Abstract
Granular sludge is a novel alternative for the treatment of wastewater and offers numerous operational and economic advantages over conventional floccular-sludge systems. The majority of research on granular sludge has focused on optimization of engineering aspects relating to reactor operation with little emphasis on the fundamental microbiology. In this study, we hypothesize two novel mechanisms for granule formation as observed in three laboratory scale sequencing batch reactors operating for biological phosphorus removal and treating two different types of wastewater. During the initial stages of granulation, two distinct granule types (white and yellow) were distinguished within the mixed microbial population. White granules appeared as compact, smooth, dense aggregates dominated by 97.5% “Candidatus Accumulibacter phosphatis,” and yellow granules appeared as loose, rough, irregular aggregates with a mixed microbial population of 12.3% “Candidatus Accumulibacter phosphatis” and 57.9% “Candidatus Competibacter phosphatis,” among other bacteria. Microscopy showed white granules as homogeneous microbial aggregates and yellow granules as segregated, microcolony-like aggregates, with phylogenetic analysis suggesting that the granule types are likely not a result of strain-associated differences. The microbial community composition and arrangement suggest different formation mechanisms occur for each granule type. White granules are hypothesized to form by outgrowth from a single microcolony into a granule dominated by one bacterial type, while yellow granules are hypothesized to form via multiple microcolony aggregation into a microcolony-segregated granule with a mixed microbial population. Further understanding and application of these mechanisms and the associated microbial ecology may provide conceptual information benefiting start-up procedures for full-scale granular-sludge reactors.
Activated sludge can be operated as an efficient biological process to treat and remove unwanted nutrients, such as carbon, nitrogen, and phosphorus, from wastewater. Two common processes applied in wastewater treatment are enhanced biological phosphorus removal (EBPR) (28), which is widely applied for carbon and phosphorus removal, and simultaneous nitrification-denitrification and phosphorus removal (SNDPR) (40), which combines carbon, phosphorus, and nitrogen removal. Conventional plants operating for EBPR and SNDPR typically operate as floccular-sludge systems, in which small aggregates (30 to 200 μm) of microorganisms make up the suspended biofilms of activated sludge (10). Aerobic granular activated sludge is a novel alternative for the treatment of wastewater and offers several operational and economic advantages over conventional floccular-sludge systems (9, 22). Granular sludges are typically larger suspended biofilm aggregates (200 to 2,000 μm) and typically contain higher biomass concentrations within the same reactor vessels than floccular-sludge systems (4, 29). However, granular sludge has yet to be applied to full-scale wastewater treatment plants (WWTP), partly due to a lack of fundamental knowledge of the microbiology associated with the granulation process.
For full-scale application of aerobic granular-sludge systems, the most likely strategy for the reactor start-up would be seeding with a conventional floccular sludge. From the floccular seed, there is a transition period with the selection and eventual formation of granules. During the start-up phase, there is typically a high degree of process performance variability, with biomass washout and large changes to the microbial community occurring due to certain changes to operational parameters applied to the system (21, 22, 39). Thus, the process performance, the start-up length, and ultimately the optimization of the start-up phase have important implications for the application of full-scale granular-sludge plants. Previous investigations of aerobic granular systems have visualized the microbial community and aggregate structure during this start-up phase (39), and mathematical models have been developed to predict granule formation and growth (31, 36). While these studies provide some models for granule formation, there has been little work on understanding the details of the specific microbial aggregation mechanisms involved in the granulation process, which needs to be optimized for full-scale operation.
Aerobic granulation is often studied using laboratory scale sequencing batch reactors (SBR) fed with synthetic wastewater (19, 20). Within laboratory scale SBR, numerous operational parameters can be manipulated to actively select for a granular sludge; these include settling time (11, 16, 29), shear force (3, 34, 35), and high organic loading rates (4, 26). In contrast to studies of engineering process parameters, the fundamental composition of the microorganisms present within these systems has been underinvestigated. Recently, however, the use of modern molecular biology techniques has provided novel insights into the formation and stability of these aerobic granular sludges within the engineered systems and is helping to bridge the gap between successful reactor operation and optimization of the microbial ecology and stability (18, 27, 32, 38, 41).
This study uses an EBPR system enriched with “Candidatus Accumulibacter phosphatis,” which is one of the most important organisms responsible for phosphate removal (12), to investigate the microbial details of granule formation from floccular sludge. In the current study, two distinct granule types segregated from within a mixed microbial community on three occasions during granulation. This granulation process was intensely investigated to deduce the mechanisms for this granule segregation. It is known that microcolony structures present within activated-sludge flocs have the potential for different microbial compositions and metabolic processes (30). Similarly, in anaerobic methanogenic granules, microcolony cluster segregation occurs within the granular structure (13). However, to our knowledge, the segregation of two different microbial granule types from within a mixed microbial system has not been previously reported. From our investigations of the microbial ecologies and structures of these two distinct granule types, two mechanisms for granulation were hypothesized. Further understanding and selection of these mechanisms and understanding of the associated microbial ecology will provide insight into facilitating the full-scale application of granular sludge.
MATERIALS AND METHODS
SBR operation.
Three laboratory scale SBR were operated for granule formation. The first SBR, called Synthetic-1, was operated for EBPR and fed with synthetic wastewater containing volatile fatty acids (VFA) and orthophosphate. The reactor had a working volume of 8 liters and a 6-h cycle time consisting of a 6-min feed period, 120-min anaerobic and 180-min aerobic phases, 4 min wasting, 40 min settling, and 10 min decanting. In each cycle, 2 liters of synthetic wastewater was fed into the reactor during the 6-min feed period, resulting in a hydraulic retention time (HRT) of 24 h. The solids retention time (SRT) was approximately 10 days. Dosing with 0.5 M HCl and 2 M NaOH controlled the pH during the anaerobic and aerobic phases at between 7.8 and 8. The sole carbon source in the synthetic feed alternated between acetate and propionate, with a switching frequency of every 2 weeks, in order to provide a selective advantage to polyphosphate-accumulating organisms (PAO) over glycogen-accumulating organisms (GAO) (23). Further details of the synthetic feed, the feeding strategy, and reactor operation can be found in Lu et al. (23). Chemical analyses were performed to detect the biochemical transformations within cycles of the SBR operation (cycle study). The orthophosphate (P-PO43−) concentrations were analyzed using a Lachat QuikChem8000 Flow Injection Analyzer (Lachat Instrument, Milwaukee, WI). VFA were measured by Perkin-Elmer gas chromatography with a DB-FFAP column (15 m by 0.53 mm by 1.0 μm [length/inside diameter/film thickness]) at 140°C.
