAGTAC | 6 |
| marq255 | CAGTACAATCTGCTCTGATGCCGCATAGTT | 6 |
| marq256 | TAGTTAAGCCAGCCCCGACACCCGCCAACA | 6 |
| marq257 | CTTACAGACAAGCTGTGACCGTCT | 6 |
| PL95 | ACATAATCAGTCCAAAGTAGATGC | 21a |
| PL102 | TATCAGACCTAACCCAAACCTTCC | 21a |
| Plasmids | ||
| pKSV7 | Temperature-sensitive integration vector; Cmr | 38a |
| pIMK2 | Integrative vector, overexpression, 6.2 kb; Kanr | 26 |
| pMC38 | mariner delivery vector | 6 |
The sequence in boldface type is the PstI restriction enzyme site.
The underlined sequence is an overlap designed to complement primer Lmo1967_SOEB.
The sequence in boldface type is the XbaI restriction enzyme site.
The sequence in boldface type is the NcoI restriction enzyme site.
The sequence in boldface type is the PstI restriction enzyme site.
Primers are 5′ to 3′.
Generation of a random mutant bank.
A mariner random mutant bank of L. monocytogenes EGDe mutants was generated as described previously by Cao et al. (6). The pMC38 vector was used to generate the bank, in preference to pMC39, due to the high transformation efficiency of this vector in electrocompetent L. monocytogenes EGDe. Plasmid retention was estimated by plating the bank separately on TSB-YE agar plates containing either 5 μg/ml erythromycin or 10 μg/ml kanamycin. Plasmid retention was calculated by dividing the number of clones that retained the plasmid by the total number of clones. The Genetix (Hampshire, United Kingdom) Qpix2 XT robotic system was utilized to pick 7,000 mariner transposon-containing L. monocytogenes colonies and stock them in TBS-YE freezing buffer [TSB-YE supplemented with K2HPO4 (36 mM), KH2PO4 (13.2 mM), sodium citrate (1.7 mM), MgSO4 (0.4 mM), (NH4)2SO4 (6.8 mM), and 4.4% glycerol] in 384-well microtiter plates. To identify nisin-susceptible clones, the entire bank was replica plated onto tryptic soy agar with yeast extract (TSA-YE) containing 2 μg/ml Nisaplin (2.5% nisin; Danisco) using the Qpix robotic system. Plates were incubated for 24 h, and nisin-susceptible clones were identified on the basis of a lack of growth.
Identification of the site of transposon insertion.
The site of transposon insertion was identified as described previously (6). Briefly, an “arbitrary” PCR was first performed by using primers Marq207 and Marq255. Reddymix Extensor PCR master mix (Therma Scientific, Waltham, MA) was utilized for PCRs according to the manufacturer's instructions. A 1/25 dilution of the product of this reaction was used as a template for a second PCR with primers Marq208 and Marq256. The resulting PCR products were sequenced by using primer Marq257 (MWG Eurofins, Ebersburg, Germany), and the location of the transposon insertion was revealed with the help of BLAST (Basic Local Alignment Search Tool) analysis (www.ncbi.nlm.nih.gov/BLAST/).
Construction of a nonpolar deletion mutant of telA.
A nonpolar deletion of telA was generated by using the temperature-sensitive shuttle vector pKSV7 coupled with splicing by overlap extension (SOE) as described previously (10). Briefly, two regions up- and downstream of the telA gene were amplified by using telASoeAB and telASoeCD, respectively. These products were cleaned, mixed in equal proportions, and spliced through a second PCR. The resultant telAsoeAD product was digested with PstI and XbaI, cloned into pKSV7, and transformed into E. coli DH10β cells. Following sequencing of the telAsoeAD insert, this plasmid was introduced into electrocompetent L. monocytogenes EGDe (26), and chloramphenicol-resistant transformants were selected. Following serial subculturing at 42°C in TSB-YE with 10 μg/ml chloramphenicol, cells into which pKSV7-telAsoeAD had integrated, via homologous recombination, into the EGDe genome were selected for by plating onto TSA-YE at 42°C. Plasmid excision, via a second recombination event, and curing occurred spontaneously after continuous subculturing in TSB-YE at 30°C. Cultures were plated onto TSA-YE at 30°C, and chloramphenicol-sensitive colonies (i.e., excisants) were identified by replica plating onto TSA-YE and TSA-YE-Cm. Successful mutagenesis was confirmed by PCR using primers Lmo1967_OutFor and Lmo1967_OutRev.
