Abstract
The ability to attach to a variety of oral surfaces is an important characteristic of Porphyromonas gingivalis. Previous studies have demonstrated that expression and production of FimA, a major subunit protein of the long fimbriae, is required for P. gingivalis colonization. Here we report that a surface protein, arginine deiminase (ArcA) of Streptococcus cristatus, represses FimA production and inhibits biofilm formation of P. gingivalis. This inhibitory function of ArcA is also observed in the formation of heterotypic P. gingivalis-Streptococcus gordonii biofilms. P. gingivalis is released from streptococcal substrates in the presence of ArcA, likely due to an inhibition of FimA production. This work suggests that ArcA may have the potential to be a specific antibiofilm agent to fight P. gingivalis infections.
Porphyromonas gingivalis is a Gram-negative anaerobe associated with several forms of periodontitis, such as chronic destructive or refractory periodontitis (4, 26, 27). The organism is also implicated in the etiology of some systemic diseases or conditions, including coronary heart disease, and the enhanced risk of a preterm infant with low birth weight (15, 25, 29). Several virulence factors of P. gingivalis have been identified and well characterized (10, 14). One of the features of the organism is its ability to bind to a variety of oral cavity surfaces, which is critical for sustaining the bacterium in supra- and subgingival environments. One of the surface components involving P. gingivalis attachment is the long (major) fimbriae. The role of the major subunit protein (FimA) of long fimbriae in the bacterium's pathogenicity has been well established. FimA appears to be capable of interacting with a number of human proteins, oral epithelial cells, and other oral bacteria (9, 18, 19, 21, 28). Indeed, several studies have shown that a FimA-deficient mutant impacts its ability to form homotypic biofilms (12, 17). FimA is also known as a critical determinant of P. gingivalis invasion, and thus a fimA mutant exhibits a lower invasion efficiency compared to the wild-type strain (11, 33). Moreover, P. gingivalis FimA has been linked to the induction of an inflammatory process in periodontal tissues through a number of mechanisms (7, 8, 38).
Expression of the fimA gene is tightly regulated in P. gingivalis in response to environmental cues. P. gingivalis appears to produce more FimA at lower temperatures or in an iron/hemin-sufficient environment (2, 34). We reported earlier that expression of the fimA gene is significantly repressed at the transcriptional level when P. gingivalis is grown in the presence of Streptococcus cristatus, an early colonizer of dental plaques (35). A surface protein of S. cristatus, arginine deiminase (ArcA), was identified as the effector protein, and the inhibitory activity of S. cristatus on fimA expression was demolished in an arcA-deficient strain (37). Arginine deiminase catalyzes the hydrolysis of l-arginine to l-citrulline and ammonia. Ammonia is believed to be important for oral biofilm pH homeostasis and caries prevention (3, 5). We further demonstrated that the hydrolytic activity of arginine deiminase is not required for its ability to induce an alteration of expression of fimA, suggesting that ArcA is likely a dual function protein (37).
Arginine deiminase is found in oral streptococci, including S. cristatus and Streptococcus gordonii. Differential expression of the arcA gene is observed in these streptococcus species (16). S. cristatus expresses a much higher level of the arcA gene than S. gordonii. This observation provides an explanation, at least in part, for why the attachment of P. gingivalis to S. gordonii readily occurs, but P. gingivalis does not bind well on S. cristatus biofilms (13, 35). It is suggested that differential expression of the arcA genes may be responsible for distinct roles of streptococcal species in biofilm formation of P. gingivalis. In this study, we examined the reduction in FimA production by P. gingivalis in the presence of purified S. cristatus ArcA. We demonstrated that biofilm formation by P. gingivalis on a saliva-coated surface and streptococcal substrates was significantly inhibited by the addition of ArcA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. P. gingivalis 33277 was grown from frozen stocks in Trypticase soy broth (TSB) or on TSB blood agar plates, supplemented with yeast extract (1 mg/ml), hemin (5 μg/ml), and menadione (1 μg/ml), at 37°C in an anaerobic chamber (85% N2, 10% H2, 5% CO2). Streptococcus strains were grown in Trypticase peptone broth (TPB) supplemented with 0.5% glucose at 37°C under aerobic conditions.
TABLE 1.