On day zero of operation, Synthetic-1 was inoculated with floccular sludge from a full-scale wastewater treatment plant. Various SBR operational changes were implemented to influence reactor performance and sludge selection during the 170-day operation. From day 63 on, the anaerobic phase was decreased from 120 min to 90 min and the aerobic phase was increased from 180 min to 210 min to allow further aerobic phosphorus uptake. From day 98 onward, the reactor sludge wasting was adjusted to intentionally waste during the first 4 min of the settle phase to selectively remove more slowly settling aggregates. From days 98 to 133, the volume of treated wastewater per cycle was increased from 2 liters to 4 liters. During this period, a diluted feed was used to maintain the same organic and nutrient load used previously. After day 133, reactor operation was returned to day 63 operation.
Two additional SBR were investigated at one time point only during the study, and these were termed Synthetic-2 (an SBR treating synthetic wastewater for EBPR) and Domestic (an SBR treating domestic wastewater for SNDPR). Synthetic-2 was operated with the same initial parameters as Synthetic-1 (described above), except for the following: a cycle time of 6 h consisted of a 6-min feed period, 124-min anaerobic and 160-min aerobic phases, 65 min settling and decanting, and a 5-min anaerobic idle period. For further details of Synthetic-2 operation, see Zhou et al. (42). The Domestic reactor was operated with a domestic wastewater feed consisting of (by volume) 90% raw domestic sewage (see Table S1 in the supplemental material for details of the composition of the domestic sewage) and 10% VFA feed containing 1.5 g NaAc·3H2O/liter. Domestic operation included two feed events with a cycle time of 6 h consisting of a 4-min feed period, 100-min anaerobic and 130-min aerobic phases, a 1-min feed period, 50-min anaerobic and 55-min aerobic phases, a 7-min anoxic phase, 5 min settling, and 5 min decanting. In each cycle, 1 liter of wastewater was fed into the reactor, resulting in an HRT of 12 h. The SRT was approximately 18 days.
Fluorescence in situ hybridization (FISH) and stereo- and light microscopy analyses.
Granule samples were fixed, and FISH was performed as previously described (1). For quantitative FISH analysis of the different granule types, between 5 and 10 granules of each type were manually homogenized before analysis to provide an accurate representation of the bacterial populations present. For cryosectioned samples, fixed granules of the two granule types were embedded in compound at the optimum cutting temperature prior to FISH (TissueTek; Sakura Finetek, Torrance, CA), as previously described (27). The embedded granules were frozen and sectioned into 10-μm-thick slices using a cryotome operated at −20°C (Kryo 1720; Leitz, Wetzler, Germany) and collected onto SuperFrost Plus microscope slides (Menzel-Glaser, Braunschweig, Germany). All other samples were manually homogenized before application onto Teflon-coated microscope slides. Samples were hybridized with fluorescein isothiocyanate (FITC)-labeled EUB Mix probes for detection of bacteria (2, 7); Cy3-labeled GAO Mix probes (for “Candidatus Competibacter phosphatis,” a known GAO) (5); and Cy5-labeled PAO Mix probes (for “Candidatus Accumulibacter phosphatis,” a known PAO, (6) (see Table S2 in the supplemental material for detailed FISH probe information). Fluorescent DNA probes were visualized and images were captured with a Zeiss LSM 510 Meta confocal laser scanning microscope (CLSM) (Carl Zeiss, Jena, Germany) using a Zeiss Neofluor ×40/1.3 oil objective. Composite images of cryosectioned granules were constructed using between 10 and 30 overlapping, consecutive images, which were reconstructed using Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA). The images were analyzed using Daime version 1.2 (8). Image segmentation was carried out using default parameters. Biovolumes were calculated from 20 to 30 images using the biovolume fraction function. The artifact rejection tool was set at a congruency threshold of 75%. For stereomicroscopic examination, whole granules were photographed using an Olympus SZH10 microscope, and light microscopic examinations were performed on an Olympus BX51 microscope with a DP70 digital camera.
Transmission electron microscopy (TEM) analysis.
Granular-sludge samples were first stabilized using 2.5% glutaraldehyde and 75 mM lysine in 0.1 M cacodylate buffer for 10 min (17), high-pressure frozen using a Leica Empact2, and then freeze substituted in 1.5% osmium tetroxide and 0.5% uranyl acetate in acetone at −85°C for 2 days. After being warmed to room temperature, samples were washed in acetone, submerged in Epon resin, and polymerized at 60°C for 2 days. Semithin 500-nm sections were stained with 1% toluidine blue and 1% borax and viewed with an Olympus BX51 light microscope. Ultrathin sections 60 nm thick were cut using a Leica Ultracut UC6 ultramicrotome and mounted on Formvar-coated copper grids, stained with 5% uranyl acetate in 5% methanol and Reynolds lead citrate, and viewed using a Jeol (Tokyo, Japan) 1010 transmission electron microscope operated at 80 kV.
Physical analysis of activated sludge.
The activated-sludge aggregate particle size distribution was measured to monitor the distribution of granule sizes. Thirty milliliters of well-mixed sludge was sampled from the SBR at the end of the aeration period and applied to a Malvern laser light-scattering instrument (Mastersizer 2000 series; Malvern Instruments, Worcestershire, United Kingdom).
DNA extraction.
White and yellow granules from the Synthetic-1 and Domestic reactors were easily distinguishable by color and were manually separated prior to DNA extraction. Genomic DNA was extracted from white and yellow granule samples using the FastDNA SPIN for Soil Kit (MP Biomedicals).
ppkI clone library construction and phylogenetic analysis.
Amplification of “Candidatus Accumulibacter phosphatis” ppkI gene fragments from the Synthetic-1 white and yellow granules was performed using the ACCppkI-254F and ACCppkI-1376R primers as previously described (25). The reaction mixtures contained 1× PCR buffer (New England Biolabs), 3 mM MgCl2, 200 μM each deoxynucleoside triphosphate (dNTP), 400 nM each forward and reverse primer, and 0.5 U/μl of DNA polymerase (New England Biolabs). The PCR was conducted using a MyCycler (Bio-Rad) with an initial 10-min denaturation step at 96°C, followed by 30 cycles of 96°C for 30 s, 64°C for 1 min, and 72°C for 2 min and a final extension at 72°C for 10 min. The PCR products (approximately 1,100 bp) were visualized by agarose gel electrophoresis, purified using the QIAquick PCR Purification Kit (Qiagen), and cloned into the pGEM-T Easy Vector (Promega) using One Shot Top10 (Invitrogen) competent cells. Twenty clones were selected for further sequencing and phylogenetic analysis. An unrooted phylogenetic tree was constructed by the neighbor-joining (22) method and evaluated by bootstrap resampling (1,000 replications) using MEGA (Molecular Evolutionary Genetics Analysis) version 4.0 (33) and CLUSTALX version 2.0.5 (37).
