Abstract
The blaOXA-51-like gene with an upstream ISAba1 (ISAba1-blaOXA-51-like gene) was originally found on the chromosomes of carbapenem-resistant or -susceptible Acinetobacter baumannii isolates. However, a plasmid-borne ISAba1-blaOXA-51-like gene has recently been identified in Acinetobacter genomic species 13TU and several A. baumannii isolates in Taiwan, and all of the isolates are carbapenem resistant. This study aimed to characterize the plasmids bearing the ISAba1-blaOXA-51-like gene and their significance in A. baumannii. Among the 117 ISAba1-blaOXA-51-like-harboring isolates collected from 10 hospitals in Taiwan, 58 isolates (49.6%) from 24 clones had the genes located on plasmids that likely originated from a common progenitor. Among the 58 isolates, four had additional copy of the ISAba1-blaOXA-51-like gene on their chromosomes. Based on the analysis of these four isolates, the plasmid-located ISAba1-blaOXA-51-like gene appeared to be acquired via one-ended transposition (Tn6080). The isolates with a plasmid bearing the ISAba1-blaOXA-51-like gene had higher rates of resistance to imipenem (98% versus 46.6%; P < 0.001) and meropenem (98% versus 69%; P = 0.019) than those with the genes chromosomally encoded, which is most likely due to increased gene dosage provided by the higher copy number of associated plasmids. Transformation with a recombinant plasmid harboring only the ISAba1-blaOXA-51-like gene was enough to confer a high level of carbapenem resistance to A. baumannii, eliminating the possible contribution of other factors on the original plasmids. This study demonstrated that the carbapenem resistance-associated plasmids carrying the ISAba1-blaOXA-51-like gene are widespread in A. baumannii strains in Taiwan.
Acinetobacter baumannii has emerged as a major pathogen of nosocomial infections in immunocompromised patients and is associated with a high mortality rate (7). The management of this pathogen has become a significant challenge due to the increased emergence of carbapenem-resistant strains (19, 22). It has been shown that most of the carbapenem resistance in A. baumannii is due to the production of carbapenemases, especially those belonging to carbapenem-hydrolyzing class D beta-lactamases (CHDLs), which are encoded by the blaOXA-51-like, blaOXA-23-like, blaOXA-24-like, and blaOXA-58-like genes (19). Among these CHDL genes, the blaOXA-51-like gene is intrinsic to and was originally located on the chromosome of A. baumannii (7, 18, 24).
The CHDLs have only a weak carbapenem-hydrolyzing activity (1, 19). However, overexpression of these CHDL genes, driven mostly by promoters provided by their upstream insertion sequences (ISs), is one of the means by which A. baumannii acquires a high level of carbapenem resistance. For example, ISAba1 upstream of the blaOXA-51-like gene is associated with the overexpression of the blaOXA-51-like gene and carbapenem resistance in A. baumannii (12, 23). However, some isolates harboring the blaOXA-51-like gene with an upstream ISAba1 (ISAba1-blaOXA-51-like gene) are still susceptible to carbapenem (9, 14, 16). The reason for this discrepancy in carbapenem susceptibility levels in isolates harboring the ISAba1-blaOXA-51-like gene has not been completely clarified. In some isolates, the high level of resistance might be due to interplay between the overexpression of the blaOXA-51-like gene and other mechanisms, such as overexpression of an efflux pump (12, 19). However, the overexpression of the CHDL genes by themselves can occasionally confer a high level of carbapenem resistance, especially when the resistance determinants are located on a plasmid, and this is likely due to the higher gene dosage provided by the higher copy number associated with plasmids (3). Recently, we have identified several A. baumannii isolates in Taiwan that harbor plasmid-borne ISAba1-blaOXA-51-like genes (ISAba1-blaOXA-82) (GenBank accession no. GQ352402). Moreover, a plasmid bearing the ISAba1-blaOXA-51-like gene (ISAba1-blaOXA-138) has also been indentified in an Acinetobacter genomic species 13TU isolate (15). The mobilization of the ISAba1-blaOXA-51-like gene to plasmids and dissemination of these plasmids will further complicate the management of A. baumannii infections. This study aimed to characterize the spread of these plasmids and their significance in A. baumannii isolates in Taiwan.
MATERIALS AND METHODS
Collection of bacteria and genomic species identification of A. baumannii.
Nonduplicate bacterial isolates that were phenotypically identified as members of the Acinetobacter calcoaceticus-A. baumannii complex were collected from 10 medical centers located in different parts of Taiwan (Fig. 1) during the period from July to October 2007. The isolates that were identified to the genomic species level as A. baumannii using a multiplex PCR method (2) were included in this study. An A. baumannii reference strain, ATCC 15151T, was used as a recipient during transformation (3). This strain does not carry carbapenemase genes other than the intrinsic blaOXA-51-like gene, which lacks an upstream ISAba1.