Strains used in this study
| Strains | Relevant characteristicsa | Source or reference |
|---|---|---|
| P. gingivalis 33277 | Type strain from ATCC, FimA type I | Lab collection |
| fimA mutant | Derivative of P. gingivalis 33277 containing an insertional mutation in fimA, Emr | 17 |
| S. cristatus CC5A | Wild-type strain | Lab collection |
| ArcAE | Derivative of S. cristatus CC5A containing an insertional mutation in arcA, Emr | 37 |
| S. gordonii DL1 | Wild-type strain | 14 |
Emr, resistance to erythromycin.
Purification of S. cristatus arginine deiminase.
Arginine deiminase was purified from surface extracts of S. cristatus CC5A using the methods described previously (37). Briefly, the surface extract of CC5A was fractionated with ammonium sulfate. The fraction precipitated with 66% saturated ammonium sulfate was purified using a Blue Sepharose column (GE Healthcare, United Kingdom).
Western blot analysis.
P. gingivalis strains were grown on TSB blood agar plates with or without purified S. cristatus CC5A ArcA (1 μg/ml) for 48 h. The surface proteins of P. gingivalis were collected by sonication and centrifugation as described previously (36). Protein concentrations of the samples were determined using a Bio-Rad protein assay. The soluble proteins (5 μg) were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to nitrocellulose membranes (Gibco BRL) with a Mini Transblot electrophoretic transfer cell (Bio-Rad Laboratories) at 100 V for 1 h. The membrane was treated with 30 ml of blocking solution (3% bovine serum albumin in phosphate-buffered saline [PBS] containing 0.1% Tween 20; pH 7.4) for 1 h and incubated for 1 h with polyclonal anti-FimA antibodies or anti-Mfa1 antibodies diluted 1:1,000 in PBS containing 0.1% Tween 20, pH 7.4. The membrane was then rinsed twice and washed three times for 15 min each with 0.1% Tween 20 in PBS. The membrane was incubated with anti-rabbit horseradish peroxidase-conjugated secondary antibodies for 1 h, rinsed, and washed. Antigen-antibody reactivity was visualized by enhanced chemiluminescence (GE Healthcare Bio-Sciences Corp., Piscataway, NJ).
Adherence assay.
Attachment of P. gingivalis to the saliva-coated surface was quantified by the method of O'Toole and Kolter (22). Bacterial strains were grown to mid-log phase (optical density at 600 nm, 1.0) and collected by centrifugation. The bacterial cells were resuspended in TSB, transferred to a six-well polystyrene plate precoated with human whole saliva (Corning Inc., Corning, NY), and incubated at 37°C. After washing, the biofilms were stained with 1% crystal violet and destained with 95% ethanol. The absorbance of ethanol solutions was determined with a spectrophotometer at 540 nm (Ultrospec 2100 Pro; Amersham Pharmacia Biotech). The assays were repeated independently three times.
Biofilm assay.
Heterotypic biofilms of P. gingivalis-streptococci were generated on a polystyrene six-well plate and quantified using quantitative PCR (qPCR). Streptococcal cells (3 × 109) were first incubated in the saliva-coated wells at 37°C aerobically for 4 h, and the unbound cells were removed by washing with PBS two times. P. gingivalis 33277 cells (3 × 109) in TSB were added to the wells, covered with streptococcal biofilms, and incubated at 37°C anaerobically for 2 h. After removal of unbound P. gingivalis cells, S. cristatus CC5A or the arcA mutant (3 × 109) suspended in TSB diluted with PBS was added to the wells, respectively. To inhibit the planktonic growth of streptococcal cells, gentamicin (50 μg/ml) was added to the TSB, since P. gingivalis cells are naturally resistant to gentamicin. In the meantime, proliferation of P. gingivalis was also limited in the diluted medium. The six-well polystyrene microtiter dishes were incubated with gentle shaking for 16 h under anaerobic conditions. The number of P. gingivalis cells bound on S. gordonii biofilms and released from the biofilms was determined by using qPCR. P. gingivalis cells were lysed with lysis solution (solution A; Invitrogen). DNA was extracted using an Easy-DNA kit (Invitrogen). The cells in biofilms were enumerated by using a QuantiTect SYBR green PCR kit with the P. gingivalis species-specific 16S rRNA gene primers 5′-TGTAGATGACTGATGGTGAAA-3′ and 5′-ACTGTTAGCAACTACCGATGT-3′ or with S. gordonii DL1 species-specific arcA gene primers 5′-ATGCCAGACTATCTCGAACG-3′ and 5′-CAATACCTTCATTGCGAAGC-3′ (30). Standards used to determine P. gingivalis cell numbers were prepared using genomic DNA from the wild-type strain 33277. A fresh culture of P. gingivalis 33277, grown in TSB, was serially diluted in PBS and plated on TSB plates to obtain the CFU per milliliter at each dilution. DNA was also isolated from the dilutions, and a qPCR assay was run, as described above, to determine cell numbers. Three trials were performed on three separate cultures.