Complementation of the ΔtelA mutation.
The complementation of the ΔtelA deletion was achieved by amplifying (with primers lmo1967_comp_Nco_f and lmo1967_comp_pst_r) and cloning telA into the pIMK2 vector to generate pIMK2Lmo1967 and integrating this vector into the EGDe ΔtelA mutant genome by using a procedure described previously (26). The integration of the vector in kanamycin-resistant colonies was confirmed by PCR using primers PL95 and PL102.
Nisin challenge assays.
The growths of L. monocytogenes strains (2% inoculum in TSB-YE) in the presence of different concentrations of nisin (0, 100, 300, and 500 μg/ml of Nisaplin [2.5% nisin]; Danisco) were compared by monitoring the optical density at 600 nm (OD600) with a Spectra Max 340 spectrophotometer (Molecular Devices, CA).
Antibiotic disk assays.
The susceptibilities of L. monocytogenes strains to a variety of antibiotics were initially determined by antibiotic disk diffusion assays as described previously (11, 19). Briefly, stationary-phase cultures (16 h) were diluted to 107 CFU/ml and swabbed onto TSA-YE. Six-millimeter antibiotic disks (Oxoid) infused with specific antibiotics were placed onto the agar plates, and following overnight growth (16 h), the zones of inhibition were measured.
MICs.
MICs in TSB-YE medium were determined as described previously (13). One-hundred-microliter volumes of 2-fold serial dilutions of the antimicrobial to be tested were added to the wells of a 96-well plate containing 100 μl of the target bacteria at a concentration of 105 CFU/ml. After 20 h (high-performance liquid chromatography [HPLC]-purified nisin A) or 16 h (other antimicrobials) of incubation, the MIC was read as the lowest concentration that resulted in an absence of visible growth.
RESULTS AND DISCUSSION
Generation and screening of a random L. monocytogenes mutant bank.
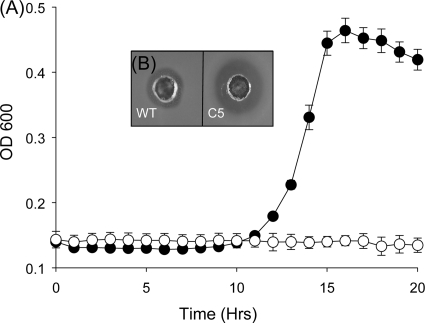
A mariner random mutant bank of L. monocytogenes EGDe was generated as described previously by Cao et al. (6). Plasmid retention within the bank was estimated to be 0.2%, thereby establishing that transposon insertion was successful. Twenty clones were picked at random, and the number of insertion events in each was determined by Southern hybridization. In each case, only single insertions were seen. The bank was arrayed by using a Qpix II colony-picking robotic system, thus facilitating the rapid screening of 7,000 random mutants for clones that were susceptible to 2 mg/ml Nisaplin (2.5% nisin) in TSA-YE. Clones that did not grow on this medium after 24 h were selected for further examination. Clone C5 was identified as a consistently nisin-susceptible mutant when growth was analyzed in the presence of Nisaplin (2.5% nisin) (Fig. 1). The location of the transposon insertion site was identified at position 595 of the 1,200-bp lmo1967 by the mariner arbitrary PCR method (6).
FIG. 1.
(A) Kinetic growth assay showing impaired growth of EGDe transposon mutant C5 (open symbols) compared to the wild type (WT) (closed symbols) in the presence of 500 μg/ml Nisaplin (2.5% nisin). (B) Deferred antagonism assay showing increased susceptibility of C5 to Nisaplin.
In silico analysis of lmo1967.
