Abstract
The molecular mechanisms of herpes simplex virus (HSV) resistance to antiviral drugs interfering with viral DNA synthesis reported so far rely on the presence of mutations within UL23 (thymidine kinase [TK]) and UL30 (DNA polymerase) genes. The interpretation of genotypic antiviral resistance assay results requires the clear distinction between resistance mutations and natural interstrain sequence variations. The objectives of this work were to describe extensively the natural polymorphism of UL23 TK and UL30 DNA polymerase among HSV-1 and HSV-2 strains and the amino acid changes potentially associated with HSV resistance to antivirals. The sequence analysis of the full-length UL23 and UL30 genes was performed. Ninety-four drug-sensitive clinical isolates (43 HSV-1 and 51 HSV-2) and 3 laboratory strains (KOS, gHSV-2, and MS2) were studied for natural polymorphism, and 25 clinical isolates exhibiting phenotypic traits of resistance to antivirals were analyzed for drug resistance mutations. Our results showed that TK and DNA polymerase are highly conserved among HSV strains, with a weaker variability for HSV-2 strains. This study provided a precise map of the natural polymorphism of both viral enzymes among HSV-1 and HSV-2 isolates, with the identification of 15 and 51 polymorphisms never previously described for TK and DNA polymerase, respectively, which will facilitate the interpretation of genotypic antiviral-resistant testing. Moreover, the genotypic characterization of 25 drug-resistant HSV isolates revealed 8 new amino acid changes located in TK and potentially accounting for acyclovir (ACV) resistance.
Herpes simplex virus type 1 (HSV-1) and HSV-2 are responsible for a variety of clinical manifestations (10). In immunocompetent individuals, the symptoms are usually self-limited, whereas severe diseases, sometimes life-threatening, may occur in immunocompromised patients (14, 17, 26). The discovery of acyclovir (ACV), almost 30 years ago, represents a milestone in the management of HSV infections. The antiviral activity and selectivity of ACV is based on the phosphorylation to its monophosphate form by the virus-encoded thymidine kinase (TK). Then ACV monophosphate is further phosphorylated by cellular thymidilate kinases to the triphosphate form and is incorporated into the growing DNA chain by the viral DNA polymerase, thereby inhibiting replication through chain termination. The decreased activity of TK confers HSV resistance to ACV, resulting in the inability of the drug to inhibit viral replication. Alternative drugs, like foscarnet (FOS), are effective without the requirement of phosphorylation by viral TK. FOS inhibits directly the viral DNA polymerase as a substrate analogue of the pyrophosphate formed during DNA synthesis (23).
HSV TK is a 376-amino-acid protein, encoded by the UL23 gene, containing an ATP binding site (codons 51 to 63), a nucleoside binding site (codons 168 to 176 for HSV-1 and 169 to 177 for HSV-2), and a highly conserved cysteine residue at position 336 for HSV-1 and 337 for HSV-2 (2, 13, 28). HSV DNA polymerase, 1,235 amino acids long for HSV-1 and 1,240 amino acids long for HSV-2, is encoded by the UL30 gene and constitutes the target of ACV and FOS. Comparison between the sequences of herpesvirus and eukaryotic DNA polymerases revealed a series of 8 conserved domains, named I to VII and delta-C (27, 54, 55).
HSV infections that are resistant to ACV and/or FOS usually occur in immunocompromised individuals, especially AIDS patients and transplant recipients. The degree of immunodepression and the prolonged therapy are considered important factors for the development of drug resistance (12, 17, 26, 36). To date, several mechanisms of resistance to ACV have been evidenced among HSV strains. The most frequent (95% of ACV-resistant isolates) results in the defective production of TK (TK-deficient virus) or in the alteration of TK substrate specificity (TK-altered virus). These phenotypes are the consequence of either single-base insertions/deletions occurring in guanosine (G) or cytidine (C) homopolymer repeats, leading to the shift of the translational reading frame of UL23 TK, or missense point mutations (21, 37, 45, 46). The third mechanism is an alteration of DNA polymerase activity due to nonsynonymous mutations within the UL30 gene. The latter mechanism may contribute to a high level of drug resistance and may induce ACV and FOS cross-resistance. Resistance mutations are located mainly in catalytic or conserved domains of TK and DNA polymerase (11, 22, 48, 51).
Currently used methods for testing the in vitro susceptibility of HSV to antivirals consist of the measurement of the 50% effective concentration (EC50) in cell culture. The plaque reduction assay (PRA) is generally considered to be the reference standard, but PRA is relatively tedious and time-consuming to perform. Genotypic assays based on the identification of mutations in the UL23 and UL30 genes represent an attractive approach to detecting drug resistance in a clinically relevant time frame. However, the interpretation of genotypic assays requires differentiating clearly resistance-associated mutations from natural interstrain sequence variations. To date, the natural polymorphism of HSV TK and DNA polymerase has not been investigated extensively, especially among HSV-2 strains (10, 31, 37).
The objectives of the work presented here were (i) to develop new tools to perform the sequencing of the entire UL23 and UL30 genes for both HSV-1 and HSV-2, (ii) to define precisely the natural polymorphism of TK and DNA polymerase among drug-sensitive clinical isolates, and (iii) to perform the genotypic study of 25 well-characterized drug-resistant clinical isolates in order to evidence new potential resistance mutations within TK and DNA polymerase.
MATERIALS AND METHODS
Viral strains, biological samples, and cell cultures.
A total of 94 HSV clinical isolates and 3 HSV laboratory strains (KOS, gHSV-2, and MS2) were included in the present study in order to describe the natural polymorphism of TK and DNA polymerase. Forty-two clinical isolates (17 HSV-1 and 25 HSV-2) were characterized as ACV sensitive and FOS sensitive by PRA (method described below), and 52 clinical isolates (26 HSV-1 and 26 HSV-2) were collected from patients who had not received any prior anti-HSV therapy. All isolates were obtained from patients suffering from HSV-induced disease. The biological samples collected were mainly mucocutaneous lesions: swabs from skin (n = 16), oro-facial (n = 14), or ano-genital (n = 48) vesicles and keratitis lesions (n = 2) suspected to be due to HSV infection. Moreover, 14 HSV isolates were collected from bronchoalveolar lavage (BAL) fluids. In addition, 25 drug-resistant clinical isolates (11 HSV-1 and 14 HSV-2) were studied in detail by genotypic antiviral resistance testing. According to PRA, 24 isolates were ACV resistant and FOS sensitive, and one isolate exhibited cross-resistance to ACV and FOS. All isolates but one were recovered from immunocompromised patients: HIV-infected patients (n = 12), bone marrow transplant (BMT) recipients (n = 8), patients with leukemia (n = 3), and patients with Wiskott-Aldrich syndrome (n = 1). The remaining isolate was obtained from an immunocompetent patient with recurrent herpetic keratitis. These resistant HSV isolates were recovered from the following biological samples: BAL fluid (n = 1), blood (n = 1), and swabs from cutaneous (n = 1), ocular (n = 1), oro-facial (n = 7), and ano-genital (n = 14) sites. The age of the patients from whom the resistant HSV isolates were recovered ranged from 8 to 77 years and from 34 to 70 years for HSV-1 and HSV-2, respectively. Antiviral treatment information was unavailable for the isolates from 13 patients sent to us by outside physicians. Regarding the remaining patients, antiviral treatment during which drug-resistant HSV was isolated consisted of either intravenous ACV and/or oral valacyclovir (n = 8) or successive courses of intravenous ACV and FOS treatments (n = 4).
HSV isolates were propagated in Vero cells cultivated in RPMI 1640 (Invitrogen, Cergy-Pontoise, France) supplemented with 2% fetal bovine serum (Invitrogen), 2 mM glutamine (Eurobio, Courtaboeuf, France), 20 μg/ml vancomycin (Lilly, Suresnes, France), and 20 μg/ml amikacin (Bristol Myers Squibb, Rueil-Malmaison, France).