RESULTS
Differential expression of FimA protein in P. gingivalis in response to ArcA.
Our previous studies demonstrated that P. gingivalis specifically represses the expression of the fimA gene at the transcriptional level in response to ArcA of S. cristatus (37). In order to determine the differential expression of fimA at the protein level, a Western blot analysis was performed. Surface protein expression in P. gingivalis was compared in the cells grown in the presence or absence of S. cristatus CC5A. As shown in Fig. 1, in the presence of purified ArcA (1 μg/ml) in the growth medium, production of FimA was inhibited to a great extent in P. gingivalis 33277. In contrast, the expression level of Mfa1, a subunit protein of short (minor) fimbriae, was not altered by the presence of ArcA. This result was consistent with our earlier reports that fimA expression in P. gingivalis grown in medium supplemented with ArcA is reduced at the transcriptional level. Our data presented here demonstrate that purified ArcA is able to inhibit FimA production in P. gingivalis by repressing fimA expression at both the transcriptional and translational levels.
FIG. 1.
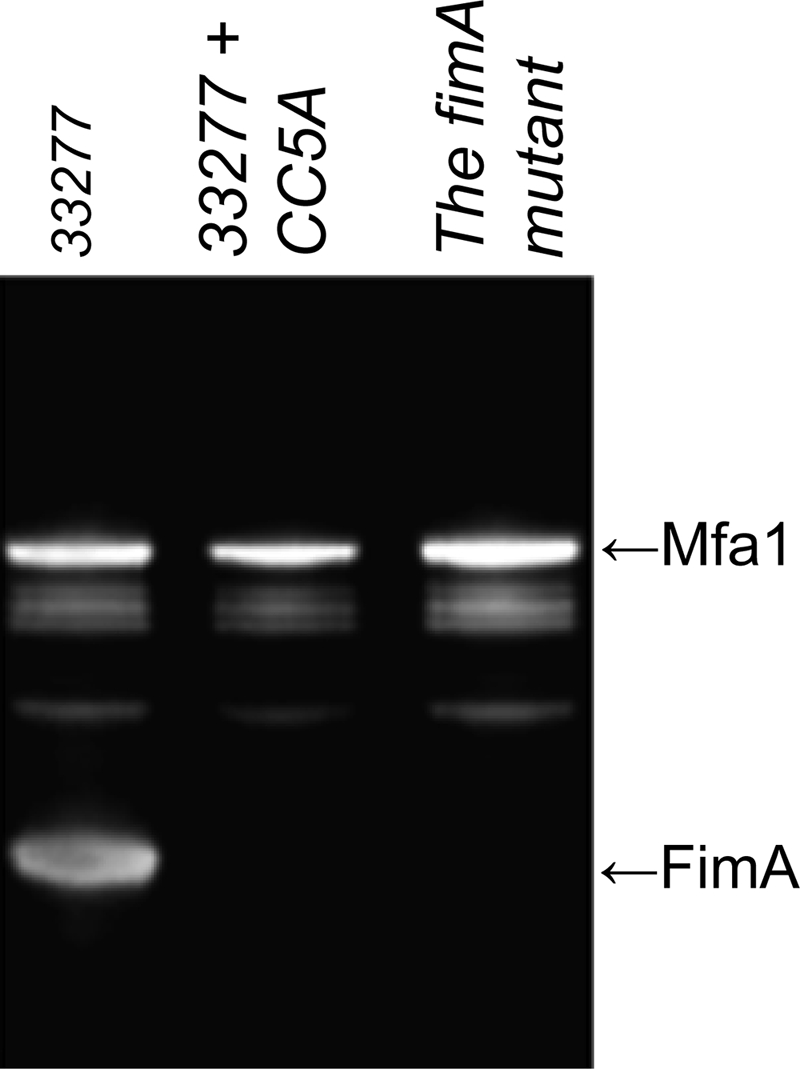
Western blot analysis of FimA and Mfa1 production in P. gingivalis strains. Shown are 18-h cultures of P. gingivalis 33277 in the presence or abersence of ArcA (1 μg/ml) and the fimA mutant grown in Trypticase soy broth. Cell extracts (5 μg) were used for the analyses.