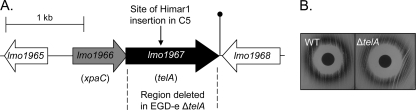
Initial in silico analysis was carried out in the form of a PSI-blast search on the 399-amino-acid (aa) predicted product of lmo1967. Hits above identities of 23% and E values of 1e−17 were considered relevant. A total of 323 bacteria were found to possess homologues, primarily those from the phyla Firmicutes and Proteobacteria. Unsurprisingly, those present in other members of the Firmicutes most closely resembled the EGDe equivalent. lmo1967 is thought to form a two-gene operon with lmo1966, which encodes a putative member of the halogen hydrol superfamily (XpaC) (Fig. 2). Indeed, the loci corresponding to lmo1966 and lmo1967 are also found in Listeria innocua, Listeria welshimeri, and Listeria grayi, while the same genetic organization is also conserved across many other members of the Firmicutes and Proteobacteria. For L. monocytogenes, transcriptomics-based studies previously revealed that the expression of this operon is upregulated in stationary phase (and is SigB dependent) and in an intestinal environment but is downregulated in blood and brain heart infusion (BHI) broth (5, 44), while proteomics has established that the Lmo1967 protein is a member of the L. monocytogenes cell wall subproteome (38). Loci corresponding to lmo1967 in other Gram-positive microorganisms are known by a variety of other names, including klaB, ynhC, yaaN, and yceH. In Bacillus subtilis, yaaN is induced under stress from the cationic antimicrobial compound poly-l-lysine (32) by both salt and alkaline shock and is part of the SigW regulon (31, 46). Notably, ynhC is also upregulated in L. lactis in response to nisin stress (21). The closest homologue of lmo1967 for which a function has been assigned is the Rhodobacter sphaeroides gene telA (38% homology), and thus, lmo1967 was redesignated telA (28). telA has been characterized on the basis of being required for full resistance to the toxic compound tellurite. Tellurite compounds were trialed as antimicrobials in the first half of the 20th century, showing efficacy against tuberculosis, leprosy, and syphilis. Presently, the main application for tellurite compounds is as an ingredient in selective media, where tellurite resistance is characterized by black color development in resistant colonies (41). A number of genes are known to be involved in tellurite resistance, including cysM, katA, sodAB, and soxS as well as a number of operons that, to date, have been associated only with tellurite resistance (teh, ter, tel, trg, and tmp) (8).
FIG. 2.
(A) Genetic organization of the point of insertion of the himar1 transposon. The dashed line shows the proportion of the gene deleted. The lollipop symbol indicates a putative terminator. All genes in the schematic are drawn to scale. (B) Disk diffusion assay showing a potassium tellurite-impregnated disk on plates spread with wild-type L. monocytogenes and the L. monocytogenes ΔtelA mutant. An increased zone of inhibition indicates increased susceptibility in the ΔtelA mutant. Lighter shades of the zone of tellurite reduction indicate an impaired ability to reduce tellurite to elemental tellurium.
Creation and characterization of the L. monocytogenes EGDe ΔtelA mutant strain.
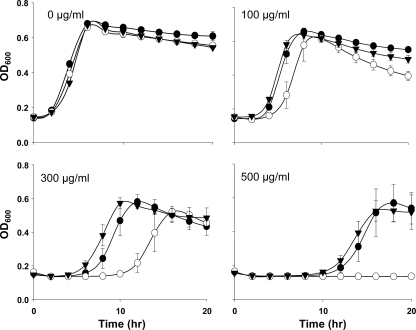
To ensure that the nisin-sensitive phenotype of mutant C5 was a consequence of the disruption of telA, a nonpolar deletion mutant of telA was created and studied. This ruled out the possibility of the nisin-sensitive phenotype being due to polar effects in the transposon mutant. The ΔtelA mutant was created by deleting 1,150 bp of the 1,200-bp gene using the SOE-PCR procedure, the temperature-sensitive shuttle vector pKSV7, and double-crossover homologous recombination. The susceptibility of the ΔtelA mutant was initially assessed by growing the strain in broth containing 100 to 500 μg/ml Nisaplin (2.5% nisin) (Fig. 3). It was apparent that the growth of the ΔtelA mutant was dramatically affected by the presence of Nisaplin, being most apparent in the presence of 500 μg/ml of the preservative when the mutant failed to grow. This contrasted with the vigorous growth of the parental strain EGDe under these conditions (Fig. 3). To ensure that this susceptibility was as a consequence of the nisin A present in Nisaplin rather than, for example, its NaCl content, MIC determination assays with pure nisin A were performed. From these experiments, it was apparent that the ΔtelA mutant was 4-fold-more susceptible (MIC = 6 μg/ml) than the parent (MIC = 25 μg/ml) after 20 h of growth (see Table 3). The ΔtelA mutation was complemented by cloning telA behind the strong promoter of plasmid pIMK2 and integrating the vector into the mutant strain (26) to generate the EGDe ΔtelA::telA strain. This complemented strain no longer exhibited a nisin-susceptible phenotype (Fig. 3).
FIG. 3.
