Abstract
The CMY-2, ACT-1, DHA-1, ACC-1, and FOX-1 enzymes are representative of five plasmid-mediated AmpC (pAmpC) β-lactamase clusters. Resistance to imipenem has been reported in Enterobacteriaceae as a result of pAmpC expression combined with decreased outer membrane permeability. The aim of this study was to determine the role of different pAmpCs in carbapenem resistance and to define the structure/activity relationship supporting carbapenemase activity. The ampC genes encoding the five pAmpCs and the chromosomal AmpC of Escherichia coli EC6, which was used as a reference cephalosporinase, were cloned and introduced into wild-type E. coli TOP10 and OmpC/OmpF porin-deficient E. coli HB4 strains. The MICs of β-lactams for the recombinant strains revealed that CMY-2, ACT-1, and DHA-1 β-lactamases conferred a high level of resistance to ceftazidime and cefotaxime once expressed in E. coli TOP10 and reduced significantly the susceptibility to imipenem once expressed in E. coli HB4. In contrast, FOX-1 and ACC-1 enzymes did not confer resistance to imipenem. Biochemical analysis showed that CMY-2 β-lactamase and, to a lesser extent, ACT-1 exhibited the highest catalytic efficiency toward imipenem and showed low Km values. A modeling study revealed that the large R2 binding site of these two enzymes may support the carbapenemase activity. Therefore, CMY-2-type, ACT-1-type, and DHA-1-type β-lactamases may promote the emergence of carbapenem resistance in porin-deficient clinical isolates.
The class C (AmpC) β-lactamases constitute a group of enzymes widely distributed in Enterobacteriaceae. They preferentially inactivate narrow-spectrum cephalosporins and, to a lesser extent, expanded-spectrum cephalosporins (ESCs), such as ceftazidime and cefotaxime. Zwitterionic cephalosporins, such as cefepime, and carbapenems, such as imipenem, ertapenem, and meropenem, which penetrate very efficiently through the native outer membrane of Gram negatives and are poor substrates of AmpC β-lactamases, remain active in vitro against enterobacterial isolates that overproduce chromosome-encoded cephalosporinase (24).
Plasmid-mediated AmpC (pAmpC) β-lactamases, which originated from chromosomal AmpC of different Gram-negative bacteria, has emerged since the 1980s (24). They can be divided into five clusters: the Citrobacter freundii cluster, represented by CMY-2, the Enterobacter cluster with MIR-1 and ACT-1, the Morganella morganii group with DHA-1, the Hafnia alvei cluster represented by ACC-1, and the Aeromonas cluster with MOX-1 (also called CMY-1) and FOX-1 enzymes, which constitute two distinct subgroups (24). pAmpC β-lactamases, whose constitutive expression often is triggered by strong promoters (29), confer a phenotype of resistance similar to that displayed by chromosomal AmpC-overproducing strains.
Modifications of the membrane permeability can markedly change the susceptibility profile of pAmpC-producing isolates. By reducing the antibiotic concentration inside the periplasm, porin change may amplify the β-lactamase effects toward weakly hydrolyzed substrates, such as cefepime and carbapenems. This combination of mechanisms supports, in part, the emergence of carbapenem resistance among Enterobacteriaceae producing pAmpCs. Interestingly, these clinical isolates produced ACT-1, DHA-1, CMY-2, or CMY-4 β-lactamase (4, 5, 15, 21, 26, 32), which is a point variant of CMY-2 conferring an identical phenotype of resistance (33). These results suggested that these pAmpC β-lactamases possess a carbapenemase activity.
The aim of this study was to test the carbapenemase activity of five representative plasmid-borne AmpC-type β-lactamases, CMY-2, ACT-1, DHA-1, ACC-1, and FOX-1, in isogenic systems containing wild-type and porin-deficient E. coli strains. A biochemical characterization and a modeling study also were performed to elucidate the structure-activity relationship accounting for carbapenemase activity.