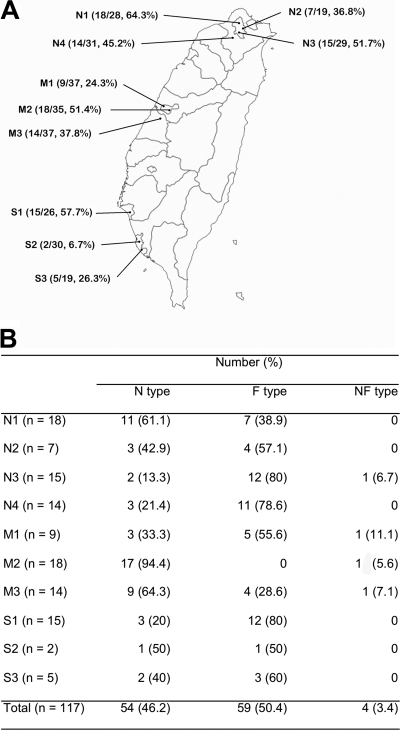
FIG. 1.
Isolates harboring the ISAba1-blaOXA-51-like gene from 10 hospitals located in different parts of Taiwan. (A) The numerators and denominators in parentheses are the number of isolates harboring the ISAba1-blaOXA-51-like gene and the total number of Acinetobacter baumannii isolates collected, respectively. (B) Percentage of isolates with either nuc (nuc-ISAba1-blaOXA-51-like structure, N type) or fxsA (fxsA-ISAba1-blaOXA-51-like structure, F type) upstream of the ISAba1-blaOXA-51-like gene. Four isolates have both genetic structures and were referred to as NF type.
Detection and sequencing of carbapenemase genes and their upstream ISs.
Genes encoding metallo-beta-lactamase (MBL) or CHDLs were detected via PCR as described by Ellington et al. (8) and Higgins et al. (11), respectively. The phenotypic assay for MBL production was carried out using a double-disk synergy test and a combined-disk test with imipenem and EDTA following protocols described by Franklin et al. (10). Detection of ISAba1, preceding the blaOXA-51-like and blaOXA-23-like genes, and IS1008, preceding the blaOXA-58-like gene, was performed by PCR mapping using a forward primer within the ISs and a reverse primer in the CHDL genes (Table 1). The partial ISAba1 and full-length blaOXA-51-like genes were amplified with LA Taq polymerase (Takara Shuzou, Otsu, Japan), TA cloned into the pCR-TOPOII vector (Invitrogen, Carlsbad, CA), and sequenced by Mission Biotech (Taipei, Taiwan).
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ → 3′) | Sitea | Purpose (reference) |
|---|---|---|---|
| ISAba1-F | CACGAATGCAGAAGTTG | 2 | PCR mapping (20) |
| IS1008-F | TCGGCACTTTCAAGGTGAAAT | PCR mapping | |
| OXA-23-like-R | AATAATATTCAGCTGTTTTAATG | PCR mapping | |
| OXA-58-like-R | TTATAAATAATGAAAAACACCCA | PCR mapping | |
| OXA-51-like-R | CTATAAAATACCTAATTKTTCTAA | PCR mapping | |
| OXA-51-F2 | ATCTTGCTCGTGCTTCG | Probe | |
| OXA-51-R1 | ATATTCCCTTGAGGCTGAAC | Probe | |
| FxsA-F | CCACTGATGAACTGTGCTAC | 1 | PCR mapping |
| SK17-Ori-F | GCTTAAAAAGACGAAAGACCG | 3 | PCR mapping |
| SK17-ORF-2-R | GTAACTCCAAATTATCTGTCTAG | 4 | PCR mapping |
| SK-17-ORF-2-F | CATGCTGGGGATTTACCGC | 5 | PCR mapping |
| SK-17-ORF-5-R | GCTTCTATTTGGGGAAACGGC | 6 | PCR mapping |
| SK-17-ORF-5-F | GATGCCGTTTCCCCAAATAG | 7 | PCR mapping |
| TMN-R | CTCCATAAGCCTGTGGCTC | 8 | PCR mapping |
| TMN-F | GGTATGGCATGGGCCTTTC | 9 | PCR mapping |
| OXA-51-like-R2 | GGATAACTAAAACACCCGTAG | 10 | PCR mapping |
| ISAba1-F-2 | GGTAAGCACTTGATGGGCAAG | 11 | PCR mapping |
| SK-17-ORF-13-R | GGAGTAAAGCAACTCTACCC | 12 | PCR mapping |
| 16SrRNA-F | AGAGTTTGATCHTGGYTYAGA | Probe (17) | |
| 16SrRNA-R | ACGGYTACCTTGTTACGACTT | ||
| TMN-F2 | GATCCAAAACAGATGAGGGTG | Probe | |
| TMN-R2 | TTGAGATTGATTCTTTCGCCAC | Probe |
As depicted in Fig. 2.
PCR mapping of the region upstream of the ISAba1-blaOXA-51-like gene.