Growth of EGDe and the ΔtelA and ΔtelA::telA mutants in the presence of antimicrobial agents. Shown is the growth of EGDe (closed circles), the EGDe ΔtelA mutant (open circles), and the EGDe ΔtelA::telA mutant (closed triangles) in TSB-YE with 100 to 500 μg/ml of Nisaplin (2.5% nisin). Error bars are standard deviations from the means of data from triplicate experiments.
Antimicrobial susceptibility assays.
Given the description of telA of other microorganisms as a tellurite resistance locus, the susceptibility of the ΔtelA mutant to potassium tellurite was also assessed. The investigations established that the ΔtelA strain is 2-fold-more susceptible to potassium tellurite (see Table 3). Notably, growth on tellurite-containing agar revealed that the mutant appeared to still be able to process tellurite to some extent, as evidenced by a characteristic black precipitate. However, the observation that the associated precipitate was less obvious than that produced by the parent strain suggests an impaired ability to reduce tellurite to elemental tellurium (Fig. 2B). L. monocytogenes EGDe was subjected to an in silico analysis to screen for the presence of homologues of the other four known tellurite resistance genetic determinants (41). However, none were identified, suggesting that the genes responsible for residual tellurite resistance mechanisms are novel. The frequency with which tellurite resistance mechanisms are located across different bacterial species is a curiosity, especially as tellurite itself is a very rare element in the environment (the 75th most abundant metal). Thus, it is anticipated that it will ultimately be established that tellurite-specific stress responses are very rare (should they exist at all) and that resistance is most frequently mediated through more-general stress response mechanisms. On this basis, the ability of the ΔtelA mutant to withstand exposure to a number of other antimicrobials, relative to the parental strain, was also assessed. In order to include as many antimicrobials as possible, this screen initially took place in the form of antimicrobial disk agar-based assays. However, in instances where differences in relative susceptibility were observed, subsequent MIC determination studies were performed. The antibiotic disks employed included cefuroxime, cefotaxime, rifampin, vancomycin, gentamicin, novobiocin, tetracycline (30 μg), trimethoprim-sulfamethoxazole (25 μg), bacitracin, erythromycin, ampicillin (10 μg), methicillin, penicillin G (5 μg), and oxacillin (1 μg). These assays revealed that the ΔtelA mutant exhibits enhanced susceptibility to oxacillin, cefuroxime, cefotaxime, methicillin, and bacitracin (Table 2). Further MIC-based analyses revealed that this corresponded to a 2-fold-greater susceptibility to the beta-lactam antibiotics cefuroxime and cefotaxime and to the non-beta-lactam antibiotic bacitracin and a dramatic 8-fold-increased susceptibility to the lantibiotic gallidermin (Table 3). MIC analysis did not reveal differences in susceptibility for oxacillin and methicillin. It is thus apparent that TelA contributes greatly to the innate resistance of L. monocytogenes EGDe to a number of important cell envelope-acting antimicrobials.
TABLE 2.
Antibiotic disk assays
| Antibiotic | Mean size of zone of inhibition (mm) ± SD |
|
|---|---|---|
| WT | TelA mutant | |
| Cefuroxime | 21.1 ± 0.2 | 24.4 ± 0.3 |
| Bacitracin | 12.5 ± 0.3 | 15.1 ± 0.4 |
| Oxacillin | 10.7 ± 0.3 | 12.3 ± 0.2 |
| Cefotaxime | 21.3 ± 0.3 | 23.7 ± 0.3 |
| Methicillin | 25.6 ± 0.2 | 26.7 ± 0.4 |
TABLE 3.
MICsa
| Antimicrobial | MIC (μg/ml) |
|
|---|---|---|
| WT | TelA mutant | |
| Nisin | 25.14 | 6.29 |
| Gallidermin | 1.97 | 0.246 |
| Bacitracin | 256 | 128 |
| Cefotaxime | 2 | 1 |
| Tellurite | 38 | 19 |
| Cefuroxime | 2.87 | 1.44 |
Results represent the averages of data from three independent experiments. As the values were identical in each case, the standard deviation is zero. MIC data are derived from a 2-fold dilution series. For nisin and gallidermin the μg/ml value is extrapolated from a μM value; e.g., a nisin concentration of 7.5 μM is equivalent to 25.14 μg/ml.