16S rRNA gene terminal restriction fragment length polymorphism (tRFLP) fingerprinting.
PCR was performed on white and yellow granules from the Domestic SBR using the bacterial (63F and 1389R) (24) primer set, except that the forward primer (63F) was labeled at the 5′ end with a phosphoramidite dye, 6-FAM (6-carboxyfluorescein). Reaction mixtures contained 1× PCR buffer (New England Biolabs), 2 mM MgCl2, 200 μM each dNTP, 400 nM each forward and reverse primer, 0.5 U/μl of DNA polymerase (New England Biolabs), and approximately 20 ng of template DNA, and reactions were performed in triplicate. PCR using the 16S rRNA gene primers was conducted in a MyCycler (Bio-Rad) with an initial 10-min denaturation step at 96°C, followed by 30 cycles at 96°C for 30 s, 56°C for 1 min, and 72°C for 2 min and a final extension at 72°C for 10 min. The PCR products were purified using the QIAquick PCR Purification Kit (Qiagen) and used in separate endonuclease restriction digestions using the enzymes AluI, HaeIII, MspI, and Sau3AI. Digestions were performed according to the manufacturer's instructions. 16S rRNA gene digests were analyzed at the Australian Genome Research Facility (Glen Osmond, SA, Australia) on an AB3730 Genetic Analyzer (Applied Biosystems, Foster City, CA) fitted with a 36-cm array and using the GS500(−250)LIZ standard. Further details of the tRFLP analysis are provided in the supplemental material.
RESULTS
Granule formation detected within the laboratory scale SBR operating for floccular EBPR sludge.
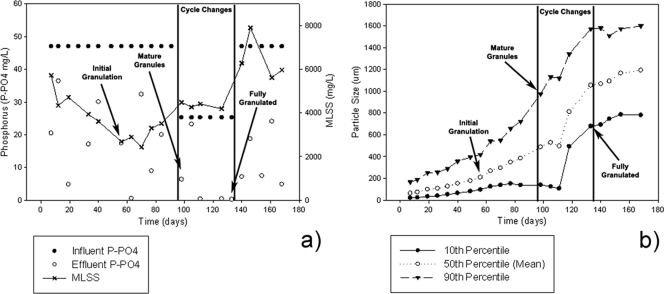
The initial objective was to operate the laboratory scale SBR, Synthetic-1, as a floccular sludge until both stable EBPR performance and a “Candidatus Accumulibacter phosphatis” enrichment of greater than 75% was achieved. The reactor operation was then to be manipulated to obtain a granular sludge while maintaining stable EBPR performance and “Candidatus Accumulibacter phosphatis” enrichment. However, granule formation occurred unexpectedly between days 50 and 60 of reactor operation. During the granulation period, approximately day 56, EBPR performance was not optimal, with only 63% P-PO43− mg liter−1 removal from the reactor (Fig. 1a) and a median particle size in the SBR of 209 μm (Fig. 1b). At this time, the mixed-liquor suspended solids (MLSS) (reactor biomass level) had decreased to 2,680 mg/liter with a “Candidatus Accumulibacter phosphatis” enrichment of 58% (below the target of 75% enrichment). By day 98, mature granules were present within the system, as exhibited by a 90th-percentile particle size of 975 μm (indicating that the upper 10% of biomass particles represented large, developed granules) and a median particle size of 487 μm. Additionally, the MLSS and EBPR performances had increased to 4,480 mg/liter and 87% removal, respectively. However, a large portion of floccular biomass was still present within the system. Cycle changes were implemented from days 98 to 133 to remove this floccular biomass (see Materials and Methods). Consequently, a fully granular-sludge system was obtained by day 133, as exhibited by a 10th-percentile particle size of 677 μm (indicating that very little floccular biomass remained in the system) and a median particle size of 1,053 μm. During this selection period, biomass levels (MLSS) were stable at around 4,190 mg/liter and the reactor had complete P-PO43− removal. After complete granulation on day 133, the cycle changes were reverted. In the remaining period (days 133 to 170), the MLSS concentration continued to increase to over 6,000 mg/liter; however, the EBPR performance was erratic.
FIG. 1.
EBPR performance (a) and particle size (b) of the Synthetic-1 SBR treating synthetic wastewater over a 168-day period. The vertical lines indicate where cycle changes were made to remove floccular material from the system and where the influent phosphorus concentration was halved due to an increase in the reactor volume exchange ratio; however, the total phosphorus treated did not change (see Materials and Methods). The “initial granulation” arrows indicate when the 50th-percentile particle size was above 200 μm, indicative of initial granule formation. The “mature granulation” arrows indicate when large, developed granules were present. The “fully granulated” arrows indicate when the majority of flocs were removed from the system. The 10th percentile indicates that 10% of particles were below this size distribution, the 50th percentile is the average (or median) particle size distribution, and the 90th percentile indicates 10% of the particles were above this size distribution.
Characterization of two distinct granule types occurring within Synthetic-1.