MATERIALS AND METHODS
Bacterial strains.
Clinical isolates Escherichia coli ECB1 and ECB2 produced ACC-1 and CMY-2 β-lactamases, respectively (17); recombinant strains E. coli DH10B (pPON-1) and E. coli EC6 produced DHA-1 enzyme and the AmpC B2 β-lactamase, respectively, which is the chromosomal wild-type cephalosporinase of E. coli (18, 25); the transformant strain E. coli (pGLK1) produced FOX-1 enzyme (10); and the strain Enterobacter asburiae CIP 105006 (Pasteur Institute, Paris, France) produced ACT-1 β-lactamase (30). All of these strains were used as sources of ampC genes.
The wild-type strain E. coli TOP10(Invitrogen, Cergy Pontoise, France) and the mutant strain E. coli HB4, which lacks porins OmpC and OmpF (19), were used as recipient strains in transformation experiments.
Cloning experiments.
The whole-cell DNAs from E. coli ECB1, E. coli ECB2, E. coli DHB10(pPON-1), E. coli EC6, E. coli(pGLK1), and E. asburiae CIP105006 were extracted as previously described (3). They were used as templates to amplify plasmid-borne ampC genes under the following PCR conditions: denaturation for 10 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C; and a final extension step of 10 min at 72°C. The set of primers, which were used to amplify the blaAmpC genes, are presented in Table 1. The PCR products, which contained the coding regions without their original promoter, subsequently were cloned into PCR-BluntII-Topo (Invitrogen), and the recombinant plasmids were transformed into E. coli strain TOP10 and E. coli HB4, as described previously (19).
TABLE 1.
Primers used in this study
| Target gene | Primer | Sequence | Reference or source |
|---|---|---|---|
| blaCMY-2 | CMY-2-A | 5′-ATGATGAAAAAATCGTTATGC-3′ | This study |
| CMY-2-B | 5′-TTATTGCAGCTTTTCAAGAATGC-3′ | ||
| blaACT-1 | ACT-1-A | 5′-ATGATGACTAAATCCCTTTGC-3′ | This study |
| ACT-1-B | 5′-CTACAGCGCGCTCAAAATACG-3′ | ||
| blaACC-1 | ACC-1-A | 5′-ATGCAGAACACATTGAAGC-3′ | This study |
| ACC-1-B | 5′-CTACTTATTCCCTTCCAATGAGC-3′ | ||
| blaDHA-1 | DHA-1-A | 5′-ATGAAAAAATCGTTATCTGC-3′ | This study |
| DHA-1-B | 5′-TTATTCCAGTGCACTCAAAATAGC-3′ | ||
| blaFOX-1 | FOX1-1-A | 5′- ATGCAACAACGACGTGCGTTCG-3′ | This study |
| FOX1-1-B | 5′-TCACTCGGCCAACTGACTCAGG-3′ | ||
| blaAmpCB2 | Int-B1 | 5′-TTTTGTATGGAACCAGACC-3′ | 19 |
| Int-HN | 5′-AAAAGCGGAGAAAAGGTCCG-3′ |
Antimicrobial agents and MIC determination.
The antibiotic agents and their sources have been described elsewhere (3). MICs were determined by an agar dilution technique on Mueller-Hinton agar (Sanofi-Diagnostics Pasteur, Paris, France) with an inoculum of 104 CFU per spot and were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (6, 7).
β-Lactamase purification.