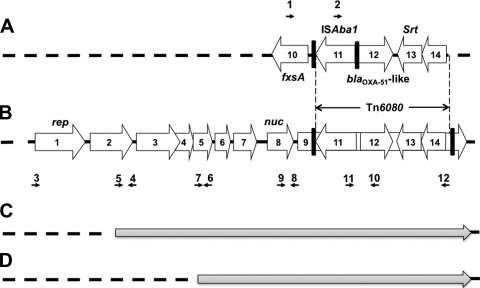
Alignment of the sequences surrounding the chromosomally encoded blaOXA-51-like gene in A. baumannii strains AB307-0294 (accession no. CP001172.1), AB0057 (CP001182.1), ACICU (CP000863.1), SDF (CU468230.2), AYE (CU459141.1), and ATCC 17978 (CP000521.1) identified a gene encoding a sortase (srt; open reading frame 13 [ORF13]) (Fig. 2 A) and another encoding a transcriptional regulator (ORF14) in the downstream region. A gene encoding a putative suppressor of F exclusion of phage T7 (fxsA) was identified immediately upstream of the blaOXA-51-like gene (ORF10). Using primers FxsA-F and OXA51-like-R, a fragment (fxsA-blaOXA-51-like sequence) of 1,303 bp was generated by PCR. The size of the amplicon increased to 1,920 bp in the presence of an insertion of ISAba1 between the fxsA and blaOXA51-like sequences (fxsA-ISAba1-blaOXA-51-like sequence) (Fig. 2A). The isolates having the fxsA-ISAba1-blaOXA-51-like genetic structure were referred to as F-type isolates.
FIG. 2.
Schematic map of the partial genetic structure flanking the ISAba1-blaOXA-51-like gene. (A) The ISAba1-blaOXA-51-like gene on the chromosome of A. baumannii with fxsA immediately upstream and a gene encoding a sortase (srt, ORF13) and another encoding a transcriptional regulator (ORF14) located downstream. The direct repeat (AAGTCTTAT) flanking ISAba1 is indicated with a thick vertical line. (B) Plasmids bearing the ISAba1-blaOXA-51-like gene and their surrounding sequences. ORF1 encodes a plasmid replicase, ORF2 encodes a protein belonging to the zeta-toxin family, and ORF8 (nuc) encodes a thermonuclease. ORF3 to ORF 7 and ORF9 encode hypothetical proteins. The detailed sequences and annotation of the ORFs can be found in GenBank (accession no. GQ352402). ORF9 was inserted by a putative transposon, Tn6080, which contained ISAba1-blaOXA-51-like-srt-ORF14. The direct repeat (AAAAAAATA) flanking Tn6080 is indicated with a thick vertical line. Regions with high similarity to that depicted in panel A are indicated by vertical dotted lines. The black horizontal arrows indicate the locations of primers listed in the Table 1. (C and D) Genetic structures of plasmids carrying the ISAba1-blaOXA-51-like gene that have partial similarity to (indicated with gray arrows) but lack some of the upstream fragments in (indicated with dotted lines) that depicted in panel B. Figures are not to scale.
The surrounding structures of plasmids bearing the ISAba1-blaOXA-51-like gene from an A. baumannii strain (pAbSK-OXA-82, accession no. GQ352402) and an Acinetobacter genomic species 13 TU strain (accession no. EU670845) were also aligned and compared. A gene encoding a thermonuclease protein (nuc; ORF8) (Fig. 2B) was identified in the upstream region of the ISAba1-blaOXA-51-like genes in these two plasmids. PCR with primers TMN-F and OXA51-like-R amplified a fragment of 2,388 bp in isolates harboring the nuc-ISAba1-blaOXA51-like structure, and these isolates were referred to N-type isolates. The gene contents of the PCR products of randomly selected isolates were verified by cloning and sequencing.
Identification of the plasmid location of the nuc-ISAba1-blaOXA-51-like sequence by Southern blot analysis.
Plasmids were purified using a plasmid DNA miniprep kit (Bioman, Taipei, Taiwan) or a plasmid maxikit (Qiagen, Valencia, CA), gel separated, transferred to nylon membranes (Perkin-Elmer, Boston, MA), and hybridized with PCR-generated probes specific for the blaOXA-51-like and nuc genes. Several representative strains were also selected for mapping the location of the nuc gene using an I-CeuI restriction enzyme mapping method (15). After transfer of the I-CeuI-cut fragments to a nylon membrane, Southern blot analysis was performed using PCR-generated probes specific for the 16S rRNA (17) or nuc genes. Southern blot analysis was performed using a digoxigenin (DIG) DNA labeling and detection kit (Roche Diagnostics, Basel, Switzerland) according to the manufacturer's instructions.
Determination of clonal relationships by PFGE.
Determination of the clonal relationship of the A. baumannii isolates was performed by pulsed-field gel electrophoresis (PFGE) as described previously (13). The restriction enzyme ApaI and a CHEF-Mapper electrophoresis system (Bio-Rad Laboratories, Richmond, CA) were used. Banding patterns were interpreted according to the criteria recommended by Tenover et al. (21) and cluster analysis using the Molecular Analyst Fingerprinting, Fingerprinting Plus, and Fingerprinting DST software (Bio-Rad Laboratories, Richmond, CA) with the unweighted-pair group method with arithmetic averages (13). Isolates with the same pulsotype were classified as clones.