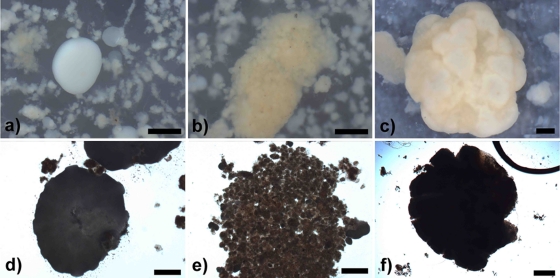
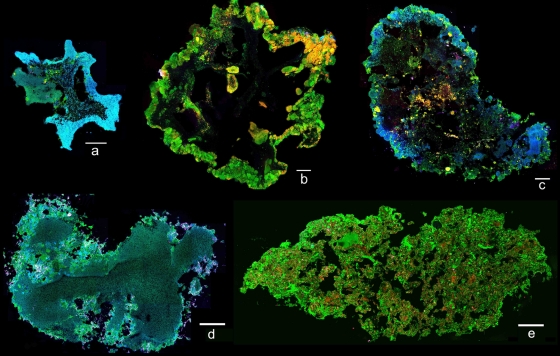
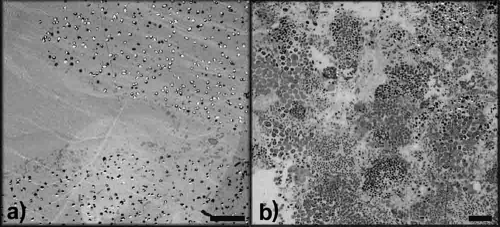
Granule formation occurred within the Synthetic-1 reactor unexpectedly following EBPR operation and initial enrichment of “Candidatus Accumulibacter phosphatis.” During the initial stages of granulation (days 56 to 98), two distinct granule types formed from the mixed microbial population. “White granules” (compact, smooth, dense, spherical aggregates) and “yellow granules” (loose, rough, irregular aggregates) were detected by stereo- and light microscopy (Fig. 2a, b, d, and e). White granules appeared as a homogeneous layer of microbial growth, while yellow granules appeared to be composed of numerous smaller microcolony structures aggregated into a larger granule. Bacterial population abundance, as assessed by quantitative FISH on numerous homogenized granules, demonstrated that the white granules were comprised of 97.5% “Candidatus Accumulibacter phosphatis” and 0.9% “Candidatus Competibacter phosphatis,” with very few other bacteria present. The yellow granules were comprised of 12.3% “Candidatus Accumulibacter phosphatis” and 57.9% “Candidatus Competibacter phosphatis,” with the remaining population consisting of other bacteria. The microbial community compositions and structures of white and yellow granules were shown to be different, as observed by cryosectioned FISH on day 98 of reactor operation (Fig. 3a and b). The outer layer of white granules formed a solid, compact outer wall dominated by “Candidatus Accumulibacter phosphatis” growing in an almost complete ring structure. In comparison, yellow granules generally formed larger granular aggregates composed of numerous compartmentalized microcolonies, which contained distinct, separate microbial communities. TEM images taken on day 98 of reactor operation showed cellular differences between white and yellow granules (Fig. 4a and b). White granules were dominated by one bacterial morphotype (cells 1 μm in diameter containing large, dark intracellular stores of polyphosphate, presumed to be “Candidatus Accumulibacter phosphatis”) in a solid, homogeneous layer on the outer edge of the granule and exhibited low cell density with large amounts of presumably extracellular polymeric substances (EPS) surrounding the “Candidatus Accumulibacter phosphatis” cells (6). Yellow granules, in comparison, exhibited large, varied numbers of bacterial morphotypes (ranging from 0.5 to 2 μm in diameter) with only a small percentage of presumed “Candidatus Accumulibacter phosphatis” morphotypes present. They also exhibited higher cell density than white granules, with noticeably less EPS surrounding the cells, and distinct, compartmentalized microcolony structures throughout the edges of the yellow granules.
FIG. 2.
Stereomicroscope images of white (a), yellow (b), and off-white (c) granules and light microscope images of white (d), yellow (e), and off-white (f) granules from the Synthetic-1 reactor. The images of white and yellow granules were taken on day 63 of Synthetic-1 operation, and the images of off-white granules were taken on day 133 of Synthetic-1 operation. White and yellow granules were present in the reactor only during the initial stages of granulation, after which a single homogeneous off-white granule population developed. All scale bars, 500 μm.
FIG. 3.
CLSM image of cryosectioned granules after FISH, with “Candidatus Accumulibacter phosphatis” in blue, “Candidatus Competibacter phosphatis” in yellow, and all other bacteria in green. (a) Synthetic-1 white granules were imaged on day 98 of reactor operation and were dominated by “Candidatus Accumulibacter phosphatis” with a solid, compact outer-wall structure. (b) Synthetic-1 yellow granules were imaged on day 98 of reactor operation and had a diverse bacterial population, were generally larger, and consisted of numerous compartmentalized microcolony structures. (c) Synthetic-1 off-white granules were imaged on day 168 of reactor operation and appeared as a mixture of the white and yellow granules with both solid outer walls and microcolony structures. (d) Domestic white granules were imaged on day 68 of reactor operation, were dominated by “Candidatus Accumulibacter phosphatis,” and exhibited a solid, compact outer-wall structure similar to that of the Synethic-1 white granules. (e) Domestic yellow granules were imaged on day 68 of reactor operation and exhibited a solid structure with fewer microcolonies and a more diverse bacterial population than the Synthetic-1 yellow granules. All scale bars, 100 μm.
FIG. 4.
TEM image of an outer edge of a white granule (a) and a yellow granule (b) taken on day 98 of Synthetic-1 reactor operation. White granules were dominated by one cellular morphotype, likely “Candidatus Accumulibacter phosphatis,” in a solid homogeneous layer. Yellow granules showed diverse numbers of bacterial morphotypes growing in distinct microcolony structures. All scale bars, 10 μm.
Comparison of “Candidatus Accumulibacter phosphatis” phylotypes associated with white and yellow granules.
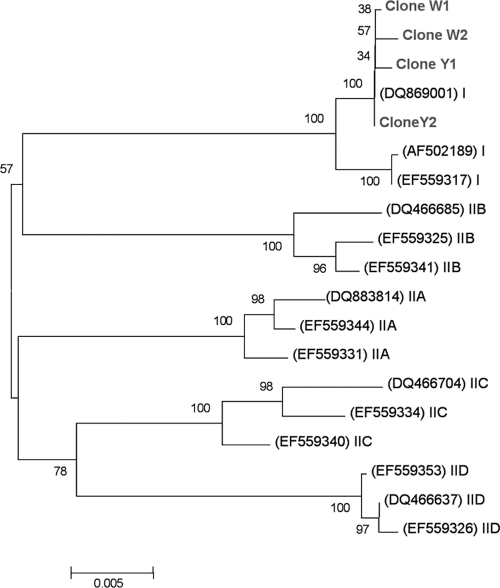
The polyphosphate kinase 1 gene (ppkI) was used as a phylogenetic marker to investigate whether different “Candidatus Accumulibacter phosphatis” phylotypes were associated with white and yellow granules and whether “Candidatus Accumulibacter phosphatis” phylotype differences were associated with the granule segregation. All the cloned ppkI gene sequences (∼1,100 bp) from both white and yellow granules grouped tightly within the “Candidatus Accumulibacter phosphatis” “type I” group (Fig. 5) (15). According to the resolution of the ppkI gene, there was no difference in “Candidatus Accumulibacter phosphatis” ppkI phylotypes associated with the white and yellow granules.
FIG. 5.
Unrooted abbreviated phylogenetic tree obtained by the neighbor-joining method, showing the positions of four ppkI clones from white (W) and yellow (Y) granules with known ppkI gene sequences from both the type I and type II “Candidatus Accumulibacter phosphatis” lineages. All cloned sequences were positioned within the type I clade branch. GenBank sequence accession numbers are given in parentheses. The numbers at the nodes show the percentage bootstrap values. The scale bar indicates the number of changes per site.
A mature granular population results in a homogeneous granule type.