Virus drug susceptibility assay.
The susceptibility of HSV isolates to antiviral drugs was determined by PRA in Vero cell culture, as previously described (53). Laboratory strains KOS (HSV-1) and gHSV2 (HSV-2) were used as susceptible controls in each assay. HSV susceptibility was tested toward ACV (Merck, Lyon, France) and FOS (AstraZeneca, Rueil-Malmaison, France). HSV isolates were considered resistant at EC50 values of ≥7 μM and 330 μM for ACV and FOS, respectively (53). The determination of HSV type was performed by means of an immunofluorescence assay using type-specific monoclonal antibodies (Trinity Biotech Plc, Wicklow, Ireland).
Genotypic antiviral resistance testing. (i) DNA extraction.
Viral DNA was extracted directly from 200 μl of viral stock (clinical isolates and laboratory strains) by using a QIAamp DNA blood minikit (Qiagen, Courtaboeuf, France), according to the manufacturer's instructions. The purified nucleic acids were eluted in 100 μl of elution buffer and stored at −20°C until used.
(ii) TK and DNA polymerase gene amplification and sequencing.
All primers used for gene amplification and sequencing are listed in Tables 1 and 2. The full-length TK (UL23) gene (1,195 bp) and UL30 (DNA polymerase) gene (3,708 bp for HSV-1 and 3,723 bp for HSV-2) from all HSV clinical isolates and laboratory strains were amplified using HSV type-specific PCR systems. For each gene, the first PCR was performed with 10 μl of DNA extract using the proofreading enzyme Expand High Fidelity (Roche, Meylan, France) in a mix containing 1× buffer, 1.5 mM MgCl2, 800 μM dNTP, and 300 μM forward and reverse outer primers. The nested PCR was carried out with 5 μl of the first PCR product using inner primers in a mix identical to that of the first PCR. UL23 amplification conditions for the first PCR included an initial denaturation step of 5 min at 94°C followed by 40 cycles of 1 min at 94°C, 1 min at 60°C, and 3 min at 72°C, with a final extension step of 10 min at 72°C. UL23 nested PCR was performed with the same conditions, but with an annealing temperature of 57°C. Cycling conditions for both UL30 first and nested PCRs were as follows: 2 min at 96°C, followed by 40 cycles of 1 min at 96°C, 1 min at 59°C, and 6 min at 68°C, with a final extension of 10 min at 68°C. Cycling conditions for UL23 and UL30 genes were the same for both HSV-1 and HSV-2. All PCRs were performed using an Eppendorf thermal cycler (Eppendorf, Le Pecq, France). Amplified products were sequenced with the Prism BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Courtaboeuf, France) and analyzed with the automated sequencer ABI PRISM 3730 genetic analyzer (Applied Biosystems).
TABLE 1.
Primers used for amplification and sequencing of HSV-1 UL23 and UL30 genes
| Target gene | Function | Name | Sequence (5′→3′)a |
|---|---|---|---|
| UL23 | First-round PCR (outer primers) | TK1-F1 | F: TAACCCCCACGAACCATAAA |
| TK1-R1 | R: CGAATTCGAACACGCAGAT | ||
| Second-round PCR (inner primers) | TK1-F2 | F: GGTGGGGTATCGACAGAGTG | |
| TK1-R2 | R: GATCTTGGTGGCGTGAAACT | ||
| Sequence reaction | TK1-A | R: TATGGGCTGCTTGCCAATAC | |
| TK1-B | F: GTTATACAGGTCGCCGTTGG | ||
| TK1-C | R: CCTCGACCAGGGTGAGATA | ||
| TK1-D | F: GGTCGAAGATGAGGGTGAGG | ||
| + TK1-F2 and TK1-R2 | |||
| UL30 | First-round PCR (outer primers) | POL1-F1 | F: CGGCTACGTCACTCTCCTGT |
| POL1-R1 | R: CGGAGATTCCGACCGTGT | ||
| Second-round PCR (inner primers) | POL1-F2 | F: GCCGACGCGAATAAACC | |
| POL1-R2 | R: GGCCGCAGACATTTATTGTAA | ||
| Sequence reaction | POL1-A | R: GCTTGAGGTGACCGTCGT | |
| POL1-B | F: CGTACTATAGCGAATGCGATGA | ||
| POL1-C | R: GAAGTGGTCCGCGGAGAT | ||
| POL1-D | F: GGTCGACAGGCACCTACAAT | ||
| POL1-E | R: ACCGGAAAGGCCAGCTC | ||
| POL1-F | F: AGGACGAGCTGGCCTTTC | ||
| POL1-G | R: CACGGCGTTGAGCTTGTAG | ||
| POL1-H | F: GTGTGGGACATAGGCCAGAG | ||
| POL1-I | R: GTCCTCCCCCTCCTCCTC | ||
| POL1-J | F: CCGACCAGAAGGGCTTTATT | ||
| POL1-K | R: GCTGCTTGTCCAGGAGCAC | ||
| POL1-L | F: CCATGCGAAAGCAGATCC | ||
| POL1-M | R: TCTTACCCCCGTAGATGACG | ||
| POL1-N | F: ATCAAACTCGAGTGCGAAAA | ||
| POL1-O | R: GCGACCGTCTCCTCTACCTC | ||
| POL1-P | F: CAGGTCCCGTCCATCAAG | ||
| + POL1-F2 and POL1-R2 |
F, forward; R, reverse.
TABLE 2.
Primers used for amplification and sequencing of HSV-2 UL23 and UL30 genes
| Target gene | Function | Name | Sequence (5′→3′)a |
|---|---|---|---|
| UL23 | First-round PCR (outer primers) | TK2-F1 | F: GTACCCACGGCCCAAAGAG |
| TK1-R1 | R: CGAATTCGAACACGCAGAT | ||
| Second-round PCR (inner primers) | TK2-F2 | F: CTATCGGAGGCCCTTTGTTG | |
| TK2-R2 | R: CGAGGTCCACTTCGCATATT | ||
| Sequence reaction | TK2-A | R: ATCGAGGACACGCTGTTTG | |
| TK2-B | F: AAGACCCAGGCAAAAATGTG | ||
| TK2-C | R: AGCGCCCAGATAACAATGAG | ||
| TK2-D | F: CCGATATGAGGAGCCAAAAC | ||
| + TK2-F2 and TK2-R2 | |||
| UL30 | First-round PCR (outer primers) | POL2-F1 | F: GACACCACACACTGGCTCTC |
| POL2-R1 | R: AGACGCAACGCAGAGAAAC | ||
| Second-round PCR (inner primers) | POL2-F2 | F: ATCCCACCCCGAGCTGTT | |
| POL2-R2 | R: GTCGGCCGCAGACATTTAT | ||
| Sequence reaction | POL2-A | R: TACACGTGGAAGACGGTGAC | |
| POL2-B | F: CGCCCCTAAGGTGTACTGC | ||
| POL2-C | R: CCCCTCGTACTTCCTGATCG | ||
| POL2-D | F: ACTTCGAGGCGGAGGTG | ||
| POL2-E | R: ATCCGAGCGAAAACAGGAG | ||
| POL2-F | F: GGAAGACCTCGTCATCCAGA | ||
| POL2-G | R: GATGTCGCGGTAGCTCAGAT | ||
| POL2-H | F: CCGACAAGGTCAAACTCTCC | ||
| POL2-I | R: AGGCTGGCAAAGTCAAACAC | ||
| POL2-J | F: GATAAGGACGACGACGAGGA | ||
| POL2-K | R: CCAGCAGCTGATCGAACTC | ||
| POL2-L | F: GGTGCAGCACGGTCTTCT | ||
| POL2-M | R: GATGGGCGTCTACGAGGA | ||
| POL2-N | F: CTGGTGCGCAAAAACAACT | ||
| POL2-O | R: CCAGCAGGTGCGAGAAGTA | ||
| + POL2-F2 and POL2-R2 |
F, forward; R, reverse.