All recombinant E. coli TOP10 strains were grown overnight at 37°C in 4 liters of trypticase soy (TS) broth containing amoxicillin (100 mg/liter) and kanamycin (30 mg/liter), resuspended in 40 ml of 100 mM phosphate buffer (pH 7), disrupted by sonication, and centrifuged at 20,000 × g for 1 h at 4°C, as described previously (3). The six AmpC β-lactamases were purified as described previously (20), except for the crude extract containing the FOX-1 enzyme, which was dialyzed overnight at 4°C against 20 mM bis-Tris (pH 6.8) at 4°C before being loaded onto a Q-Sepharose column preequilibrated with the same buffer. The FOX-1 enzyme was recovered in the flowthrough and dialyzed against 20 mM Tris-HCl buffer (pH 9) overnight at 4°C before being loaded onto a preequilibrated Q-Sepharose column. The β-lactamase activity was retained, and the proteins subsequently were eluted with a linear NaCl gradient (0 to 1 M). To assess the purity of the extracts, purified enzymes were subjected to SDS-PAGE analysis (14).
Kinetic measurements.
Purified β-lactamases were used to determine the kinetic parameters (Km and kcat) of cephaloridine, ertapenem, imipenem, and meropenem at 30°C in 100 mM sodium phosphate (pH 7.0). The rates of hydrolysis were determined with an Ultrospec 2100 spectrophotometer and were analyzed using the Swift II software (GE Healthcare). Km and kcat values for cephaloridine were determined by analyzing the ß-lactam hydrolysis under initial rate conditions by using the Eadie-Hofstee linearization of the Michaelis-Menten equation as previously described (8). Since the Km values for imipenem were low, Ki were determined instead of Km using cephaloridine as the substrate, and the kcat values were determined from initial rates at saturating substrate concentrations ([S] = 100 × Km) using 100 μl of the nondiluted enzyme extracts (8).
Modeling study.
A structural alignment of CMY-2 β-lactamase (Protein Data Bank [PDB] code 1ZC2), ACT-1 enzyme (PDB reference 2ZC7), and the AmpC β-lactamase of E. coli K-12 (PDB reference 1KVL) using the Deepview software (www.expasy/spdbv/) (11, 22, 23) was carried out. The secondary structures of the three layers were superimposed using the alternate-fit option of the software. The RMS (root mean squared) backbone deviation of each residue in the active layer from the corresponding amino acids in the two other layers was highlighted using the RMS coloring option.
RESULTS AND DISCUSSION
The PCR experiments yielded six PCR products containing the coding regions of blaCMY-2, blaACT-1, blaDHA-1, blaFOX-1, blaACC-1, and blaAmpC-B2 genes without their own promoter. These PCR products were cloned into PCR-BluntII-TOPO (Invitrogen) and transformed into E. coli TOP10 and E. coli HB4, giving rise to recombinant strains E. coli TOP10(pCMY-2), E. coli TOP10(pACT-1), E. coli TOP10(pDHA-1), E. coli TOP10(pFOX-1), E. coli TOP10(pACC-1), E. coli TOP10(pAmpC-B2), E. coli HB4(pCMY-2), E. coli HB4(pACT-1), E. coli HB4(pDHA-1), E. coli HB4(pFOX-1), E. coli HB4(pACC-1), and E. coli HB4(pAmpC-B2). The orientation of the cloned insert was the same in the recombinant plasmids, with the ampC gene under the transcriptional control of the lacZ promoter flanking the cloning site.
The MICs of β-lactams for the 12 recombinant strains are shown in Table 2. The recombinant E. coli TOP10 strains producing CMY-2, ACT-1, DHA-1, and FOX-1 β-lactamases were resistant to ceftazidime according to the CLSI criteria (6), whereas E. coli TOP10(pACC-1) was intermediate to this compound and E. coli TOP10(pAmpC-B2) remained susceptible to all ESCs (Table 2). Moreover, CMY-2, ACT-1, DHA-1, and FOX-1 conferred to E. coli TOP10 a high level of resistance to cefotaxime (128, 64, 8, and 16 μg/ml, respectively). All recombinant E. coli TOP10 strains remained susceptible to cefepime and carbapenems (6, 7).
TABLE 2.