During the initial stages of granulation (days 56 to 98), white and yellow granules appeared in a stable ratio of approximately 50:50 within the reactor, as observed via stereomicroscopy (see Fig. S1 in the supplemental material). However, once a mature granular system had been achieved, the distinction between the white and yellow granules was no longer detectable, and by day 133, a single homogeneous population of “off-white granules” persisted (see Fig. S2 in the supplemental material). The off-white granules appeared as large, dense aggregates with numerous hump-like protrusions (Fig. 2c and f) and showed a hybrid morphology from both the white and yellow granules. The microbial communities of the off-white granules were analyzed via FISH on day 168 of reactor operation (Fig. 3c). The bacterial population of the off-white granules was comprised of 68.4% “Candidatus Accumulibacter phosphatis” and 9.8% “Candidatus Competibacter phosphatis,” among other bacteria. The structure of the off-white granules also appeared to be a mixture of the white and yellow granules, with portions of the outer wall appearing as solid, homogeneous sections dominated by “Candidatus Accumulibacter phosphatis” while other sections were dominated by numerous compartmentalized microcolony structures.
Granule segregation detected within two additional SBR treating synthetic and domestic wastewater.
Two additional laboratory scale SBR showed similar signs of granule segregation. The two reactors were termed Synthetic-2 (an SBR treating synthetic wastewater for EBPR) and Domestic (an SBR treating domestic wastewater for SNDPR).
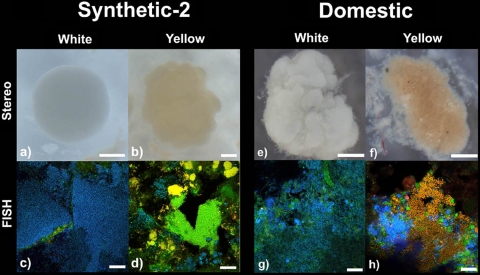
The Synthetic-2 reactor had previously been operated as a floccular EBPR system for the enrichment of “Candidatus Accumulibacter phosphatis.” The reactor had been operated for a total of 182 days prior to investigation, with initial granulation previously undetected. On day 182 of operation, Synthetic-2 achieved complete EBPR performance (see Fig. S3 in the supplemental material) and had a mean particle size of 430 μm, indicating a granular system, and MLSS was 2,830 mg/liter. Synthetic-2 granule segregation was assessed via stereomicroscopy and quantitative FISH (Fig. 6a, b, c, and d). Synthetic-2 white granules similarly appeared as dense spherical aggregates and were dominated (88.3%) by “Candidatus Accumulibacter phosphatis,” with 3.1% “Candidatus Competibacter phosphatis,” among other bacteria. Synthetic-2 yellow granules appeared as irregular aggregates, with 8.8% “Candidatus Accumulibacter phosphatis” and 65.1% “Candidatus Competibacter phosphatis” appearing in microcolony structures, among other bacteria.
FIG. 6.
Stereomicroscopy and homogenized FISH images of white and yellow granules from the Synthetic-2 reactor (a to d) on day 182 of operation and the Domestic reactor (e to h) on day 68 of operation. The FISH images show “Candidatus Accumulibacter phosphatis” in blue, “Candidatus Competibacter phosphatis” in yellow, and all other bacteria in green. Scale bars, 200 μm (a and b), 500 μm (e and f), and 20 μm (c, d, g, and h).
The Domestic SBR was purposely operated to obtain a granular sludge for SNDPR treatment of domestic wastewater. Initial granulation occurred within the Domestic reactor by day 38 of operation, with a median particle size of 220 μm. By day 68, complete phosphorous and nitrogen removal was observed (data not shown), the median particle size had increased to 1,028 μm, and MLSS was 1,250 mg/liter. Granule segregation within the Domestic SBR was assessed via stereomicroscopy and quantitative FISH on day 68 of reactor operation (Fig. 6e, f, g, and h). The Domestic white granules appeared rougher than their Synthetic counterparts, as assessed via stereomicroscopy, although they still appeared as dense spherical aggregates with bulbous outgrowths. FISH analysis showed white granules to be dominated by 72.9% “Candidatus Accumulibacter phosphatis,” with 0.2% “Candidatus Competibacter phosphatis,” among other bacteria. The “Candidatus Accumulibacter phosphatis” population abundance in the Domestic white granules was not as high as that of their synthetic-fed counterparts, with an increase in other bacteria present. The Domestic yellow granules appeared as rough, irregular aggregates with smaller microcolonies visible, as assessed via stereomicroscopy. FISH analysis exhibited a diverse bacterial population with 18.8% “Candidatus Accumulibacter phosphatis” and 20.4% “Candidatus Competibacter phosphatis,” among other bacteria.
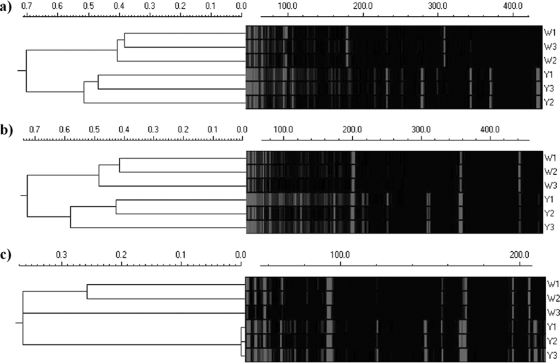
Due to the differences in wastewater feed compositions and treatment processes between the Domestic and Synthetic-1 reactors, additional analyses were carried out. Structural differences between white and yellow Domestic granules were analyzed via cryosectioning on day 68 of reactor operation (Fig. 3d and e). Domestic white-granule structure appeared as a solid, homogeneous layer of “Candidatus Accumulibacter phosphatis” growth similar to the initial Synthetic-1 white granules, although there was significantly more heterotrophic growth around the granule edge. The structure of the Domestic yellow granules appeared solid, although there were numerous channels and voids throughout the structure, and fewer distinct microcolony structures could be observed compared to the Synthetic-1 yellow granules previously described. A “snapshot” of the microbial community composition was also determined via tRFLP on the Domestic reactor granules on day 68. The 16S rRNA gene analysis was performed by tRFLP on separated white and yellow granules. Cluster analysis of the tRFLP profiles indicated there was a distinct difference between the microbial diversities of the white and yellow Domestic granules (Fig. 7). White granules exhibited a less diverse microbial community profile (fewer tRFLP peaks) than yellow granules, which exhibited a highly diverse community profile (many more tRFLP peaks), and this was supported by statistical analysis, as both the species richness number and the Shannon index were lower in white granules than in the yellow granules (data not shown).
FIG. 7.
16S rRNA gene tRFLP cluster analysis using the Domestic reactor samples. Three white (W) and three yellow (Y) PCR products were individually digested with 3 different restriction endonucleases, AluI (a), MspI (b), and Sau3AI (c). Cluster analysis of the resulting tRFLP electrograms showed samples from the same granule “type” grouped together. The Euclidean distance scale is shown for the cluster analysis, and the tRFLP fragment length (DNA base number) is indicated by a scale bar above each electrogram.