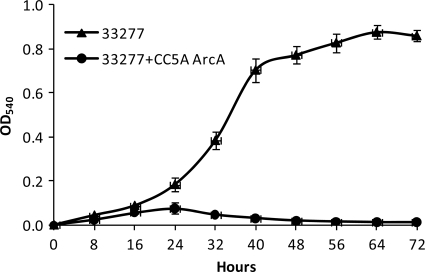
Role of S. cristatus ArcA in P. gingivalis attachment on saliva- coated surfaces.
We showed previously that S. cristatus CC5A inhibits biofilm formation of P. gingivalis (35). To further show a role of ArcA protein in P. gingivalis colonization, adherence assays were conducted in six-well polystyrene microtiter dishes. Formation of P. gingivalis biofilms in the presence or absence of ArcA was examined at 8-h intervals for a total 72 h (Fig. 2). During the first 16 h, the ability of P. gingivalis to attach to saliva-coated wells was unaffected by the presence of ArcA. After 24 h of incubation, however, the presence of ArcA markedly inhibited colonization of P. gingivalis 33277 on the saliva-coated surface, likely due to repression of fimA expression in P. gingivalis.
FIG. 2.
Quantitation of P. gingivalis attachment to a saliva-coated surface. Adherence assays were conducted in six-well polystyrene microtiter dishes. The wells were precoated with human whole saliva and inoculated with P. gingivalis 33277 at 37°C in an anaerobic chamber. The culture medium with or without ArcA (1 μg/ml) of S. cristatus CC5A was changed every 24 h during the 72-h growth period. The abilities of P. gingivalis 33277 in the presence or absence of ArcA to attach and form microcolonies on the surface were quantified by crystal violet staining. Each data point represents the mean ± standard deviation from three independent experiments.
Inhibition of P. gingivalis attachment on S. gordonii biofilms.
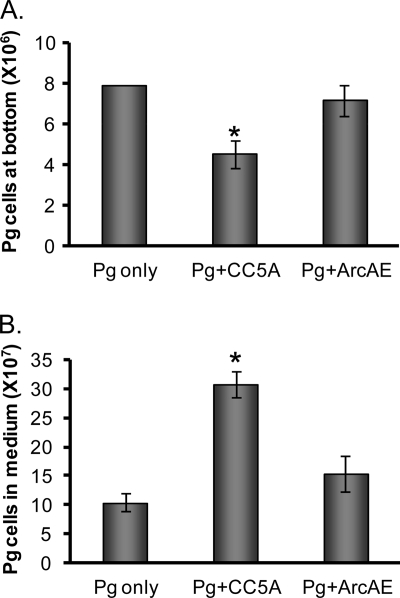
Previous studies demonstrated that earlier colonizers of dental plaques, such as S. gordonii, provide the substrata for P. gingivalis colonization in the oral cavity (19, 23). We also reported that ArcA expression is much lower in S. gordonii than in S. cristatus (16). We wondered if S. cristatus can limit P. gingivalis colonization on S. gordonii biofilms. To test this, a modified biofilm assay was used. S. gordonii DL1 was first introduced into saliva-coated wells to establish an S. gordonii biofilm. P. gingivalis 33277 was then added into the wells to form S. gordonii-P. gingivalis heterotypic biofilms. After removal of unbound P. gingivalis cells, S. cristatus CC5A or the arcA mutant (ArcAE) suspended in TSB containing gentamicin was added into the wells. Consistent with previous reports, P. gingivalis 33277 bound well on S. gordonii DL1. Introduction of S. cristatus CC5A to P. gingivalis-S. gordonii heterotypic biofilms reduced the number of P. gingivalis cells in this mixed species biofilm (Fig. 3A). In contrast, the arcA mutant (ArcAE) did not appear to affect the attachment of P. gingivalis on S. gordonii biofilms. Although the mechanism responsible for detachment of P. gingivalis from S. gordonii surfaces is not clear, one possibility is that bound P. gingivalis cells can reenter the incubation medium, but these released bacteria are unable to reattach tightly to the S. gordonii surface in the presence of CC5A. To test this hypothesis, the number of unbound P. gingivalis cells in the wells was also examined using qPCR. Significantly more P. gingivalis cells were found in the planktonic phase of the well in the presence of S. cristatus CC5A than that observed in the presence of the arcA mutant (Fig. 3B). The number of unbound P. gingivalis was negatively correlated with the number of P. gingivalis cells in the biofilms. These results provide further evidence of an antagonistic relationship between P. gingivalis and S. cristatus.