Nucleotide sequence accession numbers.
All nucleotide and amino acid sequencing results were compared with published sequences of the reference strains 17 (HSV-1) and HG52 (HSV-2) (GenBank accession numbers X14112 and Z86099, respectively) using Seqscape version 2.5 software (34, 35). TK and DNA polymerase sequences determined in this study from phenotypically characterized drug-sensitive (n = 42) and drug-resistant (n = 25) HSV clinical isolates have been deposited in the GenBank database under accession numbers HQ123046 through HQ123129 and HQ123130 through HQ123179, respectively.
RESULTS
Sensitivity of UL23 and UL30 PCRs.
The sensitivity of the UL23 and UL30 nested PCRs for both HSV-1- and HSV-2-specific PCR systems was assessed by testing serial dilutions of quantified genomes from laboratory strains KOS and gHSV2. After optimization, regarding the UL23 gene, the detection thresholds were 300 and 3,000 copies/ml for HSV-1 and HSV-2, respectively, and regarding the UL30 gene, the detection thresholds were 30,000 and 300,000 copies/ml for HSV-1 and HSV-2, respectively.
Natural polymorphism.
The natural polymorphism of HSV TK and DNA polymerase was investigated among 94 clinical isolates and 3 laboratory strains. The overall results are presented in Tables 3 and 4 and in Fig. 1.
TABLE 3.
TK and DNA polymerase comparison, at both nucleotide and amino acid levels, among 94 clinical isolates and 3 laboratory strains of HSV-1 and HSV-2
| Parametera | TK |
DNA polymerase |
||
|---|---|---|---|---|
| HSV-1 (n = 44) | HSV-2 (n = 53) | HSV-1 (n = 44) | HSV-2 (n = 53) | |
| Nucleotide identity (%) | 97.5-99.2 | 99.6-100 | 99.4-99.8 | 99.7-100 |
| Nucleotide mutations (no.) | 72 | 25 | 156 | 51 |
| Frequency per 100 bp (mean) | 0.8-2.5 (1.2) | 0-0.4 (0.2) | 0.2-0.6 (0.4) | 0-0.3 (0.1) |
| Silent mutations (%) | 61.1 | 68 | 73.7 | 51 |
| Amino acid identity (%) | 97.3-98.9 | 98.9-100 | 99.4-99.8 | 99.3-100 |
| Amino acid changes (no.) | 30 | 8 | 41 | 25 |
| Frequency per 100 aa (mean) | 1.1-2.7 (1.6) | 0-1.1 (0.4) | 0.2-0.6 (0.4) | 0-0.7 (0.3) |
| Variation of the total codons (%) | 8 | 2.1 | 3.3 | 2 |
aa, amino acids.
TABLE 4.
Natural polymorphisms within TK and DNA polymerase identified among 94 clinical isolates and 3 laboratory strains of HSV-1 and HSV-2a
| Virus | TK |
DNA polymerase |
||||
|---|---|---|---|---|---|---|
| Amino acid change | No. of isolates (%) | Reference | Amino acid change | No. of isolates (%) | Reference | |
| HSV-1 (n = 44) | C6G | 14 (31.8) | 31, 47 | G12E | 1 (2.3) | |
| A12P | 1 (2.3) | F23I | 1 (2.3) | |||
| R20S | 1 (2.3) | S33G | 40 (90.9) | 10, 47 | ||
| N23S | 44 (100) | 31, 47 | Q42K | 1 (2.3) | ||
| R30C | 1 (2.3) | E70D | 1 (2.3) | |||
| K36E | 44 (100) | 31, 47 | A102T | 1 (2.3) | ||
| K36D | 1 (2.3) | R116H | 1 (2.3) | |||
| R41H | 1 (2.3) | 10, 47 | G132C | 1 (2.3) | ||
| L42P | 15 (34.1) | 31, 47 | P137Q | 1 (2.3) | ||
| R89Q | 26 (59.1) | 31, 47 | A138V | 1 (2.3) | ||
| E111K | 1 (2.3) | A330R | 44 (100) | 10, 47 | ||
| V138I | 1 (2.3) | 15 | I352V | 1 (2.3) | ||
| E146G | 1 (2.3) | E421D | 1 (2.3) | |||
| V191A | 1 (2.3) | N425T | 5 (11.4) | |||
| A192V | 2 (4.5) | 37, 47 | T434M | 1 (2.3) | ||
| G240E | 11 (25) | 31, 47 | A646T | 3 (6.8) | 50 | |
| A243S | 1 (2.3) | 18 | D660E | 1 (2.3) | ||
| G251C | 9 (20.5) | 37, 47 | D672N | 3 (6.8) | ||
| A265T | 43 (97.7) | 31, 47 | E675A | 1 (2.3) | ||
| V267L | 8 (18.)2 | 31, 47 | D703N | 1 (2.3) | ||
| P268T | 12 (27.3) | 31, 47 | N711K | 1 (2.3) | ||
| E273Q | 1 (2.3) | G749D | 1 (2.3) | |||
| E273K | 1 (2.3) | L753M | 1 (2.3) | |||
| D286E | 9 (20.5) | 31, 47 | P875S | 1 (2.3) | ||
| H323Y | 1 (2.3) | V905M | 33 (75) | 18, 47 | ||
| A334T | 1 (2.3) | P920S | 4 (9.1) | 3, 40 | ||
| V348I | 9 (20.5) | 10, 47 | A995T | 1 (2.3) | ||
| S357C | 1 (2.3) | E1082K | 1 (2.3) | |||
| N376H | 4 (9.1) | 10, 37 | A1099T | 1 (2.3) | ||
| N376P | 3 (6.8) | 10, 31 | E1104D | 1 (2.3) | ||
| S1113C | 1 (2.3) | |||||
| S1123L | 1 (2.3) | |||||
| P1124H | 17 (38.6) | 10, 47 | ||||
| G1129V | 1 (2.3) | |||||
| E1194D | 1 (2.3) | |||||
| P1199Q | 2 (4.5) | |||||
| A1203T | 2 (4.5) | 10, 47 | ||||
| A1204T | 1 (2.3) | |||||
| T1208A | 37 (84.1) | 10, 47 | ||||
| A1209T | 1 (2.3) | |||||
| T1219M | 1 (2.3) | |||||
| HSV-2 (n = 54) | A27T | 2 (3.8) | 10 | A9T | 49 (92.5) | 1, 10 |
| G39E | 40 (73.6) | 10 | P15S | 49 (92.5) | 10, 47 | |
| N78D | 19 (34) | 10 | P37L | 1 (1.9) | ||
| E96D | 1 (1.9) | R41H | 8 (15.1) | |||
| A135P | 1 (1.9) | Q43R | 1 (1.9) | |||
| D137E | 1 (1.9) | H49Y | 1 (1.9) | |||
| L140F | 16 (28.3) | 10 | L60P | 49 (92.5) | 1 | |
| A215T | 2 (3.8) | 10 | E139K | 16 (30.2) | 10, 47 | |
| A232T | 6 (11.3) | 47 | ||||
| G324E | 2 (3.8) | |||||
| Q507H | 1 (1.9) | |||||
| D671N | 1 (1.9) | |||||
| D676N | 1 (1.9) | |||||
| D676G | 4 (7.5) | |||||
| E678G | 11 (20.8) | 16, 44 | ||||
| G680R | 1 (1.9) | |||||
| E682K | 1 (1.9) | |||||
| Q799R | 1 (1.9) | |||||
| T801P | 6 (11.3) | 10, 47 | ||||
| T903M | 1 (1.9) | |||||
| A1000T | 1 (1.9) | |||||
| A1083T | 1 (1.9) | |||||
| A1114V | 1 (1.9) | |||||
| E1149G | 1 (1.9) | |||||
| A1223V | 1 (1.9) | |||||
| del DGDE (683-686) | 2 (3.8) | |||||
| delGD (681-682) | 1 (1.9) | |||||
| del DD (676-677) | 1 (1.9) | |||||
| del DED (677-679) | 1 (1.9) | |||||
| del PGDEPA (1106-1111) | 1 (1.9) | |||||
| ins DGDE (683-686) | 1 (1.9) | |||||
del, deletion; ins, insertion. Amino acid changes never previously reported are in bold.