DISCUSSION
SBR operational parameters do not explain the occurrence of granule segregation.
The formation and segregation of two distinct types of microbial granules, white and yellow, were observed and extensively explored in our laboratory scale Synthetic-1 SBR. To our knowledge, this is the first time this has been detected in mixed-community granules treating wastewater, and it was surprising to find the same granule segregation in two additional granular reactors, Synthetic-2 (treating synthetic wastewater for EBPR) and Domestic (treating domestic wastewater for SNDPR). There was no evidence of an operational parameter employed during reactor operation for the selection of granules that resulted in the granule segregation. The SBR operational conditions for Synthetic-1 were primarily selective for floccular sludge, indicated by a long settling period, low shear force, and low organic loading (3, 11, 16, 26, 35). SBR operational changes were implemented only after granule formation had occurred and primarily served to enhance EBPR performance and remove excess floccular material from the system. Similarly, the Synthetic-2 SBR was not selectively operated for granular sludge, but similar granule formation and segregation occurred. Unlike both synthetic reactors, the Domestic reactor was operated primarily to select for granular sludge, specifically through a decreased settling time. Even though different wastewater feed, operational parameters, and biological nutrient removal process were applied to the Domestic reactor, similar signs of granule segregation were detected. This suggests that the granule segregation presented within this study was likely not a result of induced operational changes, as similar segregation was seen under both selective and nonselective pressures for granulation. Furthermore, granule segregation was observed in reactors treating two different wastewater types, indicating a possible generalized mechanism for granule segregation. However, there is a common theme of phosphorus removal and presence of PAO organisms among all the reactors presented. Consequently, it is possible that the segregation was related to the “Candidatus Accumulibacter phosphatis” communities present. This also implies that further microbial complexities, which are not yet fully understood, may be involved in the granulation process.
Detailed microscopy comparison of white and yellow granules.
The segregation of white and yellow granule types from the three reactors, treating two different types of wastewater and with different operational parameters, suggests the granule segregation was likely a systematic occurrence. Visually, the differences between white and yellow granules were quite distinct, as observed via stereo- and light microscopy. All of the white granules investigated were very solid and homogeneous in appearance, except within the Domestic reactor, where white granules did not form such smooth aggregates. All of the yellow granules investigated exhibited similar rough, irregular aggregates dominated by microcolony structures. FISH demonstrated large differences in the microbial community between white and yellow granules present within all reactors investigated. TEM analysis of the Synthetic-1 white granules further confirmed FISH analysis, as they were dominated by one bacterial morphotype, with EPS making up a large structural component. Yellow granules, in comparison, contained higher cell numbers and diversity of morphotypes growing in cluster-like arrangements with less EPS. These differences in cell densities and relative amounts of EPS within each granule type suggest that “Candidatus Accumulibacter phosphatis” may overproduce EPS, a factor that could explain some of the visual and structural differences observed between these two granule types.
As there were large differences in wastewater types, treatment parameters, and operational parameters between the Synthetic-1 and Domestic reactors, the structures of granules were compared. White granules were dominated by a solid layer of “Candidatus Accumulibacter phosphatis” within both the Synthetic-1 and Domestic reactors. However, there were slight structural differences between the yellow granules, with Synthetic-1 yellow granules exhibiting distinct microcolony structures, while these structures were not present within the Domestic yellow granules. These differences between the Synthetic-1 and Domestic granules were likely attributable to the large differences between feed and reactor operations. The Synthetic-1 reactor had a very defined feed and was operated solely for EBPR, resulting in a less complex microbial community, whereas the Domestic reactor had a highly variable, complex feed and was operated for SNDPR, resulting in a more diverse microbial community. Even with these configuration differences, the structures of the white and yellow granules from all reactors, as visualized by stereo- and light microscopy images, correlated with the structure observed from FISH cryosections and TEM. The fact that comparable granule segregation was observed in both the simplified synthetic EBPR and the rather complex Domestic SNDPR wastewater treatment systems further strengthens the likelihood of a general microbial mechanism responsible for the granule formation within laboratory scale phosphorus removal sludges.
“Candidatus Accumulibacter phosphatis” phylotype analysis and microbial diversity of white and yellow granules.
We hypothesize a microbe-associated mechanism as a reason for the granule segregation. Initially, the granule segregation was thought to be due to differences within “Candidatus Accumulibacter phosphatis” phylotypes, so that a “Candidatus Accumulibacter phosphatis” type within white granules outcompeted other bacteria or inhibited them from growing within its structure, resulting in highly enriched aggregates. However, analysis of the “Candidatus Accumulibacter phosphatis” ppkI gene phylotype associated with each granule type within the Synthetic-1 reactor found that both white and yellow granules contained type I ppkI “Candidatus Accumulibacter phosphatis” strains, indicating that granule segregation was likely not the result of “Candidatus Accumulibacter phosphatis”-associated type differences. Furthermore, due to the large differences between the Synthetic-1 and Domestic reactors, tRFLP was used to compare the microbial diversities associated with the two granule types within the Domestic reactor. This confirmed microbial community differences associated with each granule type and additionally indicated that yellow granules were more species rich/diverse than white granules, consistent with our previous findings.
Granule segregation appears to be an initial stage of the granule life cycle.
Continued observation of the Synthetic-1 granules by microscopy revealed that the distinction between white and yellow granules was noticeable only during the initial stages of granulation. Once a mature granule population was established, this distinction between white and yellow granules became less obvious, until finally a single homogeneous off-white granule population remained, which appeared to be a mixture of the initial white and yellow granules.
Our results offer new insight into granule formation in aerobic activated sludge; the granule segregation may be part of a granule life cycle, and we hypothesize two distinct mechanisms of granulation. (i) In microcolony outgrowth, the formation of white granules appears to be induced by the selection of a particular type of bacteria, in this case, “Candidatus Accumulibacter phosphatis,” which was initially enriched within the system as microcolonies or floc structures. As these bacteria grew and were selected for, outgrowths from the initial microcolony occurred, forming larger aggregates, until eventually a granular structure was achieved, comprised almost exclusively of one, likely clonal, bacterial type in a very dense, homogeneous layer (Fig. 8a). (ii) In microcolony aggregation, the formation of yellow granules appears to be an aggregation of numerous smaller microcolonies, or possibly floc structures, into a larger aggregate. These larger irregular aggregates, composed of highly mixed and physically distinct microbial populations, continue to grow, eventually forming a complete granular structure (Fig. 8b). These two distinct granule types are predominant during the initial stages of granulation. However, as the reactor operation continues, the mixing, growth, breakup, and subsequent reformation of the granules eventually results in a single homogeneous granule population containing remnants of both the initial granule types.
FIG. 8.