MICs of β-lactams for the recombinant clones, the recipient strain E. coli TOP10, and the OmpC/OmpF porin-deficient strain E. coli HB4
| E. colistrain | β-Lactam MIC (μg/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cefoxitin | Cefuroxime | Cefotaxime | Ceftazidime | Cefepime | Imipenem | Ertapenem | Meropenem | |
| TOP10(pCMY-2)a | 128 | 128 | 128 | 256 | 1 | 0.5 | 0.064 | 0.064 |
| TOP10(pACT-1)a | 512 | 512 | 64 | 128 | 0.25 | 0.25 | 0.016 | 0.032 |
| TOP10(pDHA-1)a | 32 | 128 | 8 | 64 | 0.064 | 0.25 | 0.012 | 0.032 |
| TOP10(pFOX-1)a | 512 | 256 | 16 | 64 | 0.5 | 0.25 | 0.032 | 0.125 |
| TOP10(pACC-1)a | 2 | 1 | 1 | 8 | 0.125 | 0.125 | 0.006 | 0.032 |
| TOP10(pAmpC-B2)a | 128 | 128 | 0.5 | 1 | 0.06 | 0.125 | 0.064 | 0.032 |
| TOP10 | 2 | 1 | 0.06 | 0.06 | 0.06 | 0.125 | 0.006 | 0.032 |
| HB4(pCMY-2)a | >512 | >512 | 256 | >512 | 16 | 32 | 256 | 8 |
| HB4(pACT-1)a | >512 | >512 | 64 | >512 | 4 | 16 | 256 | 4 |
| HB4(pDHA-1)a | >512 | >512 | 128 | 256 | 2 | 2 | 16 | 1 |
| HB4(pFOX-1)a | >512 | >512 | 32 | 512 | 8 | 0.5 | 4 | 0.75 |
| HB4(pACC-1)a | 256 | 128 | 4 | 16 | 4 | 0.5 | 2 | 0.5 |
| HB4(pAmpC-B2) | 512 | 512 | 4 | 8 | 1 | 0.5 | 2 | 0.032 |
| HB4 | 256 | 32 | 4 | 4 | 1 | 0.25 | 1 | 0.032 |
E. coli TOP10(pCMY-2), E. coli TOP10(pACT-1), E. coli TOP10(pDHA-1), E. coli TOP10(pFOX-1), E. coli TOP10(pACC-1), and E. coli TOP10(pAmpC-B2) produced CMY-2, ACT-1, DHA-1, FOX-1, ACC-1, and the chromosome-borne cephalosporinase of E. coli EC6. E. coli HB4(pCMY-2), E. coli HB4(pACT-1), E. coli HB4(pDHA-1), E. coli HB4(pFOX-1), E. coli HB4(pACC-1), and E. coli HB4(pAmpC-B2) produced CMY-2, ACT-1, DHA-1, FOX-1, ACC-1, and the chromosome-borne cephalosporinase of E. coli EC6.
AmpC-producing E. coli HB4 recombinant strains displayed higher levels of resistance than E. coli TOP10 recombinant strains. The weak hydrolytic activity of cephalosporinases toward poor substrates was magnified by the simultaneous lack of OmpC and OmpF porins. E. coli HB4 recombinant strains producing CMY-2, ACT-1, DHA-1, FOX-1, and ACC-1 were resistant to ceftazidime (MIC ≥ 32 μg/ml) and cefotaxime (MIC ≥ 4 μg/ml) at high levels (Table 2). The E. coli HB4 recombinant strains were susceptible to cefepime, except for E. coli HB4 (pCMY-2), which was intermediate to this compound (16 μg/ml). The CMY-2 and ACT-1 β-lactamases conferred a high level of resistance to imipenem in E. coli HB4 (32 and 16 μg/ml, respectively), whereas DHA-1 enzyme conferred a reduced susceptibility to this compound (MIC of 2 μg/ml). In contrast, E. coli HB4(pACC-1), E. coli HB4(pFOX-1), and E. coli HB4(pAmpC-B2) remained fully susceptible to imipenem. All E. coli HB4 recombinant strains were susceptible to meropenem, except E. coli HB4(pCMY-2) and E. coli HB4(pACT-1), which were resistant and intermediate, respectively (8 and 4 μg/ml, respectively).