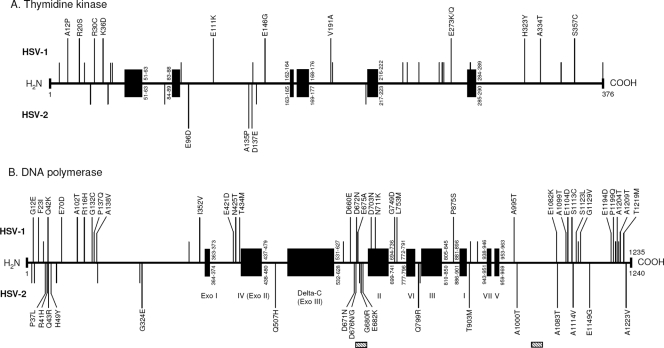
FIG. 1.
Natural polymorphism map (to scale) of thymidine kinase (TK) (A) and DNA polymerase (B) among HSV-1 and HSV-2 strains. For each viral enzyme, conserved regions and functional domains are indicated by the black boxes, and natural polymorphisms are represented separately for HSV-1 (top) and HSV-2 (bottom). Amino acid changes related to natural polymorphism are indicated by vertical bars: short vertical bars correspond to changes previously reported, long vertical bars labeled with amino acid change correspond to changes newly described in this study. Regarding HSV-2 DNA polymerase, hatched boxes indicate amino acid insertions or deletions related to natural polymorphism.
(i) TK analysis.
At the nucleotide level, the interstrain identity of the UL23 gene ranged from 97.5% to 99.2% for HSV-1 and from 99.6 to 100% for HSV-2. In comparison with the gene sequences from the corresponding reference strains, 72 and 25 nucleotide mutations were identified for the HSV-1 and HSV-2 strains, respectively, within the 1,195-bp coding sequence. For each strain, the number of nucleotide mutations ranged from 0.8 to 2.5 per 100 bp (mean of 1.2) for HSV-1 and from 0 to 0.4 per 100 bp (mean of 0.2) for HSV-2 (Table 3). At the amino acid level, TK revealed >97.3% peptide sequence identity. Overall, the number of amino acid changes related to natural polymorphism was 30 for HSV-1 and 8 for HSV-2, which represents 8% and 2.1% of the total codons of the protein, respectively (Table 3). The amino acid changes within the TK related to natural polymorphism and identified in this study are represented in Fig. 1A. All these changes were located within nonconserved domains of the viral protein, except the previously identified natural polymorphism D286E, which was detected in 9/44 (20.5%) HSV-1 clinical isolates. A total of 12 and 3 new polymorphisms never previously described were identified within HSV-1 and HSV-2 TK, respectively (Table 4).
(ii) DNA polymerase analysis.
The analysis of the UL30 gene evidenced an interstrain identity ranging from 99.4% to 99.8% and from 99.7 to 100% for HSV-1 and HSV-2, respectively. Surprisingly, with HSV-1 isolates, 156 nucleotide mutations, of which 73.7% produced silent mutations, were evidenced, whereas in HSV-2 isolates, only 51 nucleotide mutations were identified, and of them, 51% produced silent mutations. The average number of nucleotide mutations per strain was 0.4 and 0.1 per 100 bp for HSV-1 and HSV-2, respectively (Table 3). The overall amino acid identity of DNA polymerase ranged from 99.3% to 100%. Forty-one and twenty-five amino acid changes distributed along the DNA polymerase were identified for HSV-1 and HSV-2, respectively, corresponding to 3.3% and 2% of the total codons of the protein (Table 3). Of note, 7 (13.2%) HSV-2 isolates harbored amino acid insertions or deletions in two distinct regions of the UL30 DNA polymerase (codons 679 to 686 and codons 1106 to 1111) that accounted for natural polymorphism (Table 4). This natural polymorphism was unevenly distributed alongside the viral protein. Indeed, amino acid changes were located mainly outside the defined conserved domains, except D703N and N711K changes, which were located in conserved catalytic region II of the DNA polymerase (Fig. 1B). For HSV-1 isolates, the greatest numbers of polymorphisms were strongly clustered at both the UL30 N terminus (codons 12 to 138) and C terminus (codons 1082 to 1219). In this study, 33 and 18 novel polymorphisms unreported before were identified in the DNA polymerase of HSV-1 and HSV-2 clinical isolates, respectively (Table 4).
Genotypic characterization of drug-resistant HSV clinical isolates.
The genotypic characterization of TK and DNA polymerase from 24 ACV-resistant/FOS-sensitive isolates and one ACV- and FOS-resistant isolate of HSV-1 (n = 11) and HSV-2 (n = 14) was performed (Table 5).
TABLE 5.
Genotypic characterization of TK and DNA polymerase from 25 drug-resistant HSV clinical isolatesc
| Isolate no. | Patient age | Clinical context | Sample | HSV type | Phenotypic susceptibility (EC50 [μM]) |
TK mutations |
DNA polymerase mutations |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACV | FOS | Natural polymorphism | Resistancea | Unknownb | Natural polymorphism | Resistancea | Unknownb | |||||
| 1 | 21 | BMT | Mouth | 1 | >50 | <66 | C6G N23S K36E L42P G240E A265T | No mutation | A207P | S33G A330R N425T V905M | ||
| 2 | 18 | BMT | Blood | 1 | 11 | <66 | C6G N23S K36E R41H A192V A265T | No mutation | L170P | S33G A330R V905M T1208A | ||
| 3 | 77 | IC | Eye | 1 | 35 | <66 | N23S K36E R89Q A265T | No mutation | Y53D | S33G A330R V905M E1005K | ||
| 4 | 51 | BMT | Anal | 1 | 16 | <66 | N23S K36E | −1C 553 = frameshift | S33G A330R A899V | L1188F | ||
| 5 | 8 | Wiskott-Aldrich | Lip | 1 | 16 | <66 | N23S K36E A192V G251C A265T V267L P268T | −1G 856 = frameshift | A330R V905M T1208A | H98Y | ||
| 6 | 37 | BMT | Skin | 1 | 26 | <66 | N23S K36E R89Q | +1G 437-438 = Stop 225 | S33G A330R A646T S1123L P1124H T1208A | R1229I | ||
| 7 | 59 | CLL | Gingiva | 1 | 31 | <66 | N23S K36E R89Q | +1C 464-465 = Stop 228 | S33G A330R P1124H T1208A | A870G | ||
| 8 | 39 | BMT | Mouth | 1 | 10 | <66 | N23S K36E R89Q | +1G 437-438 = Stop 228 | A330R P875S V905M P1124H T1208A | |||
| 9 | 52 | BMT | Palate | 1 | >50 | 106 | N23S K36E R89Q | +1G 437-438 = Stop 228 | S33G A330R V905M A1203T T1208A | |||
| 10 | 8 | ALL | BAL fluid | 1 | 22 | <66 | N23S K36E R89Q A265T | C336Y | S33G A330R V905M | |||
| 11 | 8 | BMT | Mouth | 1 | >50 | 644 | N23S K36E G251C A265T P268T | No mutation | R176W | S33G A330R P1124H T1208A | S724N | V117L L267M |
| 12 | 47 | HIV | Genital | 2 | 43 | <66 | G39E N78D L140F | No mutation | S66P A72S | A9T P15S L60P | ||
| 13 | 45 | HIV | Genital | 2 | 33 | <66 | None | No mutation | M183I | A9T P15S R41H L60P E139K A232T | ||
| 14 | 43 | HIV | Genital | 2 | 24 | 92 | G39E N78D | No mutation | M70R | None | A724V | |
| 15 | 53 | HIV | Anal | 2 | 11 | 99 | None | R221H | A9T P15S L60P | |||
| 16 | 52 | HIV | Genital | 2 | >50 | <66 | None | D137Stop | A9T P15S L60P | |||
| 17 | 34 | HIV | Genital | 2 | >50 | <66 | G39E N78D | Q222Stop | A9T P15S L60P T801P | |||
| 18 | 37 | HIV | Anal | 2 | 18 | <66 | A27T G39E N78D | −1C 556 = Stop 263 | S29A | A9T P15S L60P E678G del DD676-677 | ||
| 19 | 55 | HIV | Anal | 2 | >50 | <66 | G39E | +2G 439-440 = Stop 184 | A9T P15S L60P del DGDE683-686 | |||
| 20 | 63 | HIV | Anal | 2 | >50 | <66 | G39E | −1C 467 = Stop 183 | A9T P15S L60P E139K T801P | |||
| 21 | 39 | HIV | Lip | 2 | >50 | 89 | G39E N78D L140F | −1C 452 = Stop 183 | P15S L60P E678G | |||
| 22 | 44 | HIV | Genital | 2 | >50 | 76 | G39E N78D L140F | +1G 439-440 = Stop 229 | A9T P15S L60P E678G | Y823C | ||
| 23 | 44 | BMT | Anal | 2 | >50 | <66 | G39E N78D L140F | +1G 439-440 = Stop 229 | A9T P15S L60P | I291V | ||
| 24 | 70 | CLL | Anal | 2 | >50 | <66 | G39E | −1G 440 = Stop 229 | A9T P15S L60P E139K | H837R | ||
| 25 | 39 | HIV | Genital | 2 | 42 | 145 | None | No mutation | I101S | None | D785N | V544A |
Previously described resistance mutations.
