Abstract
Both bacteria and thaumarchaea contribute to ammonia oxidation, the first step in nitrification. The abundance of putative ammonia oxidizers is estimated by quantification of the functional gene amoA, which encodes ammonia monooxygenase subunit A. In soil, thaumarchaeal amoA genes often outnumber the equivalent bacterial genes. Ecophysiological studies indicate that thaumarchaeal ammonia oxidizers may have a selective advantage at low ammonia concentrations, with potential adaptation to soils in which mineralization is the major source of ammonia. To test this hypothesis, thaumarchaeal and bacterial ammonia oxidizers were investigated during nitrification in microcosms containing an organic, acidic forest peat soil (pH 4.1) with a low ammonium concentration but high potential for ammonia release during mineralization. Net nitrification rates were high but were not influenced by addition of ammonium. Bacterial amoA genes could not be detected, presumably because of low abundance of bacterial ammonia oxidizers. Phylogenetic analysis of thaumarchaeal 16S rRNA gene sequences indicated that dominant populations belonged to group 1.1c, 1.3, and “deep peat” lineages, while known amo-containing lineages (groups 1.1a and 1.1b) comprised only a small proportion of the total community. Growth of thaumarchaeal ammonia oxidizers was indicated by increased abundance of amoA genes during nitrification but was unaffected by addition of ammonium. Similarly, denaturing gradient gel electrophoresis analysis of amoA gene transcripts demonstrated small temporal changes in thaumarchaeal ammonia oxidizer communities but no effect of ammonium amendment. Thaumarchaea therefore appeared to dominate ammonia oxidation in this soil and oxidized ammonia arising from mineralization of organic matter rather than added inorganic nitrogen.
Autotrophic nitrification, the sequential oxidation of ammonia to nitrite and nitrate, leads to significant losses of ammonium-based fertilizers applied to soil, through leaching and denitrification of nitrate, and contributes significantly to production of the greenhouse gas nitrous oxide (48). Ammonia oxidation usually limits rates of soil nitrification and involves initial conversion of ammonia by ammonia monooxygenase to hydroxylamine in ammonia-oxidizing bacteria (AOB) and potentially to an uncharacterized intermediate in ammonia-oxidizing archaea (AOA). Until recently, AOB belonging to the Betaproteobacteria were considered responsible for all autotrophic ammonia oxidation in soil (25, 47). This belief was challenged by the discovery of homologues of genes encoding subunits of the key functional enzyme ammonia monooxygenase, associated with the Thaumarchaeota lineage (5, 46) in soil (51) and marine (52) metagenome studies. Subsequent isolation of a thaumarchaeal chemolithoautotroph, Nitrosopumilus maritimus, with a cell yield and specific growth rate similar to bacterial ammonia oxidizers (24), confirmed the existence of thaumarchaeal ammonia oxidizers and raised questions regarding their role in nitrification in the environment.
Thaumarchaeal amoA genes are ubiquitous in soil, where several lineages are found, including groups 1.1a, 1.1b, 1.1c, and 1.3, the first two lineages being associated with ammonia monooxygenase subunit genes (42). Thaumarchaeal amoA genes frequently outnumber bacterial amoA genes, often by more than 2 orders of magnitude (16, 27, 31), suggesting that thaumarchaea play an important role in soil nitrification. Further evidence for soil ammonia oxidation by archaea comes from microcosm studies of soils with low ammonium concentrations. Offre et al. (35) demonstrated that the abundance of archaeal, but not bacterial amoA genes, increased during nitrification in a mineral soil, and the growing AOA populations were sensitive to acetylene inhibition. Tourna et al. (50) observed changes in (transcript-defined) AOA community structure associated with microcosms incubated at different temperatures. In contrast, Jia and Conrad (20) found that only AOB increased in abundance and assimilated inorganic carbon during nitrification in agricultural soil microcosms receiving regular amendments of ammonia.