Despite its broad distribution, this is the first occasion upon which a telA homologue in a Gram-positive bacterium has been investigated in depth. Even more importantly, while an analysis of transcriptomics data reveals that the level of expression of telA homologues is increased in a variety of bacteria in response to exposure to nisin, poly-l-lysine, vancomycin, and daptomycin (7, 21, 27, 32), this is the first time that antimicrobial susceptibility has been noted for a ΔtelA mutant.
Here TelA has been identified as a novel mediator of resistance to a number of cell envelope-acting antimicrobials, including the lantibiotics nisin and gallidermin, the beta-lactams cefuroxime and cefotaxime, as well as bacitracin. Both lantibiotics are inhibitors of peptidoglycan synthesis as a consequence of binding lipid II, and in addition, nisin and, on occasion, gallidermin can also form pores in the cell membrane of target microorganisms (2). The beta-lactam antibiotics inhibit the cross-linking of peptidoglycan chains, while bacitracin affects the dephosphorylation of bactophenol during peptidoglycan synthesis (39, 43). While bacitracin is not usually employed as an anti-Listeria agent, the identification of a locus that contributes to the innate resistance of L. monocytogenes to nisin and cephalosporins is notable in light of their respective use as food preservatives and clinical antibiotics. Nisin is the only natural antibacterial preservative that has been approved for food applications, and thus, its frequently limited activity against L. monocytogenes is problematic; however, its efficacy increases when integrated as part of a hurdle approach to food protection. Cephalosporins are often used as the antibiotics of first choice when treating infections of unknown etiology, and thus, the high innate resistance of L. monocytogenes to this class of antibiotics may also have serious consequences. Curiously, this is not the first occasion upon which nisin and cephalosporin resistance have been linked, as the L. monocytogenes ΔlisK mutant exhibits enhanced nisin resistance but greater susceptibility to cephalosporins (11), while mutants that are spontaneously resistant to the lantibiotic lacticin 3147 have also shown increased cephalosporin susceptibility (17). A further understanding of this phenomenon could lead to the development of strategies to disrupt these mechanisms with a view to increasing the susceptibility of the pathogen to these antimicrobials.
Acknowledgments
We acknowledge Nicola Curtis for her assistance in the screening of the L. monocytogenes mutant bank. We thank Hélène Marquis for providing plasmid pMC38, Joss Delves-Broughton (Danisco) for Nisaplin powder, and Friedrich Götz for purified gallidermin.
This work was supported by the Irish Department of Agriculture by a Food Institutional Research Measure (FIRM) grant (Funlac).
Footnotes
Published ahead of print on 16 August 2010.
REFERENCES
- 1.Begley, M., C. Hill, and R. P. Ross. 2006. Tolerance of Listeria monocytogenes to cell envelope-acting antimicrobial agents is dependent on SigB. Appl. Environ. Microbiol. 72:2231-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonelli, R. R., T. Schneider, H. G. Sahl, and I. Wiedemann. 2006. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob. Agents Chemother. 50:1449-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breukink, E., I. Wiedemann, C. van Kraaij, O. P. Kuipers, H. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 4.Brotz, H., M. Josten, I. Wiedemann, U. Schneider, F. Gotz, G. Bierbaum, and H. G. Sahl. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30:317-327. [DOI] [PubMed] [Google Scholar]
- 5.Camejo, A., C. Buchrieser, E. Couve, F. Carvalho, O. Reis, P. Ferreira, S. Sousa, P. Cossart, and D. Cabanes. 2009. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 5:e1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, M., A. P. Bitar, and H. Marquis. 2007. A mariner-based transposition system for Listeria monocytogenes. Appl. Environ. Microbiol. 73:2758-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, M., T. Wang, R. Ye, and J. D. Helmann. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Mol. Microbiol. 45:1267-1276. [DOI] [PubMed] [Google Scholar]