FIG. 3.
Inhibition of the formation of S. gordonii-P. gingivalis heterotypic biofilms by S. cristatus. P. gingivalis 33277 was introduced into wells covered with S. gordonii biofilms and incubated in the absence of S. cristatus (Pg only) or in the presence of S. cristatus CC5A (Pg+CC5A) or the arcA mutant (Pg+ArcAE) for 16 h. The numbers of P. gingivalis cells bound on S. gordonii biofilms (A) or in the planktonic phase (B) were determined by using qPCR. The variances among the groups were analyzed by Student's t test. The asterisk indicates a significant difference from the Pg-only control (P < 0.05).
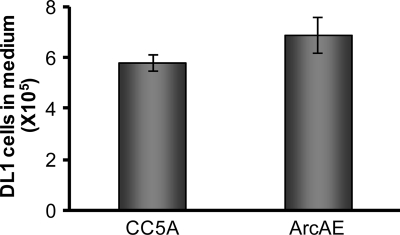
Moreover, to exclude the possibility that the enhanced number of P. gingivalis cells in the planktonic phase seen in the presence of S. cristatus CC5A was due to a reduced stability of S. gordonii biofilms, we examined the number of S. gordonii DL1 in the planktonic phase 16 h after S. cristatus CC5A or its arcA mutant was introduced and incubated with S. gordonii biofilms. We found that there was no significant difference in the number of S. gordonii released from the biofilms in the presence of wild-type strain CC5A or the arcA mutant (ArcAE), suggesting that S. cristatus CC5A functions as a specific inhibitor of P. gingivalis biofilms but has no apparent effect on S. gordonii biofilm stability (Fig. 4).
FIG. 4.
Stability of S. gordonii biofilms in the presence of S. cristatus. S. gordonii DL1 biofilms were established on polystyrene surfaces covered with human whole saliva. S. cristatus CC5A or the arcA mutant (ArecAE) was introduced into the wells and incubated for 16 h. The unbound DL1 was determined using qPCR. Error bars indicate standard deviations.
DISCUSSION
P. gingivalis is one of the primary periodontal pathogens involved in periodontitis. The majority of P. gingivalis strains isolated from subgingival plaques of patients with periodontitis carry the fimA gene (6). The fimA gene is classified into six types (genotypes I, Ib, II, III, IV, and V) based on the nucleotide sequence. We previously demonstrated that ArcA of S. cristatus inhibits expression of types I and II of fimA at the transcriptional level. Here, we provide evidence that production of the FimA protein is also significantly lower for P. gingivalis 33277 (type I fimA) grown in a medium containing purified ArcA. A comparison of biofilm formation of P. gingivalis grown in the absence or presence of S. cristatus ArcA showed a critical role of ArcA in the inhibition of P. gingivalis attachment to saliva-coated surfaces. This observation is similar to our earlier finding in a fimA mutant (17), suggesting that ArcA prevents biofilm formation of P. gingivalis by repressing expression of FimA. The ability to form biofilms is considered an important pathogenic feature of P. gingivalis, as bacteria in biofilms often have a lower susceptibility to antibiotic treatments. Therefore, this work raises the intriguing possibility of interfering with P. gingivalis colonization with indigenous oral bacteria, which could be translated into a therapeutic application to combat P. gingivalis infection.