These in vitro results agree with the in vivo emergence of the imipenem resistance among AmpC-producing enterobacterial isolates, which mainly harbored the blaCMY-2 gene, its derivative blaCMY-4 gene, or the blaACT-1 gene. The MICs of carbapenems for the E. coli HB4(pCMY-2), E. coli HB4(pACT-1), and E. coli HB4(DHA-1) transformants are identical or closely related (1-fold dilution difference) to those displayed by the clinical enterobacterial isolates (4, 5, 15, 21, 26, 32). The porin-deficient E. coli HB4 strain constitutes a reliable in vitro model that could predict the selection of imipenem-resistant strains from clinical isolates producing β-lactamases with carbapenemase properties.
The CMY-2, ACT-1, DHA-1, FOX-1, ACC-1, and AmpC-B2 β-lactamases were extracted from the E. coli TOP10 recombinant strains. The concentration of enzymes was similar among the six crude extracts. The purification yielded six extracts containing 1.56, 2.1, 0.27, 0.17, 0.3, and 1.4 mg/ml of proteins, respectively. The comparison of specific activities before and after purification showed purification factors of 75, 78, 15, 6.5, 5.8, and 70, respectively. AmpC enzymes were purified to near homogeneity as deduced from the SDS-PAGE analysis (data not shown).
The Km and kcat values for imipenem and cephaloridine are presented in Table 3. The kcat values of the purified enzymes for cephaloridine were similar to those described previously (1, 9). The overall catalytic efficiencies of CMY-2 and ACT-1 β-lactamases for imipenem were higher than the values of the FOX-1 and AmpC-B2 enzymes, which could be related to lower Km values. Bauvois et al. already reported the increased catalytic efficiency of CMY-2 β-lactamase against ESCs compared to that of other pAmpCs, which resulted from a higher affinity (1). The kinetic parameters of DHA-1 and ACC-1 enzymes were not determinable due to the very slow hydrolysis rates displayed by their corresponding extract, which could be attributable, in part, to the very small amount of proteins recovered after the purification step. Unfortunately, despite several attempts, the protein concentration in these extracts could not be further increased.
TABLE 3.
Kinetic parameters of the five pAmpC β-lactamases
| β-Lactamase | Cephaloridine |
Imipenem |
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (mM·s−1) | kcat (s−1) | Km (μM) | kcat/Km (mM·s−1) | |
| CMY-2 | 412 ± 25 | 125 ± 30 | 3,300 | 0.04 ± 0.01 | 1 ± 0.5 | 40 |
| ACT-1 | 481 ± 18 | 420 ± 30 | 1,145 | 0.02 ± 0.005 | 3 ± 1 | 7 |
| DHA-1 | 196 ± 30 | 330 ± 30 | 595 | <0.001 | NDa | |
| FOX-1 | 228 ± 20 | 630 ± 40 | 360 | 0.01 ± 0.003 | 35 ± 4 | 0.3 |
| ACC-1 | 140 ± 32 | 420 ± 35 | 330 | <0.001 | ND | |
| AmpC-B2 | 240 ± 15 | 950 ± 10 | 250 | 0.08 ± 0.002 | 35 ± 2 | 2 |
ND, not determined.
It is noteworthy that the hydrolysis of ertapenem and meropenem was not detectable for all six purified AmpC extracts, which contrasted with the increased MIC values of ertapenem for the recombinant E. coli HB4(pCMY-2) and E. coli HB4(pACT-1) strains. This discrepancy between phenotypic and biochemical results could result from the low-but-not-zero deacylation rate of AmpC β-lactamases for those compounds (13).