- 8.Chasteen, T. G., D. E. Fuentes, J. C. Tantalean, and C. C. Vasquez. 2009. Tellurite: history, oxidative stress, and molecular mechanisms of resistance. FEMS Microbiol. Rev. 33:820-832. [DOI] [PubMed] [Google Scholar]
- 9.Collins, B., N. Curtis, P. D. Cotter, C. Hill, and R. P. Ross. 2010. The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various β-lactam antibiotics. Antimicrob. Agents Chemother. 54:4416-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotter, P. D., N. Emerson, C. G. Gahan, and C. Hill. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 181:6840-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter, P. D., C. M. Guinane, and C. Hill. 2002. The LisRK signal transduction system determines the sensitivity of Listeria monocytogenes to nisin and cephalosporins. Antimicrob. Agents Chemother. 46:2784-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter, P. D., C. Hill, and R. P. Ross. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777-788. [DOI] [PubMed] [Google Scholar]
- 13.Draper, L. A., K. Grainger, L. H. Deegan, P. D. Cotter, C. Hill, and R. P. Ross. 2009. Cross-immunity and immune mimicry as mechanisms of resistance to the lantibiotic lacticin 3147. Mol. Microbiol. 71:1043-1054. [DOI] [PubMed] [Google Scholar]
- 14.Fabretti, F., C. Theilacker, L. Baldassarri, Z. Kaczynski, A. Kropec, O. Holst, and J. Huebner. 2006. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 74:4164-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez, L., S. Delgado, H. Herrero, A. Maldonado, and J. M. Rodriguez. 2008. The bacteriocin nisin, an effective agent for the treatment of staphylococcal mastitis during lactation. J. Hum. Lact. 24:311-316. [DOI] [PubMed] [Google Scholar]
- 15a.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 16.Gravesen, A., K. Sorensen, F. M. Aarestrup, and S. Knochel. 2001. Spontaneous nisin-resistant Listeria monocytogenes mutants with increased expression of a putative penicillin-binding protein and their sensitivity to various antibiotics. Microb. Drug Resist. 7:127-135. [DOI] [PubMed] [Google Scholar]
- 17.Guinane, C. M., P. D. Cotter, C. Hill, and R. P. Ross. 2006. Spontaneous resistance in Lactococcus lactis IL1403 to the lantibiotic lacticin 3147. FEMS Microbiol. Lett. 260:77-83. [DOI] [PubMed] [Google Scholar]
- 18.Hasper, H. E., N. E. Kramer, J. L. Smith, J. D. Hillman, C. Zachariah, O. P. Kuipers, B. de Kruijff, and E. Breukink. 2006. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313:1636-1637. [DOI] [PubMed] [Google Scholar]
- 19.Kallipolitis, B. H., H. Ingmer, C. G. Gahan, C. Hill, and L. Sogaard-Andersen. 2003. CesRK, a two-component signal transduction system in Listeria monocytogenes, responds to the presence of cell wall-acting antibiotics and affects beta-lactam resistance. Antimicrob. Agents Chemother. 47:3421-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovacs, M., A. Halfmann, I. Fedtke, M. Heintz, A. Peschel, W. Vollmer, R. Hakenbeck, and R. Bruckner. 2006. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in Gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 188:5797-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer, N. E., S. A. van Hijum, J. Knol, J. Kok, and O. P. Kuipers. 2006. Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob. Agents Chemother. 50:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lubelski, J., R. Rink, R. Khusainov, G. N. Moll, and O. P. Kuipers. 2008. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol. Life Sci. 65:455-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandin, P., H. Fsihi, O. Dussurget, M. Vergassola, E. Milohanic, A. Toledo-Arana, I. Lasa, J. Johansson, and P. Cossart. 2005. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol. Microbiol. 57:1367-1380. [DOI] [PubMed] [Google Scholar]
- 24.Margolles, A., A. B. Florez, J. A. Moreno, D. van Sinderen, and C. G. de los Reyes-Gavilan. 2006. Two membrane proteins from Bifidobacterium breve UCC2003 constitute an ABC-type multidrug transporter. Microbiology 152:3497-3505. [DOI] [PubMed] [Google Scholar]
- 25.May, J. J., R. Finking, F. Wiegeshoff, T. T. Weber, N. Bandur, U. Koert, and M. A. Marahiel. 2005. Inhibition of the D-alanine:D-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium's susceptibility to antibiotics that target the cell wall. FEBS J. 272:2993-3003. [DOI] [PubMed] [Google Scholar]