More than 600 bacterial species have been identified in the oral cavity, although some of them have not been cultured in vitro (24). Distinctive microbial profiles are found in healthy individuals compared to patients with different forms of periodontitis (1, 4). For example, significantly higher numbers of P. gingivalis, Tannerella forsythia, and Treponema spp. are detected in periodontitis patient samples. Apparently, the complexity and diversity of oral microbial communities have a major impact on oral health. In highly choreographed oral microbial communities, a successful colonization of P. gingivalis largely depends on its selective interaction with earlier colonizers, such as streptococci. There is strong evidence for the involvement of two sets of protein-protein interactions between P. gingivalis and S. gordonii (20, 23). FimA of P. gingivalis interacts with glyceraldehyde-3-phosphate dehydrogenase of S. gordonii, while Mfa1, a short fimbrial protein, interacts with streptococcal SspB, a member of the antigen I/II family. It is not clear if these two sets of protein-protein interactions play distinct roles in the colonization of P. gingivalis on streptococcal substrates. We have shown here that in the presence of S. cristatus, approximately 50% of P. gingivalis cells are pulled out from the heterotypic P. gingivalis-S. gordonii biofilms after anaerobic incubation for 16 h. We have further demonstrated a critical role of ArcA expression in this process. These results suggest that the interaction of FimA and glyceraldehyde-3-phosphate dehydrogenase is required for stabilization of P. gingivalis colonization on streptococcal substrates. Moreover, the results also provide a strong molecular basis for our earlier clinical observation of a negative correlation between the distributions of P. gingivalis and S. cristatus (32). Therefore, streptococcal ArcA may be utilized as a specific antagonistic agent against colonization of P. gingivalis.
Acknowledgments
This work was supported by Public Health Service grant R01 DE014699 (H.X.) from the National Institute of Dental and Craniofacial Research.
Footnotes
Published ahead of print on 26 July 2010.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano, A., A. Sharma, H. T. Sojar, H. K. Kuramitsu, and R. J. Genco. 1994. Effects of temperature stress on expression of fimbriae and superoxide dismutase by Porphyromonas gingivalis. Infect. Immun. 62:4682-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burne, R. A., and R. E. Marquis. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 193:1-6. [DOI] [PubMed] [Google Scholar]
- 4.Colombo, A. P., S. K. Boches, S. L. Cotton, J. M. Goodson, R. Kent, A. D. Haffajee, S. S. Socransky, H. Hasturk, T. E. Van Dyke, F. Dewhirst, and B. J. Paster. 2009. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J. Periodontol. 80:1421-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curran, T. M., J. Lieou, and R. E. Marquis. 1995. Arginine deiminase system and acid adaptation of oral streptococci. Appl. Environ. Microbiol. 61:4494-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enersen, M., I. Olsen, O. Kvalheim, and D. A. Caugant. 2008. FimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J. Clin. Microbiol. 46:31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajishengallis, G., M. Wang, E. Harokopakis, M. Triantafilou, and K. Triantafilou. 2006. Porphyromonas gingivalis fimbriae proactively modulate β2 integrin adhesive activity and promote binding to and internalization by macrophages. Infect. Immun. 74:5658-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harokopakis, E., M. H. Albzreh, M. H. Martin, and G. Hajishengallis. 2006. TLR2 transmodulates monocyte adhesion and transmigration via Rac1- and PI3K-mediated inside-out signaling in response to Porphyromonas gingivalis fimbriae. J. Immunol. 176:7645-7656. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto, M., S. Ogawa, Y. Asai, Y. Takai, and T. Ogawa. 2003. Binding of Porphyromonas gingivalis fimbriae to Treponema denticola dentilisin. FEMS Microbiol. Lett. 226:267-271. [DOI] [PubMed] [Google Scholar]
- 10.Holt, S. C., L. Kesavalu, S. Walker, and C. A. Genco. 1999. Virulence factors of Porphyromonas gingivalis. Periodontology 2000 20:168-238. [DOI] [PubMed] [Google Scholar]
- 11.Kato, T., S. Kawai, K. Nakano, H. Inaba, M. Kuboniwa, I. Nakagawa, K. Tsuda, H. Omori, T. Ooshima, T. Yoshimori, and A. Amano. 2007. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell. Microbiol. 9:753-765. [DOI] [PubMed] [Google Scholar]