Not previously described as known polymorphism or associated with resistance.
BMT, bone marrow transplantation; IC, immunocompetent individual; ALL, acute lymphoblastic leukemia; CLL, chronic lymphocytic leukemia; BAL, bronchoalveolar lavage. The EC50 cutoff values for resistance to ACV and FOS were 7 and 330 μM, respectively.
Among 18 isolates, the reduced susceptibility to antivirals evidenced by PRA was confirmed by the genotypic analysis (HSV-1 isolates 4 to 11 and HSV-2 isolates 15 to 25). Regarding TK, 13 clinical isolates harbored a C or G insertion/deletion in homopolymer repeats, leading to the creation of either a frameshift (isolates 4 and 5) or a premature stop codon (isolates 6 to 9 and 18 to 24). Isolates 16 and 17 presented nucleotide mutations producing stop codons (D137Stop and Q222Stop, respectively). Isolates 10 and 15 harbored amino acid changes (C336Y and R221H, respectively) previously associated with ACV resistance. Isolate 11 contained an S724N change within DNA polymerase associated with cross-resistance to ACV and FOS. This isolate was recovered from a patient who received successive courses of intravenous ACV and FOS treatments. Isolate 25 harbored a D785N change in conserved domain VI of the DNA polymerase which is involved in the acquisition of drug resistance.
Apart from the mutational changes surely conferring drug resistance, several amino acid changes associated with natural polymorphism, either previously reported in the literature or newly identified in the first part of this study, were detected in both TK and DNA polymerase from all those 18 HSV clinical isolates. Moreover, new potential natural polymorphisms located outside conserved domains were evidenced in TK (S29A, isolate 18) and in DNA polymerase (H98Y, isolate 5; V117L and L267M, isolate 11; A870G, isolate 7; L1188F, isolate 4; R1229I, isolate 6). Regarding HSV-2 DNA polymerase, new polymorphisms were located within both nonconserved regions (I291V, isolate 23) and conserved domains (Exo III motif [V544A, isolate 25] and region III [Y823C, isolate 22; H837R, isolate 24]). Furthermore, 2 amino acid deletions, del DD 676 and 677 and del DGDE 683 to 686, located between domains delta-C and II of the DNA polymerase in isolates 18 and 19, respectively, were evidenced. These modifications have been associated previously in our study with natural polymorphism among susceptible HSV-2 clinical isolates.
Several HSV-1 clinical isolates harbored amino acid changes not previously described in the literature and potentially conferring drug resistance. Indeed, the 3 novel changes A207P, L170P, and Y53D of unknown phenotype were evidenced in TK from ACV-resistant HSV-1 isolates (isolates 1, 2 and 3, respectively), whereas no additional mutation in DNA polymerase could be found. Moreover, in addition to the S724N change in DNA polymerase, isolate 11 contained the change R176W in TK. This change likely confers ACV resistance since the R177W change in HSV-1 TK has been identified previously in strains exhibiting ACV resistance. Potential novel amino acid changes related to drug resistance have also been identified among HSV-2 isolates. S66P, A72S (isolate 12), and M183I (isolate 14) were the only changes located within TK likely to confer ACV resistance. In HSV-2 isolate 15, the A724V change in DNA polymerase might be associated with ACV resistance. Therefore, the M70R change, located outside conserved domains of the TK, presumably reflected natural polymorphism.
DISCUSSION
In this study, new techniques for the sequencing of the entire UL23 and UL30 genes from HSV-1 and HSV-2 clinical isolates for rapid genotypic antiviral resistance testing were developed. This molecular approach shortens the time for the detection of drug resistance mutations compared to the PRA. In this work, gene sequencing was performed from viral stocks obtained after propagation in cell culture. However, in accordance with previous results, the detection thresholds allow the amplification and sequencing of the UL23 gene directly from most of the clinical samples (18). Conversely, the higher detection thresholds obtained for the UL30 gene (30,000 and 300,000 copies/ml for HSV-1 and HSV-2, respectively), related to the size of the gene (about 3,700 bp), deserve some modifications to improve the sensitivity of these techniques for direct gene sequencing from clinical samples.
The genetic variability of the HSV genome has been described previously (6, 7, 25, 42, 43). Our results concerning 94 drug-sensitive clinical isolates and 3 laboratory strains (KOS, gHSV2, and MS2) confirmed that UL23 and UL30 genes are highly conserved. The interstrain identity was >97.5% at the nucleotide and the amino acid levels for both genes among all HSV strains investigated. Nevertheless, a weaker variability was evidenced for HSV-2. Thus, the total number of nucleotide mutations was about 3-fold higher in UL23 and UL30 genes in HSV-1 strains than in HSV-2 strains, and the same trend was observed when considering the amino acid changes. Moreover, the amino acid variation of the total codons of TK and DNA polymerase in HSV-1 strains was about 4-fold and 2-fold higher, respectively, than in HSV-2 strains (Table 3). Our results are in agreement with previous reports (9). A higher GC content could account for higher polymorphism (38). Nevertheless, HSV-1 and HSV-2 genomes harbor similar GC contents, around 70% (16). Moreover, UL23 and UL30 genes exhibit nearly identical GC contents for the GenBank reference strains 17 (HSV-1) and HG52 (HSV-2) (data not shown). The explanation of the higher variability of the HSV-1 genome compared to the HSV-2 genome remains unclear and needs further investigation.