- 26.Monk, I. R., C. G. Gahan, and C. Hill. 2008. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl. Environ. Microbiol. 74:3921-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muthaiyan, A., J. A. Silverman, R. K. Jayaswal, and B. J. Wilkinson. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 52:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Gara, J. P., M. Gomelsky, and S. Kaplan. 1997. Identification and molecular genetic analysis of multiple loci contributing to high-level tellurite resistance in Rhodobacter sphaeroides 2.4.1. Appl. Environ. Microbiol. 63:4713-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peschel, A., R. W. Jack, M. Otto, L. V. Collins, P. Staubitz, G. Nicholson, H. Kalbacher, W. F. Nieuwenhuizen, G. Jung, A. Tarkowski, K. P. van Kessel, and J. A. van Strijp. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with L-lysine. J. Exp. Med. 193:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 31.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pietiainen, M., M. Gardemeister, M. Mecklin, S. Leskela, M. Sarvas, and V. P. Kontinen. 2005. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology 151:1577-1592. [DOI] [PubMed] [Google Scholar]
- 33.Piper, C., L. A. Draper, P. D. Cotter, R. P. Ross, and C. Hill. 2009. A comparison of the activities of lacticin 3147 and nisin against drug-resistant Staphylococcus aureus and Enterococcus species. J. Antimicrob. Chemother. 64:546-551. [DOI] [PubMed] [Google Scholar]
- 34.Popowska, M., and Z. Markiewicz. 2006. Characterization of Listeria monocytogenes protein Lmo0327 with murein hydrolase activity. Arch. Microbiol. 186:69-86. [DOI] [PubMed] [Google Scholar]
- 35.Poyart, C., E. Pellegrini, M. Marceau, M. Baptista, F. Jaubert, M. C. Lamy, and P. Trieu-Cuot. 2003. Attenuated virulence of Streptococcus agalactiae deficient in D-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol. Microbiol. 49:1615-1625. [DOI] [PubMed] [Google Scholar]
- 36.Samant, S., F. F. Hsu, A. A. Neyfakh, and H. Lee. 2009. The Bacillus anthracis protein MprF is required for synthesis of lysylphosphatidylglycerols and for resistance to cationic antimicrobial peptides. J. Bacteriol. 191:1311-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sass, P., and G. Bierbaum. 2009. Native graS mutation supports the susceptibility of Staphylococcus aureus strain SG511 to antimicrobial peptides. Int. J. Med. Microbiol. 299:313-322. [DOI] [PubMed] [Google Scholar]
- 38.Schaumburg, J., O. Diekmann, P. Hagendorff, S. Bergmann, M. Rohde, S. Hammerschmidt, L. Jansch, J. Wehland, and U. Karst. 2004. The cell wall subproteome of Listeria monocytogenes. Proteomics 4:2991-3006. [DOI] [PubMed] [Google Scholar]
- 38a.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 39.Stone, K. J., and J. L. Strominger. 1971. Mechanism of action of bacitracin: complexation with metal ion and C 55-isoprenyl pyrophosphate. Proc. Natl. Acad. Sci. U. S. A. 68:3223-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun, Z., J. Zhong, X. Liang, J. Liu, X. Chen, and L. Huan. 2009. Novel mechanism for nisin resistance via proteolytic degradation of nisin by the nisin resistance protein NSR. Antimicrob. Agents Chemother. 53:1964-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor, D. E. 1999. Bacterial tellurite resistance. Trends Microbiol. 7:111-115. [DOI] [PubMed] [Google Scholar]
- 42.Thedieck, K., T. Hain, W. Mohamed, B. J. Tindall, M. Nimtz, T. Chakraborty, J. Wehland, and L. Jansch. 2006. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol. 62:1325-1339. [DOI] [PubMed] [Google Scholar]
- 43.Tipper, D. J. 1985. Mode of action of beta-lactam antibiotics. Pharmacol. Ther. 27:1-35. [DOI] [PubMed] [Google Scholar]
- 44.Toledo-Arana, A., O. Dussurget, G. Nikitas, N. Sesto, H. Guet-Revillet, D. Balestrino, E. Loh, J. Gripenland, T. Tiensuu, K. Vaitkevicius, M. Barthelemy, M. Vergassola, M. A. Nahori, G. Soubigou, B. Regnault, J. Y. Coppee, M. Lecuit, J. Johansson, and P. Cossart. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950-956. [DOI] [PubMed] [Google Scholar]
- 45.Wiedemann, I., R. Benz, and H. G. Sahl. 2004. Lipid II-mediated pore formation by the peptide antibiotic nisin: a black lipid membrane study. J. Bacteriol. 186:3259-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiegert, T., G. Homuth, S. Versteeg, and W. Schumann. 2001. Alkaline shock induces the Bacillus subtilis sigma(W) regulon. Mol. Microbiol. 41:59-71. [DOI] [PubMed] [Google Scholar]