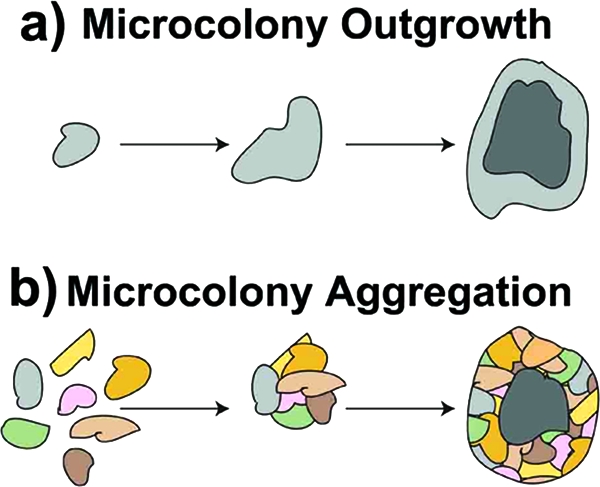
Two hypothesized mechanisms for granulation. (a) Microcolony outgrowth (white granules), where a small microcolony composed of one bacterial type is selected for and grows outward, eventually forming its own dense homogeneous granule. (b) Microcolony aggregation (yellow granules), where numerous small microcolonies composed of different bacterial types aggregate and grow as an entity, eventually forming a rough, highly segregated granule. (Courtesy of Frances Slater.)
Biofilm growth models for flocculation and granulation have been previously described (31, 36). These models support the hypothesis of microcolony outgrowth and microcolony aggregation occurring as two separate mechanisms for granulation and explain the structural, microbial, and microscopic differences between the two granule types present within the SBR. This is the first occasion we are aware of where evidence for both these models was detected simultaneously in mixed-culture reactor biofilms. While our microscopic and ecological analyses provide evidence and insight here, a more conclusive understanding would be obtained by functional studies to reveal specific proteins or genes involved in these hypothesized growth models, which would possibly elucidate critical steps in the granulation process.
Relevance of hypothesized granulation mechanisms to full-scale treatment systems.
The granule segregation detected here illustrates two hypothesized mechanisms for the formation of the initial granular aggregates in laboratory scale SBR. However, granule segregation and these formation mechanisms have yet to be examined in full-scale wastewater treatment systems. It is likely that the microcolony outgrowth mechanism is a result of the high laboratory “Candidatus Accumulibacter phosphatis” enrichments achieved; thus, in full-scale systems, this granule growth model may not be prevalent. Probably more plausible in full-scale systems is the occurrence of microcolony aggregation. While there are lower abundances of “Candidatus Accumulibacter phosphatis” or other crucial organisms present during microcolony aggregation, full-scale systems typically have between 5 and 20% “Candidatus Accumulibacter phosphatis” and are still capable of successful nutrient removal (14). Further research is required to fully understand the fundamental microbial aspects related to both potential granulation mechanisms and whether they occur at full scale. Specifically, there is an opportunity here to study EPS components and gene expression associated with both granule formation and life cycle, thus improving our understanding of the entire granulation process.
Supplementary Material
Acknowledgments
This work was funded by the Environmental Biotechnology Cooperative Research Centre (EBCRC), which was established and funded by the Australian Government, together with industry and university partners. J.J.B. acknowledges EBCRC for funding of a Ph.D. scholarship. P.L.B. acknowledges EBCRC, Waste Technologies of Australia, The University of Queensland, and the Queensland Government Smart State Fellowship Program for funding of a senior research fellowship. This project is supported by International Science Linkages established under the Australian Government's innovation statement, Backing Australia's Ability, and was linked to the European Union INNOWATECH program.
We acknowledge Rick Webb from the Centre for Microscopy and Microanalysis (CMM) of The University of Queensland for help with TEM procedures and imaging; Marta Coma, Maite Pijuan, and Marieska Verawaty from the Advanced Water Management Centre (AWMC) for operation of and data from the Domestic reactor; and Frances Slater from the AWMC for production of Fig. 8.
Footnotes
Published ahead of print on 17 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R. I. 1995. Fluorescently labeled, ribosomoal-RNA-targeted oligonucleotide probes in the study of microbial ecology Mol. Ecol. 4:543-553. [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S ribosomal-RNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrojo, B., A. Mosquera-Corral, J. M. Garrido, and R. Mendez. 2004. Aerobic granulation with industrial wastewater in sequencing batch reactors. Water Res. 38:3389-3399. [DOI] [PubMed] [Google Scholar]
- 4.Beun, J. J., A. Hendriks, M. C. M. Van Loosdrecht, E. Morgenroth, P. A. Wilderer, and J. J. Heijnen. 1999. Aerobic granulation in a sequencing batch reactor. Water Res. 33:2283-2290. [DOI] [PubMed] [Google Scholar]
- 5.Crocetti, G. R., J. F. Banfield, J. Keller, P. L. Bond, and L. L. Blackall. 2002. Glycogen-accumulating organisms in laboratory-scale and full-scale wastewater treatment processes. Microbiology 148:3353-3364. [DOI] [PubMed] [Google Scholar]
- 6.Crocetti, G. R., P. Hugenholtz, P. L. Bond, A. Schuler, J. Keller, D. Jenkins, and L. L. Blackall. 2000. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantification. Appl. Environ. Microbiol. 66:1175-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 8.Daims, H., S. Lucker, and M. Wagner. 2006. Daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8:200-213. [DOI] [PubMed] [Google Scholar]
- 9.De Bruin, L. M. M., M. K. De Kreuk, H. F. R. Van Der Roest, and M. C. M. Van Loosdrecht. 2004. Aerobic granular sludge technology: an alternative to activated sludge? Water Sci. Technol. 49:1-7. [PubMed] [Google Scholar]
- 10.de Kreuk, M. K., N. Kishida, and M. C. M. van Loosdrecht. 2007. Aerobic granular sludge— state of the art. Water Sci. Technol. 55:75-81. [DOI] [PubMed] [Google Scholar]
- 11.Etterer, T., and P. A. Wilderer. 2001. Generation and properties of aerobic granular sludge. Water Sci. Technol. 43:19-26. [PubMed] [Google Scholar]
- 12.García Martín, H., N. Ivanova, V. Kunin, F. Warnecke, K. W. Barry, A. C. McHardy, C. Yeates, S. M. He, A. A. Salamov, E. Szeto, E. Dalin, N. H. Putnam, H. J. Shapiro, J. L. Pangilinan, I. Rigoutsos, N. C. Kyrpides, L. L. Blackall, K. D. McMahon, and P. Hugenholtz. 2006. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 24:1263-1269. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Gil, G., P. N. L. Lens, A. van Aelst, H. van As, A. I. Versprille, and G. Lettinga. 2001. Cluster structure of anaerobic aggregates of an expanded granular sludge bed reactor. Appl. Environ. Microbiol. 67:3683-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu, A. Z., A. M. Saunders, J. B. Neethling, H. D. Stensel, and L. L. Blackall. 2008. Functionally relevant microorganisms to enhanced biological phosphorus removal performance at full-scale wastewater treatment plants in United States. Water Environ. Res. 80:688-698. [DOI] [PubMed] [Google Scholar]
- 15.He, S., D. L. Gall, and K. D. McMahon. 2007. “Candidatus accumulibacter” population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes. Appl. Environ. Microbiol. 73:5865-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heijnen, J. J., and M. C. M. Van Loosdrecht. May 1998. Method for acquiring grain-shaped growth of a microorganism in a reactor. U.S. patent 6,566,119.