One objective of this work was to determine the natural polymorphism of TK and DNA polymerase among drug-sensitive HSV strains. Thus, to date, few studies have described extensively the degree of HSV DNA polymerase polymorphism, especially concerning HSV-2 (10, 18, 47). In the present study, 12 and 3 novel polymorphisms within TK and 33 and 18 novel polymorphisms within DNA polymerase have been evidenced among HSV-1 and HSV-2 isolates, respectively (Table 4). The precise role of some mutations remains unclear. The R41H change has been reported previously in TK from both ACV-sensitive and ACV-resistant HSV-1 strains (5, 10, 15, 47, 49, 51). Here, this change was evidenced in only one sensitive isolate. Despite that this change is located outside conserved regions, its role regarding TK activity must be clarified. In DNA polymerase, the E678G change has been described previously in resistant HSV-2 strains (16, 44). In our study, this change has been found among 5 different ACV- and FOS-sensitive HSV-2 isolates. Moreover, it is located in a nonconserved region of the DNA polymerase, between δ-region C and region II. All together, these data indicate that the E678G change is likely to be related to natural polymorphism. The D672N change has been identified among 3 HSV-1 isolates obtained from patients without any previous antiviral treatment, whereas it was recently evidenced among 3 phenotypically characterized ACV- and FOS-resistant HSV-1 isolates (47). This discrepancy deserves further studies to assess the exact role of the D672N change within HSV-1. Amino acid deletions and insertions located within 2 different regions of the DNA polymerase were identified among phenotypically characterized drug-sensitive HSV-2 isolates. This phenomenon has never been reported previously. With regard to the crystal structure of HSV DNA polymerase, these amino acid insertions or deletions might not interfere with the catalytic and functional sites of the enzyme (33). Thus, they are likely related to the HSV-2 DNA polymerase natural polymorphism. Overall, this part of our study permitted us to draw a more detailed natural polymorphism map of TK and DNA polymerase for HSV-1 and HSV-2 isolates (Fig. 1).
In order to identify potential new mutations within TK and DNA polymerase leading to HSV resistance, the genotypic characterization of 24 ACV-resistant/FOS-sensitive isolates and one ACV- and FOS-resistant isolate was performed. Regarding TK, C or G insertions/deletions in homopolymer repeats, evidenced in 13 isolates, and the changes R221H and C336Y have previously been implicated in many ACV-resistant HSV strains (21, 24, 37, 40, 41, 47). Moreover, this study described new amino acid changes in TK potentially related to ACV resistance: A207P, L170P, and Y53D in HSV-1 isolates 1, 2, and 3, respectively. Only the Y53D change was located in a conserved domain. Since no other resistance mutation was evidenced either in TK or DNA polymerase, these 3 changes are likely to be related to ACV resistance. To date, none of them has been described in the literature. However, according to Sauerbrei et al. (47), the Y53H and Y53N changes, located in the ATP-binding site, were most likely implicated in ACV resistance. Similarly, in TK from HSV-2 isolates 12 and 13, amino acid changes whose role in ACV resistance needs to be investigated were evidenced: S66P, A72S, and M183I. Even if these changes are located outside conserved regions, they might have led to lower TK catalytic activity resulting in ACV resistance since there was no evidence for an involvement of the DNA polymerase.
DNA polymerase sequencing of isolate 11 revealed the S724N change, located in conserved region II, which has been involved previously in HSV-1 cross-resistance to ACV and FOS (3). However, this isolate contained also the R176W change within the TK. To date, the latter amino acid change has been reported only in an HSV-1 laboratory strain selected after serial passages in the presence of ACV. But this strain harbored mutations in both TK and DNA polymerase, making it difficult to interpret the respective roles of each mutation regarding ACV resistance (52). Moreover, the changes R176Q for HSV-1 and R177W for HSV-2 have been associated with ACV resistance (21, 24, 30, 39). The involvement of the TK R176W change in HSV-1 ACV resistance requires assessment. The ACV-resistant/FOS-sensitive isolate 25 contained the D785N change, located in the conserved region VI of DNA polymerase. This change has been related previously to the ACV-sensitive/FOS-resistant HSV-2 phenotype (4, 48). Nevertheless, one can consider that this change may account for the elevation of the FOS EC50 (145 μM), above the cutoff value for resistance (330 μM), leading to the possible involvement of the TK I101S change in ACV resistance. The putative role of both changes remains to be elucidated. HSV-2 isolate 15, harboring the ACV-resistant/FOS-sensitive phenotype, contained the A724V change within conserved region II of DNA polymerase. This change has never been reported so far. Nevertheless, the A724T change has previously been reported to be related to the ACV-sensitive/FOS-resistant phenotype in HSV-2 clinical isolates (4, 48). Additionally, the A719V and A719T changes, the counterparts in HSV-1, are known to confer resistance to both ACV and FOS (3, 29, 32). In conclusion, the A724V change may account for resistance to ACV in HSV-2 isolate 15.
Considering all the previously reported mutations related to natural polymorphism and resistance to antivirals, and the potential resistance mutations described above, we have identified in this study some new potential natural polymorphisms among drug-resistant HSV isolates, either within TK (S29A for HSV-2) or DNA polymerase (H98Y, V117L, L267M, A870G, L1188F, and R1229I for HSV-1; I291V, V544A, Y823C, and H837R for HSV-2) (Table 5).
In conclusion, this study has provided new data concerning the natural polymorphism of TK and DNA polymerase for both HSV-1 and HSV-2, which will facilitate the interpretation of genotypic antiviral resistance testing. Moreover, 8 novel amino acid changes in TK potentially related to ACV resistance have been identified: Y53D, I101S, L170P, and A207P for HSV-1, and S66P, A72S, R176W, and M183I for HSV-2. The exact role of these different changes in the acquisition of resistance to ACV is now going to be investigated using site-directed mutagenesis, as previously described (19, 20). Together with previous studies, these results may allow us to provide a large database of HSV UL23 TK and UL30 DNA polymerase mutations, in order to develop new tools for a fast interpretation of genotypic resistance testing, as recently performed for human cytomegalovirus (8).
Acknowledgments
We thank Françoise Conan, Bruno Le Labousse, Fabienne Mousnier, and Zaïna Aït-Arkoub for helpful advice and excellent technical assistance.
This work was supported in part by the Association pour la Recherche sur les Infections Virales (ARIV).
Footnotes
Published ahead of print on 23 August 2010.