- 12.Kuboniwa, M., A. Amano, E. Hashino, Y. Yamamoto, H. Inaba, N. Hamada, K. Nakayama, G. D. Tribble, R. J. Lamont, and S. Shizukuishi. 2009. Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis. BMC Microbiol. 9:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 14.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, D., M. A. Smith, J. Elter, C. Champagne, C. L. Downey, J. Beck, and S. Offenbacher. 2003. Porphyromonas gingivalis infection in pregnant mice is associated with placental dissemination, an increase in the placental Th1/Th2 cytokine ratio, and fetal growth restriction. Infect. Immun. 71:5163-5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, X., R. J. Lamont, J. Wu, and H. Xie. 2008. Role of differential expression of streptococcal arginine deiminase in inhibition of fimA expression in Porphyromonas gingivalis. J. Bacteriol. 190:4367-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, X., J. Wu, and H. Xie. 2006. Porphyromonas gingivalis minor fimbriae are required for cell-cell interactions. Infect. Immun. 74:6011-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeda, K., H. Nagata, M. Kuboniwa, K. Kataoka, N. Nishida, M. Tanaka, and S. Shizukuishi. 2004. Characterization of binding of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase to Porphyromonas gingivalis major fimbriae. Infect. Immun. 72:5475-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeda, K., H. Nagata, A. Nonaka, K. Kataoka, M. Tanaka, and S. Shizukuishi. 2004. Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae. Microbes Infect. 6:1163-1170. [DOI] [PubMed] [Google Scholar]
- 20.Maeda, K., H. Nagata, Y. Yamamoto, M. Tanaka, J. Tanaka, N. Minamino, and S. Shizukuishi. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect. Immun. 72:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura, T., A. Amano, I. Nakagawa, and S. Hamada. 1999. Specific interactions between Porphyromonas gingivalis fimbriae and human extracellular matrix proteins. FEMS Microbiol. Lett. 175:267-272. [DOI] [PubMed] [Google Scholar]
- 22.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 23.Park, Y., M. R. Simionato, K. Sekiya, Y. Murakami, D. James, W. Chen, M. Hackett, F. Yoshimura, D. R. Demuth, and R. J. Lamont. 2005. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect. Immun. 73:3983-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paster, B. J., and F. E. Dewhirst. 2009. Molecular microbial diagnosis. Periodontol. 2000 51:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seymour, G. J., P. J. Ford, M. P. Cullinan, S. Leishman, and K. Yamazaki. 2007. Relationship between periodontal infections and systemic disease. Clin. Microbiol. Infect. 13(Suppl. 4):3-10. [DOI] [PubMed] [Google Scholar]
- 26.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 27.Socransky, S. S., C. Smith, and A. D. Haffajee. 2002. Subgingival microbial profiles in refractory periodontal disease. J. Clin. Periodontol. 29:260-268. [DOI] [PubMed] [Google Scholar]
- 28.Sojar, H. T., A. Sharma, and R. J. Genco. 2002. Porphyromonas gingivalis fimbriae bind to cytokeratin of epithelial cells. Infect. Immun. 70:96-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song, H., M. Belanger, J. Whitlock, E. Kozarov, and A. Progulske-Fox. 2005. Hemagglutinin B is involved in the adherence of Porphyromonas gingivalis to human coronary artery endothelial cells. Infect. Immun. 73:7267-7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran, S. D., and J. D. Rudney. 1999. Improved multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans, Bacteroides forsythus, and Porphyromonas gingivalis. J. Clin. Microbiol. 37:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reference deleted.
- 32.Wang, B. Y., J. Wu, R. J. Lamont, X. Lin, and H. Xie. 2009. Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J. Clin. Microbiol. 47:3902-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie, H., S. Cai, and R. J. Lamont. 1997. Environmental regulation of fimbrial gene expression in Porphyromonas gingivalis. Infect. Immun. 65:2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie, H., G. S. Cook, J. W. Costerton, G. Bruce, T. M. Rose, and R. J. Lamont. 2000. Intergeneric communication in dental plaque biofilms. J. Bacteriol. 182:7067-7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie, H., N. Kozlova, and R. J. Lamont. 2004. Porphyromonas gingivalis genes involved in fimA regulation. Infect. Immun. 72:651-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie, H., X. Lin, B. Y. Wang, J. Wu, and R. J. Lamont. 2007. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology 153:3228-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou, Q., and S. Amar. 2006. Identification of proteins differentially expressed in human monocytes exposed to Porphyromonas gingivalis and its purified components by high-throughput immunoblotting. Infect. Immun. 74:1204-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]