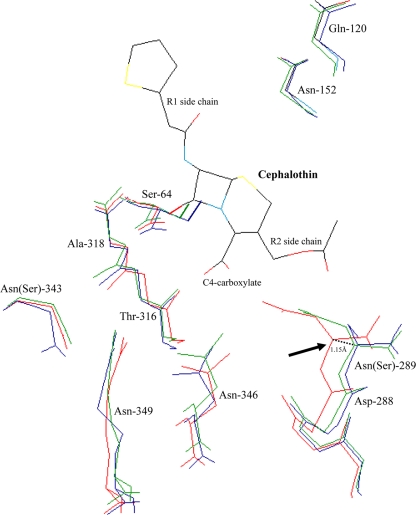
The modeling study showed that the overall structures of CMY-2, ACT-1, and the chromosomal AmpC β-lactamase of E. coli K-12 were homologous. In particular, the location and geometry of the catalytic serine residue (Ser-64) and the main-chain nitrogen atoms of Ser-64 and Ala(Ser)318 that form the oxyanion hole were well conserved. The location of residues Gln-120 and Asn-152, which also supply amide groups to the hydrogen bond to the acylamide carbonyl group of β-lactams, also were well conserved. Despite the overall similarity in structure, CMY-2, ACT-1, and AmpC-B2 had noticeable conformational differences in the binding site (Fig. 1). The residues of CMY-2 and ACT-1 that constitute the short coil located downstream of the helix H-9 at the edge of the R2 binding site (residues 287 to 289) presented a 0.55- to 1.15-Å shift compared to that of the AmpC β-lactamase of E. coli K-12. This structural discrepancy might improve the accommodation of the antibiotic inside the catalytic pocket by reducing the steric hindrances between the R2 substituents of the β-lactam rings and the top of the R2 binding site, which is constituted by the residues 287 to 289 (12, 16). Similarly, the crystallographic and biochemical study of the CMY-10 β-lactamase, which was derived from the plasmid-borne CMY-1 enzyme by an Asn-to-Ile substitution at position 346 and which exhibited increased catalytic efficiency against imipenem, revealed an open gap in the R2 binding site between the helices H-9, H-10, and the adjacent helix H-11 (12). Other structural differences in the R2 binding site also may contribute to the hydrolytic activity discrepancies. Indeed, some amino acids, such as the Asn-289 that is involved in the substrates binding in the AmpC β-lactamase of E. coli (16, 27, 28), are not well conserved in some pAmpCs enzymes (Ser-289 in CMY-2).
FIG. 1.
Superimposition of the crystallographic structures of CMY-2 β-lactamase (green; PDB code 1ZC2), ACT-1 enzyme (blue; PDB code 2ZC7) 31, and the AmpC β-lactamase of E. coli K-12 (red; PDB code 1KVL). Amino acids that are involved in the substrate binding (Gln-120, Asn-152, Ser-287, Asp-288, Ser-289, Thr-316, Asn-346, and Arg-349) and that constitute the oxyanion binding pocket (Ser-64 and Ser-318) are represented (2, 16, 28). The lateral side chains of Asn-289, Asn-346, and Arg-349, which contribute to the C4-carboxylate β-lactam binding (2, 16, 28), are shown. The side chain of the reactive Ser-64, which attacks the carbonyl carbon of the β-lactam ring, is shown in boldface.
This study reveals that the imipenem resistance may occur mostly among pAmpC producers of the CMY-2, ACT-1, and DHA-1 types. The carbapenemase property of the CMY-2 β-lactamase may be more important, since this cephalosporinase is widely distributed throughout the world among humans and animals (24).
Acknowledgments
This work was funded by a grant from the INSERM, the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, France, and by grants from the European Community (LSHM-CT-2005-018705) and TROCAR (HEALTH-F3-2008-223031).
Footnotes
Published ahead of print on 23 August 2010.