REFERENCES
- 1.Andrei, G., P. Fiten, M. Froeyen, E. De Clercq, G. Opdenakker, and R. Snoeck. 2007. DNA polymerase mutations in drug-resistant herpes simplex virus mutants determine in vivo neurovirulence and drug-enzyme interactions. Antivir. Ther. 12:719-732. [PubMed] [Google Scholar]
- 2.Balasubramaniam, N. K., V. Veerisetty, and G. A. Gentry. 1990. Herpesviral deoxythymidine kinases contain a site analogous to the phosphoryl-binding arginine-rich region of porcine adenylate kinase; comparison of secondary structure predictions and conservation. J. Gen. Virol. 71(12):2979-2987. [DOI] [PubMed] [Google Scholar]
- 3.Bestman-Smith, J., and G. Boivin. 2003. Drug resistance patterns of recombinant herpes simplex virus DNA polymerase mutants generated with a set of overlapping cosmids and plasmids. J. Virol. 77:7820-7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bestman-Smith, J., and G. Boivin. 2002. Herpes simplex virus isolates with reduced adefovir susceptibility selected in vivo by foscarnet therapy. J. Med. Virol. 67:88-91. [DOI] [PubMed] [Google Scholar]
- 5.Bestman-Smith, J., I. Schmit, B. Papadopoulou, and G. Boivin. 2001. Highly reliable heterologous system for evaluating resistance of clinical herpes simplex virus isolates to nucleoside analogues. J. Virol. 75:3105-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchman, T. G., T. Simpson, C. Nosal, B. Roizman, and A. J. Nahmias. 1980. The structure of herpes simplex virus DNA and its application to molecular epidemiology. Ann. N. Y. Acad. Sci. 354:279-290. [DOI] [PubMed] [Google Scholar]
- 7.Chaney, S. M., K. G. Warren, J. Kettyls, A. Zbitnue, and J. H. Subak-Sharpe. 1983. A comparative analysis of restriction enzyme digests of the DNA of herpes simplex virus isolated from genital and facial lesions. J. Gen. Virol. 64(2):357-371. [DOI] [PubMed] [Google Scholar]
- 8.Chevillotte, M., J. von Einem, B. M. Meier, F. M. Lin, H. A. Kestler, and T. Mertens. 2010. A new tool linking human cytomegalovirus drug resistance mutations to resistance phenotypes. Antiviral Res. 85:318-327. [DOI] [PubMed] [Google Scholar]
- 9.Chiba, A., T. Suzutani, M. Saijo, S. Koyano, and M. Azuma. 1998. Analysis of nucleotide sequence variations in herpes simplex virus types 1 and 2, and varicella-zoster virus. Acta Virol. 42:401-407. [PubMed] [Google Scholar]
- 10.Chibo, D., J. Druce, J. Sasadeusz, and C. Birch. 2004. Molecular analysis of clinical isolates of acyclovir resistant herpes simplex virus. Antiviral Res. 61:83-91. [DOI] [PubMed] [Google Scholar]
- 11.Chibo, D., A. Mijch, R. Doherty, and C. Birch. 2002. Novel mutations in the thymidine kinase and DNA polymerase genes of acyclovir and foscarnet resistant herpes simplex viruses infecting an immunocompromised patient. J. Clin. Virol. 25:165-170. [DOI] [PubMed] [Google Scholar]
- 12.Danve-Szatanek, C., M. Aymard, D. Thouvenot, F. Morfin, G. Agius, I. Bertin, S. Billaudel, B. Chanzy, M. Coste-Burel, L. Finkielsztejn, H. Fleury, T. Hadou, C. Henquell, H. Lafeuille, M. E. Lafon, A. Le Faou, M. C. Legrand, L. Maille, C. Mengelle, P. Morand, F. Morinet, E. Nicand, S. Omar, B. Picard, B. Pozzetto, J. Puel, D. Raoult, C. Scieux, M. Segondy, J. M. Seigneurin, R. Teyssou, and C. Zandotti. 2004. Surveillance network for herpes simplex virus resistance to antiviral drugs: 3-year follow-up. J. Clin. Microbiol. 42:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darby, G., H. J. Field, and S. A. Salisbury. 1981. Altered substrate specificity of herpes simplex virus thymidine kinase confers acyclovir-resistance. Nature 289:81-83. [DOI] [PubMed] [Google Scholar]
- 14.Dignani, M. C., A. Mykietiuk, M. Michelet, D. Intile, L. Mammana, P. Desmery, G. Milone, and S. Pavlovsky. 2002. Valacyclovir prophylaxis for the prevention of herpes simplex virus reactivation in recipients of progenitor cells transplantation. Bone Marrow Transplant. 29:263-267. [DOI] [PubMed] [Google Scholar]
- 15.Duan, R., R. D. de Vries, J. M. van Dun, F. B. van Loenen, A. D. Osterhaus, L. Remeijer, and G. M. Verjans. 2009. Acyclovir susceptibility and genetic characteristics of sequential herpes simplex virus type 1 corneal isolates from patients with recurrent herpetic keratitis. J. Infect. Dis. 200:1402-1414. [DOI] [PubMed] [Google Scholar]
- 16.Duffy, K. E., M. R. Quail, T. T. Nguyen, R. J. Wittrock, J. O. Bartus, W. M. Halsey, J. J. Leary, T. H. Bacon, and R. T. Sarisky. 2002. Assessing the contribution of the herpes simplex virus DNA polymerase to spontaneous mutations. BMC Infect. Dis. 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Englund, J. A., M. E. Zimmerman, E. M. Swierkosz, J. L. Goodman, D. R. Scholl, and H. H. Balfour, Jr. 1990. Herpes simplex virus resistant to acyclovir. A study in a tertiary care center. Ann. Intern. Med. 112:416-422. [DOI] [PubMed] [Google Scholar]
- 18.Frobert, E., J. C. Cortay, T. Ooka, F. Najioullah, D. Thouvenot, B. Lina, and F. Morfin. 2008. Genotypic detection of acyclovir-resistant HSV-1: characterization of 67 ACV-sensitive and 14 ACV-resistant viruses. Antiviral Res. 79:28-36. [DOI] [PubMed] [Google Scholar]
- 19.Frobert, E., T. Ooka, J. C. Cortay, B. Lina, D. Thouvenot, and F. Morfin. 2005. Herpes simplex virus thymidine kinase mutations associated with resistance to acyclovir: a site-directed mutagenesis study. Antimicrob. Agents Chemother. 49:1055-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frobert, E., T. Ooka, J. C. Cortay, B. Lina, D. Thouvenot, and F. Morfin. 2007. Resistance of herpes simplex virus type 1 to acyclovir: thymidine kinase gene mutagenesis study. Antiviral Res. 73:147-150. [DOI] [PubMed] [Google Scholar]
- 21.Gaudreau, A., E. Hill, H. H. Balfour, Jr., A. Erice, and G. Boivin. 1998. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J. Infect. Dis. 178:297-303. [DOI] [PubMed] [Google Scholar]
- 22.Gibbs, J. S., H. C. Chiou, K. F. Bastow, Y. C. Cheng, and D. M. Coen. 1988. Identification of amino acids in herpes simplex virus DNA polymerase involved in substrate and drug recognition. Proc. Natl. Acad. Sci. U. S. A. 85:6672-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert, C., J. Bestman-Smith, and G. Boivin. 2002. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist. Updat. 5:88-114. [DOI] [PubMed] [Google Scholar]
- 24.Graham, D., B. A. Larder, and M. M. Inglis. 1986. Evidence that the ‘active centre’ of the herpes simplex virus thymidine kinase involves an interaction between three distinct regions of the polypeptide. J. Gen. Virol. 67(4):753-758. [DOI] [PubMed] [Google Scholar]
- 25.Hayward, G. S., R. J. Jacob, S. C. Wadsworth, and B. Roizman. 1975. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc. Natl. Acad. Sci. U. S. A. 72:4243-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill, E. L., G. A. Hunter, and M. N. Ellis. 1991. In vitro and in vivo characterization of herpes simplex virus clinical isolates recovered from patients infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 35:2322-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang, C. B., K. L. Ruffner, and D. M. Coen. 1992. A point mutation within a distinct conserved region of the herpes simplex virus DNA polymerase gene confers drug resistance. J. Virol. 66:1774-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kit, S., M. Kit, H. Qavi, D. Trkula, and H. Otsuka. 1983. Nucleotide sequence of the herpes simplex virus type 2 (HSV-2) thymidine kinase gene and predicted amino acid sequence of thymidine kinase polypeptide and its comparison with the HSV-1 thymidine kinase gene. Biochim. Biophys. Acta 741:158-170. [DOI] [PubMed] [Google Scholar]