Antimicrobial susceptibility tests.
MICs were determined using the agar dilution method according to the recommendations of the Clinical and Laboratory Standards Institute (4). Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 were used as quality control strains during MIC tests. The breakpoint concentrations for all antibiotics were as previously defined (5).
Electrotransformation of recombinant plasmids harboring ISAba1-blaOXA-51-like or blaOXA-51-like genes.
The amplified ISAba1-blaOXA-51-like or blaOXA-51-like gene was cloned into an E. coli-A. baumannii shuttle vector, pYMAb3 (3). pYMAb3 is derived from E. coli plasmid pET-28a (Novagen, Madison, WI) and has a plasmid replicon from A. baumannii plasmid pMAC (3). A. baumannii ATCC 15151T and F-type isolates susceptible to carbapenem were randomly selected as recipient cells. Transformation of the empty shuttle vector was also performed as a control. Electroporation was performed using an ECM 630 electroporator (BTX, San Diego, CA) and 2-mm electrode gap cuvettes as previously described (3). The transformants were selected on agar plates containing imipenem (8 μg/ml). Transformants carrying an empty shuttle vector or a shuttle vector with the blaOXA-51-like gene were also selected on agar plates containing kanamycin (40 μg/ml). Successful transformations were verified by plasmid analysis and PCR mapping of the recombinant plasmid.
Statistical analysis.
The antimicrobial resistance rates of N-type and F-type isolates were compared using the chi-square test with Yates correction or Fisher's exact test with SPSS version 17 for Windows (SPSS, Chicago, IL). A P value of <0.05 was considered significant.
Nucleotide sequence accession numbers.
The nucleotide sequences of blaOXA-172 to blaOXA-177 were assigned accession no. HM113558 to HM113563 in the GenBank database.
RESULTS
Bacterial isolates harboring the ISAba1-blaOXA-51-like gene with different upstream genes.
A total of 291 A. baumannii isolates were collected from 10 hospitals during the study period. One hundred seventeen isolates (40.2%) had ISAba1 next to the blaOXA-51-like gene (ISAba1-blaOXA-51-like gene). The prevalence of the isolates harboring the ISAba1-blaOXA-51-like gene ranged from 6.7 to 64.3% among the different hospitals (Fig. 1A). The sequence upstream of the ISAba1-blaOXA-51-like gene could be PCR amplified with primers for either nuc (nuc-ISAba1-blaOXA-51-like sequence, N type) or fxsA (fxsA-ISAba1-blaOXA-51-like sequence, F type) in 54 and 59 isolates, respectively. Four isolates had both the nuc and fxsA upstream sequences, indicating that they had at least two copies of the ISAba1-blaOXA-51-like gene, and were referred to as NF type. The percentages of N-type and F-type isolates were different among the 10 hospitals (Fig. 1B). Most of the isolates having the ISAba1-blaOXA-51-like gene did not carry other CHDL genes. Among the N-type isolates, one (from hospital M3) also had an IS1008-blaOXA-58 gene and three (all from hospital M2) had ISAba1-blaOXA-23-like genes. Among the F-type isolates, one (from hospital N4) also had a blaOXA-24-like gene. No MBL production was detected in the isolates.
Location of the nuc-ISAba1-blaOXA-51-like structure in N- and NF-type isolates.
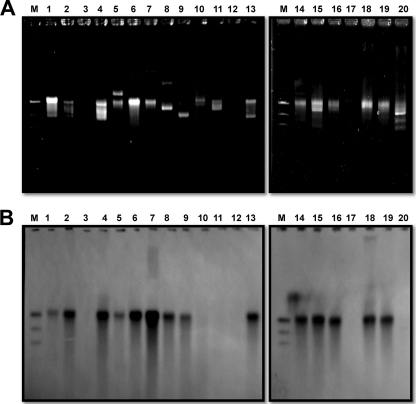
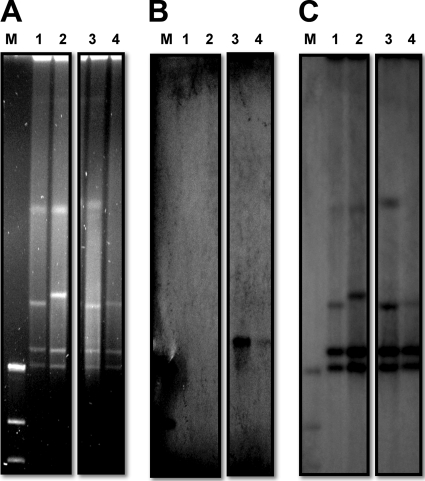
Previous studies have shown that nuc-ISAba1-blaOXA-51-like structures can be found in Acinetobacter plasmids (GenBank accession no. GQ352402 and EU670845). Hence, we determined whether the N-type or NF-type isolates also had plasmids harboring the nuc-ISAba1-blaOXA-51-like structure. The results demonstrated that the N-type and NF-type isolates had different plasmid patterns (Fig. 3 A), but Southern blot analysis revealed that all of them had a plasmid of similar size that harbored the blaOXA-51-like (Fig. 3B) and nuc (data not shown) genes. The sizes of the plasmids were comparable to that of pAbSK-OXA-82. In addition, based on the I-CeuI mapping method, nuc was localized to a plasmid (Fig. 4 B, lanes 3 and 4) that also contained the blaOXA-51-like gene (data not shown) in N-type and NF-type isolates.