REFERENCES
- 1.Bauvois, C., A. S. Ibuka, A. Celso, J. Alba, Y. Ishii, J. M. Frère, and M. Galleni. 2005. Kinetic properties of four plasmid-mediated AmpC β-lactamases. Antimicrob. Agents Chemother. 49:4240-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beadle, B. M., and B. K. Shoichet. 2002. Structural basis for imipenem inhibition of class C β-lactamases. Antimicrob. Agents Chemother. 46:3978-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellais, S., D. Aubert, T. Naas, and P. Nordmann. 2000. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing β-lactamases in Chryseobacterium meningosepticum. Antimicrob. Agents Chemother. 44:1878-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, V. T., G. Arlet, B. M. Ericsson, A. Tammelin, P. Courvalin, and T. Lambert. 2000. Emergence of imipenem resistance in Klebsiella pneumoniae owing to combination of plasmid-mediated CMY-4 and permeability alteration. J. Antimicrob. Chemother. 46:895-900. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial. Susceptibility testing: 20th informational supplement. M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial. Susceptibility testing: 20th informational supplement (June 2010 Update). M100-S20-U. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Cornish-Bowden, A. 1995. Fundamentals of enzyme kinetics, p. 30-37. Portland Press, Seattle, WA.
- 9.Fosse, T., C. Giraud-Morin, I. Madinier, and R. Labia. 2003. Sequence analysis and biochemical characterization of chromosomal CAV-1 (Aeromonas caviae), the parental cephalosporinase of plasmid-mediated AmpC ‘FOX’ cluster. FEMS Microbiol. Lett. 222:93-98. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez Leiza, M., J. C. Perez-Diaz, J. Ayala, J. M. Casellas, J. Martinez-Beltran, K. Bush, and F. Baquero. 1994. Gene sequence and biochemical characterization of FOX-1 from Klebsiella pneumoniae, a new AmpC-type plasmid-mediated β-lactamase with two molecular variants. Antimicrob. Agents Chemother. 38:2150-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 12.Kim, J. Y., H. I. Jung, Y. J. An, J. H. Lee, S. J. Kim, S. H. Jeong, K. J. Lee, P. G. Suh, H. S. Lee, S. H. Lee, and S. S. Cha. 2006. Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C β-lactamase. Mol. Microbiol. 60:907-916. [DOI] [PubMed] [Google Scholar]
- 13.Lakaye, B., A. Dubus, S. Lepage, S. Groslambert, and J.-M. Frère. 1999. When drug inactivation renders the target irrelevant to antibiotic resistance: a case story with β-lactams. Mol. Microbiol. 31:89-101. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli, U. K., and M. Favre. 1973. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 80:575-599. [DOI] [PubMed] [Google Scholar]
- 15.Lee, K., D. Yong, Y. S. Choi, J. H. Yum, J. M. Kim, N. Woodford, D. M. Livermore, and Y. Chong. 2007. Reduced imipenem susceptibility in Klebsiella pneumoniae clinical isolates with plasmid-mediated CMY-2 and DHA-1 β-lactamases co-mediated by porin loss. Int. J. Antimicrob. Agents 29:201-206. [DOI] [PubMed] [Google Scholar]
- 16.Lobkovsky, E., P. C. Moews, H. Liu, H. Zhao, J. M. Frere, and J. R. Knox. 1993. Evolution of an enzyme activity: crystallographic structure at 2-Å resolution of cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc. Natl. Acad. Sci. U. S. A. 90:11257-11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mammeri, H., F. Eb, A. Berkani, and P. Nordmann. 2008. Molecular characterization of AmpC-producing Escherichia coli clinical isolates recovered in a French hospital. J. Antimicrob. Chemother. 61:498-503. [DOI] [PubMed] [Google Scholar]