- 17.Jacques, M., and L. Graham. 1989. Improved preservation of bacterial capsule for electron-microscopy J. Electron Microsc. Tech. 11:167-169. [DOI] [PubMed] [Google Scholar]
- 18.Lemaire, R., R. I. Webb, and Z. Yuan. 2008. Micro-scale observations of the structure of aerobic microbial granules used for the treatment of nutrient-rich industrial wastewater. Int. Soc. Microb. Ecol. 1:1-14. [DOI] [PubMed] [Google Scholar]
- 19.Lin, Y. M., Y. Liu, and J. H. Tay. 2003. Development and characteristics of phosphorus-accumulating microbial granules in sequencing batch reactors. Appl. Microbiol. Biotechnol. 62:430-435. [DOI] [PubMed] [Google Scholar]
- 20.Liu, W. T., A. T. Nielsen, J. H. Wu, C. S. Tsai, Y. Matsuo, and S. Molin. 2001. In situ identification of polyphosphate- and polyhydroxyalkanoate-accumulating traits for microbial populations in a biological phosphorus removal process. Environ. Microbiol. 3:110-122. [DOI] [PubMed] [Google Scholar]
- 21.Liu, X. W., H. Q. Yu, B. J. Ni, and G. P. Sheng. 2009. Characterization, modeling and application of aerobic granular sludge for wastewater treatment. Adv. Biochem. Eng. Biotechnol. 113:275-303. [DOI] [PubMed] [Google Scholar]
- 22.Liu, Y., and J. H. Tay. 2004. State of the art of biogranulation technology for wastewater treatment. Biotechnol. Adv. 22:533-563. [DOI] [PubMed] [Google Scholar]
- 23.Lu, H. B., A. Oehmen, B. Virdis, J. Keller, and Z. G. Yuan. 2006. Obtaining highly enriched cultures of Candidatus accumulibacter phosphates through alternating carbon sources. Water Res. 40:3838-3848. [DOI] [PubMed] [Google Scholar]
- 24.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMahon, K. D., S. Yilmaz, S. M. He, D. L. Gall, D. Jenkins, and J. D. Keasling. 2007. Polyphosphate kinase genes from full-scale activated sludge plants. Appl. Microbiol. Biotechnol. 77:167-173. [DOI] [PubMed] [Google Scholar]
- 26.McSwain, B. S., R. L. Irvine, and P. A. Wilderer. 2004. The effect of intermittent feeding on aerobic granule structure. Water Sci. Technol. 49:19-25. [PubMed] [Google Scholar]
- 27.Meyer, R. L., A. M. Saunders, R. J. X. Zeng, J. Keller, and L. L. Blackall. 2003. Microscale structure and function of anaerobic-aerobic granules containing glycogen accumulating organisms. FEMS Microbiol. Ecol. 45:253-261. [DOI] [PubMed] [Google Scholar]
- 28.Mino, T., M. C. M. Van Loosdrecht, and J. J. Heijnen. 1998. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 32:3193-3207. [Google Scholar]
- 29.Morgenroth, E., T. Sherden, M. C. M. Van Loosdrecht, J. J. Heijnen, and P. A. Wilderer. 1997. Aerobic granular sludge in sequencing batch reactor. Water Res. 31:3191-3194. [Google Scholar]
- 30.Mueller, E., K. Kriebitzsch, P. A. Wilderer, and S. Wuertz. 2002. Community structure of micro- and macroflocs in pin-point sludge and the influence of sludge age and potassium addition on microfloc formation. Water Sci. Technol. 46:405-412. [PubMed] [Google Scholar]
- 31.Picioreanu, C., B. J. Xavier, and M. C. M. van Loosdrecht. 2004. Advances in mathematical modeling of biofilm structure. Biofilm 1:337-349. [DOI] [PubMed] [Google Scholar]
- 32.Sekiguchi, Y., H. Takahashi, Y. Kamagata, A. Ohashi, and H. Harada. 2001. In situ detection, isolation, and physiological properties of a thin filamentous microorganism abundant in methanogenic granular sludges: a novel isolate affiliated with a clone cluster, the green non-sulfur bacteria, subdivision I. Appl. Environ. Microbiol. 67:5740-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 34.Tay, J. H., Q. S. Liu, and Y. Liu. 2004. The effect of upflow air velocity on the structure of aerobic granules cultivated in a sequencing batch reactor. Water Sci. Technol. 49:35-40. [PubMed] [Google Scholar]
- 35.Tay, J. H., Q. S. Liu, and Y. Liu. 2001. The effects of shear force on the formation, structure and metabolism of aerobic granules. Appl. Microbiol. Biotechnol. 57:227-233. [DOI] [PubMed] [Google Scholar]
- 36.Thomas, D. N., S. J. Judd, and N. Fawcett. 1999. Flocculation modelling: a review. Water Res. 33:1579-1592. [Google Scholar]
- 37.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner, M., R. Erhart, W. Manz, R. Amann, H. Lemmer, D. Wedi, and K. H. Schleifer. 1994. Development of a ribosomal-RNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl. Environ. Microbiol. 60:792-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber, S. D., W. Ludwig, K. H. Schleifer, and J. Fried. 2007. Microbial composition and structure of aerobic granular sewage biofilms. Appl. Environ. Microbiol. 73:6233-6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng, R. J., R. Lemaire, Z. G. Yuan, and J. Keller. 2003. Simultaneous nitrification, denitrification and phosphorus removal in a lab-scale sequencing batch reactor. Biotechnol. Bioeng. 84:170-178. [DOI] [PubMed] [Google Scholar]
- 41.Zheng, Y. M., and H. Q. Yu. 2007. Determination of the pore-size distribution and porosity of aerobic granules using size-exclusion chromatography. Water Res. 41:39-46. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, Y., M. Pijuan, R. J. Zeng, and Z. G. Yuan. 2009. Involvement of the TCA cycle in the anaerobic metabolism of polyphosphate accumulating organisms (PAOs). Water Res. 43:1330-1340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.