FIG. 3.
Southern blot analysis depicting the plasmid location of the blaOXA-51-like gene. (A) Gel migration patterns of the isolates. (B) Hybridization with the blaOXA-51-like probe. The plasmids in lanes 3, 10, 11, 12, 17, and 20 were from isolates harboring the fxsA-ISAba1-blaOXA-51-like structure (F type), and those in lanes 1, 2, 5, 6, 7, 8, 9, 13, 14, 15, 16, 18, and 19 were from isolates harboring the nuc-ISAba1-blaOXA-51-like structure (N type). The plasmid in lane 4 was from an NF-type isolate.
FIG. 4.
Identification of the plasmid location of nuc by I-CeuI mapping. (A) Pulsed-field gel electrophoresis of I-CeuI-restricted genomic fragments. (B an C) Hybridization of the fragments with nuc (B) or 16S rRNA (C) probes. Lanes M, DNA molecular weight marker (DIG labeled); lanes 1, I-CeuI-restricted fragments from an isolate harboring only the blaOXA-51-like gene without an upstream ISAba1 sequence; lanes 2, I-CeuI-restricted fragments from an isolate harboring the fxsA-ISAba1-blaOXA-51-like structure (F type); lanes 3, I-CeuI-restricted fragments from an isolate harboring the nuc-ISAba1-blaOXA-51-like structure (N type); lanes 4, I-CeuI-restricted fragments from an isolate harboring both genetic structures (NF type).
Partial genetic structures around the ISAba1-blaOXA-51-like gene.
PCR mapping with different primer sets (locations are depicted in Fig. 2A and B) showed that all the F-type and NF-type isolates had a genetic structure as depicted in Fig. 2A, and a direct repeat (AAGTCTTAT) was generated upon the insertion of ISAba1 between the fxsA and blaOXA-51-like genes. For the N-type and NF-type isolates, most (89.7%) had a genetic arrangement around the nuc-ISAba1-blaOXA-51-like structure that was similar to that found in pAbSK-OXA-82 (Fig. 2B). The srt gene (Fig. 2B, ORF13) and a gene encoding a transcriptional regulator (Fig. 2B, ORF14) were also located downstream of a plasmid bearing ISAba1-blaOXA-51-like genes. The ISAba1-blaOXA-51-like-srt-ORF14 genetic structure was found inserted into ORF9 on the plasmid and flanked by a direct repeat (AAAAAAATA), indicating the mobilization of these structures from the chromosome by one-ended transposition. The putative transposon carrying ISAba1-blaOXA-51-like-srt-ORF14 was designated Tn6080. Six isolates lacked some of the upstream structure, as depicted in Fig. 2C (five isolates) and D (one isolate).
Clonal relationship of N-type and NF-type isolates.
The N-type and NF-type isolates belonged to 24 clones (Table 2). Nine of the clones were found in at least two hospitals. This indicated clonal dissemination between hospitals, with clone VII being the most widely disseminated.
TABLE 2.
Characteristics of the Acinetobacter baumannii isolates harboring plasmids bearing the ISAba1-blaOXA-51-like genea
| Isolate group and hospital | Pulsotype (no. of isolates and plasmid typeb) | CHDL gene | Amino acid at positionc: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 42 (K) | 106 (G) | 129 (I) | 167 (L) | 199 (E) | 222 (W) | 226 (P) | 270 (Q) | |||
| N type | ||||||||||
| N1 | IV (2B), VI (2B), VII (2B, 2C), XII (1B), XIII (1B), XXIV(1B) | blaOXA-82 | V | |||||||
| N2 | IX (1B), XIV (1B), XXII (1D) | blaOXA-176 | S | V | ||||||
| N3 | IV (1B) | blaOXA-174 | V | K | ||||||
| N3 | X (1B) | blaOXA-172 | V | L | ||||||
| N4 | I (1B), IV (1B) | blaOXA-82 | V | |||||||
| N4 | II (1B) | blaOXA-177 | V | G | ||||||
| M1 | III (1B), XV (1B), XVI (1C) | blaOXA-172 | V | L | ||||||
| M2 | V (2B), VII (3B), IX (8B), XIV (1B), XV (1B), XVII (1B) | blaOXA-172 | V | L | ||||||
| M2 | IX (1B) | blaOXA-173 | V | L | L | |||||
| M3 | XVIII (1B), XIX (2B), XX (1B), XXI (3B), XXIII (1B) | blaOXA-82 | V | |||||||
| M3 | XI (1B) | blaOXA-175 | R | V | ||||||
| S1 | VII (3B) | blaOXA-82 | V | |||||||
| S2 | XIII (1B) | blaOXA-82 | V | |||||||
| S3 | VII (1B), VIII(1B) | blaOXA-82 | V | |||||||
| NF type | ||||||||||
| N3 | I (1C) | blaOXA-82 | V | |||||||
| M1 | XI (1B) | blaOXA-82 | V | |||||||
| M2 | IX (1C) | blaOXA-82 | V | |||||||
| M3 | XXII (1B) | blaOXA-82 | V | |||||||
Abbreviations: CHDL, carbapenem-hydrolyzing class D beta-lactamase; N type, isolates harboring the nuc-ISAba1-blaOXA-51-like structure; NF type, isolates having both the nuc-ISAba1-blaOXA-51-like and fxsA-ISAba1-blaOXA-51-like structures (both blaOXA-51-like genes in NF-type isolates are blaOXA-82).