- 18.Mammeri, H., M. Galleni, and P. Normann. 2009. Role of the Ser-287-Asn replacement in the hydrolysis spectrum extension of AmpC β-lactamases in Escherichia coli. Antimicrob. Agents Chemother. 53:323-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mammeri, H., P. Nordmann, A. Berkani, and F. Eb. 2008. Contribution of extended-spectrum AmpC (ESAC) β-lactamases to carbapenem resistance in Escherichia coli. FEMS Microbiol. Lett. 282:238-240. [DOI] [PubMed] [Google Scholar]
- 20.Mammeri, H., L. Poirel, and P. Nordmann. 2007. Extension of the hydrolysis spectrum of AmpC β-lactamase of Escherichia coli due to amino acid insertion in the H-10 helix. J. Antimicrob. Chemother. 60:490-494. [DOI] [PubMed] [Google Scholar]
- 21.Oteo, J., A. Delgado-Iribarren, D. Vega, V. Bautista, M. C. Rodriguez, M. Velasco, J. M. Saavedra, M. Perez-Vazquez, S. Garcia-Cobos, L. Martinez-Martinez, and J. Campos. 2008. Emergence of imipenem resistance in clinical Escherichia coli during therapy. Int. J. Antimicrob. Agents 32:534-537. [DOI] [PubMed] [Google Scholar]
- 22.Peitsch, M. C. 1995. Protein modeling by e-mail. Biotechnology 13:658-660. [Google Scholar]
- 23.Peitsch, M. C. 1996. ProMod and Swiss-Model: internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 24.Philippon, A., G. Arlet, and G. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel, L., M. Guibert, D. Girlich, T. Naas, and P. Nordmann. 1999. Cloning, sequence analyses, expression, and distribution of ampC-ampR from Morganella morgannii clinical isolates. Antimicrob. Agents Chemother. 43:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel, L., C. Héritier, C. Spicq, and P. Nordmann. 2004. In vivo acquisition of high-level resistance to imipenem in Escherichia coli. J. Clin. Microbiol. 42:3831-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powers, R. A., E. Caselli, P. J. Focia, F. Prati, and B. K. Shoichet. 2001. Structures of ceftazidime and its transition-state analogue in complexe with AmpC β-lactamase: implications for resistance mutations and inhibitor design. Biochemistry 40:9207-9214. [DOI] [PubMed] [Google Scholar]
- 28.Powers, R. A., and B. K. Shoichet. 2002. Structure based approach for binding site identification on AmpC β-lactamase. J. Med. Chem. 45:3222-3234. [DOI] [PubMed] [Google Scholar]
- 29.Reisbig, M. D., and N. Hanson. 2004. Promoter sequences necessary for high-level expression of the plasmid-associated ampC β-lactamase gene blaMIR-1. Antimicrob. Agents Chemother. 48:4177-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rottman, M., Y. Benzerara, B. Hanau-Berçot, C. Bizet, A. Philippon, and G. Arlet. 2002. Chromosomal ampC genes in Enterobacter species other than Enterobacter cloacae, and ancestral association of the ACT-1 plasmid-encoded cephalosporinase to Enterobacter asburiae. FEMS Microbiol. Lett. 210:87-92. [DOI] [PubMed] [Google Scholar]
- 31.Shimizu-Ibuka, A., C. Bauvois, H. Sakai, and M. Galleni. 2008. Structure of the plasmid-mediated class C β-lactamase ACT-1. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64:334-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stapleton, P. D., K. P. Shannon, and G. L. French. 1999. Carbapenem resistance in Escherichia coli associated with plasmid-determined CMY-4 β-lactamase production and loss of an outer membrane protein. Antimicrob. Agents Chemother. 43:1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verdet, C., G. Arlet, S. Ben Redjeb, A. Ben Hassen, P. H. Lagrange, and A. Philippon. 1998. Characterisation of CMY-4, an AmpC-type plasmid-mediated β-lactamase in a Tunisian clinical isolate of Proteus mirabilis. FEMS Microbiol. Lett. 169:235-240. [DOI] [PubMed] [Google Scholar]