- 29.Knopf, C. W. 1987. The herpes simplex virus type 1 DNA polymerase gene: site of phosphonoacetic acid resistance mutation in strain Angelotti is highly conserved. J. Gen. Virol. 68(5):1429-1433. [DOI] [PubMed] [Google Scholar]
- 30.Kost, R. G., E. L. Hill, M. Tigges, and S. E. Straus. 1993. Brief report: recurrent acyclovir-resistant genital herpes in an immunocompetent patient. N. Engl. J. Med. 329:1777-1782. [DOI] [PubMed] [Google Scholar]
- 31.Kudo, E., H. Shiota, T. Naito, K. Satake, and M. Itakura. 1998. Polymorphisms of thymidine kinase gene in herpes simplex virus type 1: analysis of clinical isolates from herpetic keratitis patients and laboratory strains. J. Med. Virol. 56:151-158. [DOI] [PubMed] [Google Scholar]
- 32.Larder, B. A., S. D. Kemp, and G. Darby. 1987. Related functional domains in virus DNA polymerases. EMBO J. 6:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, S., J. D. Knafels, J. S. Chang, G. A. Waszak, E. T. Baldwin, M. R. Deibel, Jr., D. R. Thomsen, F. L. Homa, P. A. Wells, M. C. Tory, R. A. Poorman, H. Gao, X. Qiu, and A. P. Seddon. 2006. Crystal structure of the herpes simplex virus 1 DNA polymerase. J. Biol. Chem. 281:18193-18200. [DOI] [PubMed] [Google Scholar]
- 34.McGeoch, D. J., A. Dolan, S. Donald, and F. J. Rixon. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 181:1-13. [DOI] [PubMed] [Google Scholar]
- 35.McGeoch, D. J., H. W. Moss, D. McNab, and M. C. Frame. 1987. DNA sequence and genetic content of the HindIII l region in the short unique component of the herpes simplex virus type 2 genome: identification of the gene encoding glycoprotein G, and evolutionary comparisons. J. Gen. Virol. 68(1):19-38. [DOI] [PubMed] [Google Scholar]
- 36.Morfin, F., K. Bilger, A. Boucher, A. Thiebaut, F. Najioullah, N. Bleyzac, N. Raus, S. Bosshard, M. Aymard, M. Michallet, and D. Thouvenot. 2004. HSV excretion after bone marrow transplantation: a 4-year survey. J. Clin. Virol. 30:341-345. [DOI] [PubMed] [Google Scholar]
- 37.Morfin, F., G. Souillet, K. Bilger, T. Ooka, M. Aymard, and D. Thouvenot. 2000. Genetic characterization of thymidine kinase from acyclovir-resistant and -susceptible herpes simplex virus type 1 isolated from bone marrow transplant recipients. J. Infect. Dis. 182:290-293. [DOI] [PubMed] [Google Scholar]
- 38.Morfin, F., and D. Thouvenot. 2003. Herpes simplex virus resistance to antiviral drugs. J. Clin. Virol. 26:29-37. [DOI] [PubMed] [Google Scholar]
- 39.Nugier, F., J. N. Colin, M. Aymard, and M. Langlois. 1992. Occurrence and characterization of acyclovir-resistant herpes simplex virus isolates: report on a two-year sensitivity screening survey. J. Med. Virol. 36:1-12. [DOI] [PubMed] [Google Scholar]
- 40.Prichard, M. N., D. C. Quenelle, C. B. Hartline, E. A. Harden, G. Jefferson, S. L. Frederick, S. L. Daily, R. J. Whitley, K. N. Tiwari, J. A. Maddry, J. A. Secrist III, and E. R. Kern. 2009. Inhibition of herpesvirus replication by 5-substituted 4′-thiopyrimidine nucleosides. Antimicrob. Agents Chemother. 53:5251-5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rechtin, T. M., M. E. Black, F. Mao, M. L. Lewis, and R. R. Drake. 1995. Purification and photoaffinity labeling of herpes simplex virus type-1 thymidine kinase. J. Biol. Chem. 270:7055-7060. [DOI] [PubMed] [Google Scholar]
- 42.Rojas, J. M., J. Dopazo, E. Martin-Blanco, C. Lopez-Galindez, and E. Tabares. 1993. Analysis of genetic variability of populations of herpes simplex viruses. Virus Res. 28:249-261. [DOI] [PubMed] [Google Scholar]
- 43.Rojas, J. M., J. Dopazo, M. Santana, C. Lopez-Galindez, and E. Tabares. 1995. Comparative study of the genetic variability in thymidine kinase and glycoprotein B genes of herpes simplex viruses by the RNase A mismatch cleavage method. Virus Res. 35:205-214. [DOI] [PubMed] [Google Scholar]
- 44.Sarisky, R. T., T. T. Nguyen, K. E. Duffy, R. J. Wittrock, and J. J. Leary. 2000. Difference in incidence of spontaneous mutations between herpes simplex virus types 1 and 2. Antimicrob. Agents Chemother. 44:1524-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarisky, R. T., M. R. Quail, P. E. Clark, T. T. Nguyen, W. S. Halsey, R. J. Wittrock, J. O'Leary Bartus, M. M. Van Horn, G. M. Sathe, S. Van Horn, M. D. Kelly, T. H. Bacon, and J. J. Leary. 2001. Characterization of herpes simplex viruses selected in culture for resistance to penciclovir or acyclovir. J. Virol. 75:1761-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sasadeusz, J. J., F. Tufaro, S. Safrin, K. Schubert, M. M. Hubinette, P. K. Cheung, and S. L. Sacks. 1997. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J. Virol. 71:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sauerbrei, A., S. Deinhardt, R. Zell, and P. Wutzler. 2010. Phenotypic and genotypic characterization of acyclovir-resistant clinical isolates of herpes simplex virus. Antiviral Res. 86:246-252. [DOI] [PubMed] [Google Scholar]
- 48.Schmit, I., and G. Boivin. 1999. Characterization of the DNA polymerase and thymidine kinase genes of herpes simplex virus isolates from AIDS patients in whom acyclovir and foscarnet therapy sequentially failed. J. Infect. Dis. 180:487-490. [DOI] [PubMed] [Google Scholar]
- 49.Schulte, E. C., A. Sauerbrei, D. Hoffmann, C. Zimmer, B. Hemmer, and M. Mühlau. 2010. Acyclovir resistance in herpes simplex encephalitis. Ann. Neurol. 67:830-833. [DOI] [PubMed] [Google Scholar]
- 50.Stránská, R., R. Schuurman, D. R. Scholl, J. A. Jollick, C. J. Shaw, C. Loef, M. Polman, and A. M. van Loon. 2004. ELVIRA HSV, a yield reduction assay for rapid herpes simplex virus susceptibility testing. Antimicrob. Agents Chemother. 48:2331-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stránská, R., A. M. van Loon, M. Polman, M. F. Beersma, R. G. Bredius, A. C. Lankester, E. Meijer, and R. Schuurman. 2004. Genotypic and phenotypic characterization of acyclovir-resistant herpes simplex viruses isolated from haematopoietic stem cell transplant recipients. Antivir. Ther. 9:565-575. [PubMed] [Google Scholar]
- 52.Suzutani, T., K. Ishioka, E. De Clercq, K. Ishibashi, H. Kaneko, T. Kira, K. Hashimoto, M. Ogasawara, K. Ohtani, N. Wakamiya, and M. Saijo. 2003. Differential mutation patterns in thymidine kinase and DNA polymerase genes of herpes simplex virus type 1 clones passaged in the presence of acyclovir or penciclovir. Antimicrob. Agents Chemother. 47:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thi, T. N., C. Deback, I. Malet, P. Bonnafous, Z. Ait-Arkoub, and H. Agut. 2006. Rapid determination of antiviral drug susceptibility of herpes simplex virus types 1 and 2 by real-time PCR. Antiviral Res. 69:152-157. [DOI] [PubMed] [Google Scholar]
- 54.Tsurumi, T., K. Maeno, and Y. Nishiyama. 1987. Nucleotide sequence of the DNA polymerase gene of herpes simplex virus type 2 and comparison with the type 1 counterpart. Gene 52:129-137. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, J., D. W. Chung, C. K. Tan, K. M. Downey, E. W. Davie, and A. G. So. 1991. Primary structure of the catalytic subunit of calf thymus DNA polymerase delta: sequence similarities with other DNA polymerases. Biochemistry 30:11742-11750. [DOI] [PubMed] [Google Scholar]