The plasmid type refers to the genetic structures depicted in Fig. 2, with type B having all the known partial sequences as depicted in Fig. 2B, type C as depicted in Fig. 2C, and type D as depicted in Fig. 2D.
The reference amino acids (in parentheses) and their positions are from OXA-66.
It was found that most hospitals frequently contained isolates belonging to multiple clones, except hospital S1, in which all three isolates belonged to clone VII. The four NF-type isolates each belonged to different clones. F-type isolates that had the same pulsotype as the NF-type isolate were found in hospitals N3 and M1 (data not shown).
Antimicrobial MICs for the N-type and F-type isolates.
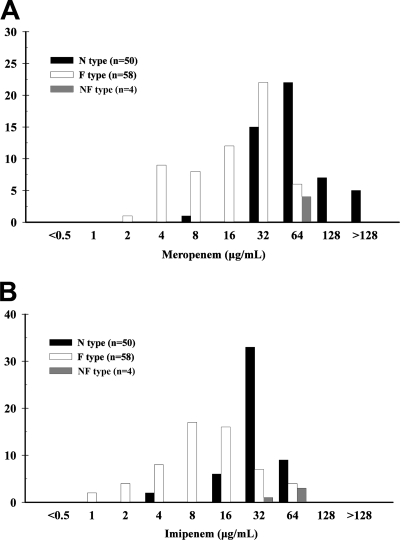
After excluding the NF-type isolates and those that encoded other CHDL genes, the antimicrobial MICs for N-type (50 isolates) and F-type isolates (58 isolates) were compared. The carbapenem MICs for the N-type isolates were higher than those for the F-type isolates (Fig. 5). The N-type isolates had a higher imipenem resistance rate (98% versus 46.6%, P < 0.001) and a higher meropenem resistance rate (98% versus 69%, P = 0.019) than the F-type isolates. Unexpectedly, the N-type isolates also had a higher rate of resistance to sulbactam (80% versus 58.6%, P = 0.029). The N-type and F-type isolates had similar rates of resistance to cefepime (90% versus 98.3%, P = 0.094), ceftazidime (96 versus 98.3%, P = 0.595), piperacillin-tazobactam (90% versus 84.5%, P = 0.573), amikacin (98% versus 98.3%, P = 1.000), and ciprofloxacin (98% versus 98.3%, P = 1.000). All the isolates were susceptible to colistin.
FIG. 5.
Meropenem and imipenem MICs for isolates harboring plasmids bearing the nuc-ISAba1-blaOXA-51-like structure (N type), the chromosomally encoded fxsA-ISAba1-blaOXA-51-like structure (F type), or both genetic structures (NF type).
Sequences of ISAba1-blaOXA-51-like genes.
Among the 54 N-type isolates, most of the blaOXA-51-like genes were blaOXA-82 (27 isolates [50%]), followed by blaOXA-172 (20 isolates [37%]). Other blaOXA-51-like genes included blaOXA-176 (three isolates) and blaOXA-173, blaOXA-174, blaOXA-175, and blaOXA-177 (one isolate each). OXA-172 was different from OXA-82 at three amino acids (Table 2). blaOXA-82 could be found in isolates from different hospitals, but blaOXA-172 was found mostly in isolates from two hospitals in central Taiwan. Among 23 randomly selected F-type isolates, their blaOXA-51-like genes were blaOXA-82 (39.1%), blaOXA-66 (34.8%), blaOXA-80 (21.7%), and blaOXA-83 (4.3%). Interestingly, the two copies of the blaOXA-51-like gene in the four NF-type isolates were all blaOXA-82. The putative promoter sequences and their distances from blaOXA-51-like were identical in the N, F, and NF-type isolates (data not shown).
Electrotransformation of a plasmid carrying the ISAba1-blaOXA-51-like or blaOXA-51-like gene.
To determine whether a plasmid carrying only the ISAba1-blaOXA-51-like gene (without the presence of other factors in the original plasmid) can confer a high level of carbapenem resistance, a recombinant plasmid harboring ISAba1-blaOXA-82 was electrotransformed into ATCC 15151T and two randomly selected carbapenem-susceptible F-type isolates. The transformants demonstrated an increase in imipenem MIC in recipient A. baumannii ATCC 15151T cells (from 0.5 to 32 μg/ml) and the F-type isolates (from 4 to 64 μg/ml). Transformation of the shuttle vector carrying blaOXA-82 (without ISAba1) only mildly increased the imipenem MIC for A. baumannii ATCC 15151T (from 0.5 to 2 μg/ml). Transformation of the empty shuttle vector failed to increase carbapenem resistance levels.
DISCUSSION
In addition to the dissemination of extrinsic CHDL genes, including the blaOXA-23-like, blaOXA-24-like, and blaOXA-58-like genes, by plasmids in A. baumannii (18), it is now evident that the blaOXA-51-like gene and its upstream ISAba1 sequence have disseminated in Taiwan via plasmids. The widespread distribution of isolates harboring plasmids bearing the ISAba1-blaOXA-51-like gene in Taiwan is mostly due to plasmids spreading among different clones, as the isolated plasmids were all similar in size, contained the same plasmid replicase gene (ORF1) (Fig. 2B), and had similar structures around the ISAba1-blaOXA-51-like gene. Clonal spreading of A. baumannii also contributed to some extent of the emergence of isolates harboring plasmids bearing ISAba1-blaOXA-51-like genes, especially clone VII.
The origin of the ISAba1-blaOXA-82 plasmid might be deduced by analysis of F-type and NF-type isolates. Presumably, the ISAba1 sequence was first transposed at the upstream region of chromosomal blaOXA-82, as indicated by the flanking direct repeat (AAGTCTTAT) (Fig. 2A). Later, the genetic structure Tn6080 (ISAba1-blaOXA-82-srt-ORF14) was copied and integrated into the plasmid in ORF9 via one-ended transposition, as evident by the flanking direct repeat (AAAAAAATA) (Fig. 2B). The one-ended transposition mechanism has also been suggested to be responsible for ISAba4-mediated mobilization of blaOXA-23 as transposon Tn2007 (6). The origin of the blaOXA-172 plasmid might have resulted from an independent transposition event, but it could have also originated from mutation of a blaOXA-82 plasmid, as these plasmids were similar; their CHDL genes differed at only a few nucleotides (data not shown), and the corresponding CHDLs differed at three amino acids (Table 2). Other blaOXA-51-like genes were most likely derived from mutation of blaOXA-82 or blaOXA-172.
Compared with the variation of carbapenem MICs in F-type isolates, some of which were below the resistance breakpoint, all but one of the N-type and all NF-type isolates carrying an ISAba1-blaOXA-51-like plasmid were not susceptible to carbapenem. The higher carbapenem MIC in N-type relative to F-type isolates did not have any association with their blaOXA-51-like promoters, which were identical among the isolates. Increase gene dosage via high-copy plasmids is the most likely reason for the carbapenem resistance in isolates harboring plasmids bearing the ISAba1-blaOXA-51-like gene. Another possibility was that the high level of carbapenem resistance resulted from a synergistic effect from other determinants located on the same plasmid. This was unlikely, as transformation of a plasmid harboring only the ISAba1-blaOXA-51-like gene (ISAba1-blaOXA-82) was enough to confer a high level of carbapenem resistance. This phenomenon has also been recently demonstrated (16). Interestingly, the N-type isolates also had a higher sulbactam resistance rate, the mechanism of which is undetermined.
The detection of plasmids bearing the ISAba1-blaOXA-51-like gene is crucial, as it is central in the treatment of patients and control of plasmid spreading. However, it is not convenient to detect these plasmids by Southern blot analysis. PCR mapping might be a more feasible method for first-step screening. It has been shown that chromosomally carried blaOXA-51-like genes are preceded by fxsA, based on the analysis of all available A. baumannii genomes. In isolates harboring the ISAba1-blaOXA-51-like gene, the possibility of mobilization of the ISAba1-blaOXA-51-like gene should be noted if PCR mapping fails to identify the upstream fxsA. In Taiwan, mapping of nuc upstream of the ISAba1-blaOXA-51-like gene indicates localization of the ISAba1-blaOXA-51-like gene to a plasmid.
In conclusion, plasmids bearing the ISAba1-blaOXA-51-like gene are widespread in carbapenem-resistant A. baumannii isolates in Taiwan. The origin of the plasmid-borne ISAba1-blaOXA-51-like gene is likely a result of transposition. The widespread distribution of these plasmids is due primarily to their spread among different A. baumannii clones, although spreading of the clones may also play a role. The plasmids bearing the ISAba1-blaOXA-51-like gene are associated with a high level of carbapenem resistance, indicating that detection of these genetic structures and prevention of their further spreading is warranted.
Acknowledgments
We thank Chien-Pei Chen for her technical assistance. The isolates used in this study were from part of a collection from the Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART) program, 2007. All the contributors of the isolates are highly appreciated.
The study was supported by grants from Taipei Veterans General Hospital (V99C1-014 and V99S5-006) and the National Science Council (NSC97-2314-B-010-006-MY3 and NSC98-2314-B-010-010-MY3).
There is no conflict of interest with any commercial company.
Footnotes
Published ahead of print on 16 August 2010.
REFERENCES
- 1.Brown, S., and S. Amyes. 2006. OXA (beta)-lactamases in Acinetobacter: the story so far. J. Antimicrob. Chemother. 57:1-3. [DOI] [PubMed] [Google Scholar]
- 2.Chen, T. L., L. K. Siu, R. C. Wu, M. F. Shaio, L. Y. Huang, C. P. Fung, C. M. Lee, and W. L. Cho. 2007. Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:801-806. [DOI] [PubMed] [Google Scholar]
- 3.Chen, T. L., R. C. Wu, M. F. Shaio, C. P. Fung, and W. L. Cho. 2008. Acquisition of a plasmid-borne blaOXA-58 gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob. Agents Chemother. 52:2573-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed. CLSI document M7-A8. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Corvec, S., L. Poirel, T. Naas, H. Drugeon, and P. Nordmann. 2007. Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939-951. [DOI] [PubMed] [Google Scholar]
- 8.Ellington, M. J., J. Kistler, D. M. Livermore, and N. Woodford. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J. Antimicrob. Chemother. 59:321-322. [DOI] [PubMed] [Google Scholar]
- 9.Figueiredo, S., L. Poirel, J. Croize, C. Recule, and P. Nordmann. 2009. In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1-mediated overexpression of the natural bla(OXA-66) oxacillinase gene. Antimicrob. Agents Chemother. 53:2657-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franklin, C., L. Liolios, and A. Y. Peleg. 2006. Phenotypic detection of carbapenem-susceptible metallo-beta-lactamase-producing gram-negative bacilli in the clinical laboratory. J. Clin. Microbiol. 44:3139-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins, P. G., M. Lehmann, and H. Seifert. 2010. Inclusion of OXA-143 primers in a multiplex polymerase chain reaction (PCR) for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents. 35:305. [DOI] [PubMed] [Google Scholar]
- 12.Hu, W. S., S. M. Yao, C. P. Fung, Y. P. Hsieh, C. P. Liu, and J. F. Lin. 2007. An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3844-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, L. Y., T. L. Chen, P. L. Lu, C. A. Tsai, W. L. Cho, F. Y. Chang, C. P. Fung, and L. K. Siu. 2008. Dissemination of multidrug-resistant, class 1 integron-carrying Acinetobacter baumannii isolates in Taiwan. Clin. Microbiol. Infect. 14:1010-1019. [DOI] [PubMed] [Google Scholar]
- 14.Lee, Y. T., L. Y. Huang, D. H. Chiang, C. P. Chen, T. L. Chen, F. D. Wang, C. P. Fung, L. K. Siu, and W. L. Cho. 2009. Differences in phenotypic and genotypic characteristics among imipenem-non-susceptible Acinetobacter isolates belonging to different genomic species in Taiwan. Int. J. Antimicrob. Agents. 34:580-584. [DOI] [PubMed] [Google Scholar]
- 15.Lee, Y. T., J. F. Turton, T. L. Chen, R. C. Wu, W. C. Chang, C. P. Fung, C. P. Chen, W. L. Cho, L. Y. Huang, and L. K. Siu. 2009. First identification of blaOXA-51-like in non-baumannii Acinetobacter spp. J. Chemother. 21:514-520. [DOI] [PubMed] [Google Scholar]
- 16.Lin, Y. C., K. C. Hsia, Y. C. Chen, W. H. Sheng, S. C. Chang, M. H. Liao, and S. Y. Li. 2010. Genetic basis of multidrug resistance in Acinetobacter spp. clinical isolates in Taiwan. Antimicrob. Agents Chemother. 54:2078-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mammeri, H., L. Poirel, N. Mangeney, and P. Nordmann. 2003. Chromosomal integration of a cephalosporinase gene from Acinetobacter baumannii into Oligella urethralis as a source of acquired resistance to beta-lactams. Antimicrob. Agents Chemother. 47:1536-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 20.Segal, H., S. Garny, and B. G. Elisha. 2005. Is IS(ABA-1) customized for Acinetobacter? FEMS. Microbiol. Lett. 243:425-429. [DOI] [PubMed] [Google Scholar]
- 21.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsakris, A., A. Ikonomidis, S. Pournaras, L. S. Tzouvelekis, D. Sofianou, N. J. Legakis, and A. N. Maniatis. 2006. VIM-1 metallo-beta-lactamase in Acinetobacter baumannii. Emerg. Infect. Dis. 12:981-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turton, J. F., M. E. Ward, N. Woodford, M. E. Kaufmann, R. Pike, D. M. Livermore, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS. Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]
- 24.Turton, J. F., N. Woodford, J. Glover, S. Yarde, M. E. Kaufmann, and T. L. Pitt. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44:2974-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]