The limited number of cultivated representatives, particularly of archaeal ammonia oxidizers, limits ecophysiological studies, but it appears that archaeal ammonia oxidizers, including N. maritimus (24, 29), Nitrosocaldus yellowstonii (9), and Nitrososphaera gargensis (15), may be adapted to growth at low ammonia concentration. Similar physiology in soil thaumarchaeal ammonia oxidizers might be beneficial where the ammonia concentration is low. For example, NH3 availability will be low in acid soils, due to ionization to ammonium (31), or where ammonia is generated continuously at low concentrations, e.g., through mineralization of organic nitrogen, rather than being supplied at high concentration in fertilizer or animal waste. There is also evidence for assimilation of organic carbon by archaea (17, 37) or growth without incorporating inorganic carbon (20), leading to suggestions that archaeal ammonia oxidizers may be mixotrophic or heterotrophic (32). These two factors might lead to dominance of thaumarchaeal, rather than bacterial, ammonia oxidizers in unfertilized organic soils where ammonia is derived mainly from mineralization of organic matter rather than input of inorganic nitrogen. To test this hypothesis, we investigated net nitrification and ammonia oxidizer communities in an acidic organic forest soil derived from peat from the Ljubljana marsh, Slovenia. Bacterial and archaeal ammonia oxidizer communities were investigated by determining the abundance and diversity of respective amoA genes during nitrification in soil microcosms.
MATERIALS AND METHODS
Soil microcosms.
Net nitrification rate was determined in a preliminary experiment (data not shown) using soil collected in September 2008 from Ljubljana marsh, Slovenia. The dynamics of ammonia oxidizer communities were studied in microcosms containing soil collected from the same site in February 2009. This acidic soil has a high organic carbon content (45%), high C:N ratio (16.5), and high water-holding capacity (WHC; 8 g H2O g of soil−1) (1). Soil was sampled from the upper 30-cm soil layer at three locations, approximately 30 cm apart, and equal quantities of soil from each location were combined, sieved (mesh size, 8 mm), and stored at 4°C prior to use in microcosms. The pH of soil suspensions in distilled water (1:2, soil:water) was measured in triplicate with a glass electrode (pH meter inoLab pH 720; Weilheim, Germany) and was 4.0 (standard error [SE], 0.02) and 4.1 (SE, 0.01) in September and February, respectively.
The effect of acetylene on nitrification was studied in triplicate microcosms consisting of 330-ml serum bottles containing 10 g wet soil, adjusted to 60% WHC, and closed with a butyl rubber stopper. Acetylene was added to the headspace, resulting in final concentrations of 0 Pa (control), 10 Pa (0.01%), or 100 Pa (0.1%). Microcosms were incubated at 28°C for 30 days and were opened every 3 days to maintain aerobic conditions, with addition of distilled water (if required) to replace that lost through evaporation (as determined by weight loss). The acetylene headspace concentration was reestablished after resealing the microcosms. Microcosms were destructively sampled at 0, 5, 10, 15, 20, and 30 days.
The effects of ammonium concentration on nitrification rate and ammonia oxidizer communities were determined in triplicate microcosms consisting of 60-ml flasks containing 10 g wet soil. Soil was amended with ammonium sulfate to a final concentration of 0 (control), 10, or 100 μg NH4+-N g of dry soil−1, and water content was maintained at 70% WHC, as described above. Flasks were incubated at 22°C and were destructively sampled after incubation for 0, 4, 10, and 20 days.
After destructive sampling, a portion of soil from each microcosm was processed immediately (within 30 min) for analysis of ammonium and combined nitrite and nitrate concentrations, and the remainder was stored at −20°C for molecular analysis. Ammonium and nitrite plus nitrate concentrations were determined colorimetrically by flow injection analysis (FLOW SYS [Alliance Instruments, Salzburg, Austria] and FIA Star 5010 analyzer [Tecator]) after extraction from 5 g soil in 40 ml of 1 M KCl. Nitrite rarely accumulates in soil, and preliminary studies showed the nitrite concentration to be negligible (in relation to nitrate concentration) or below the detection limit during nitrification in this soil. For convenience, therefore, concentrations are subsequently referred to in terms of nitrate only. Total nucleic acids were extracted from 0.5-g soil samples as described by Griffiths et al. (14) with some modifications (33), and nucleic acid extracts were subdivided into two aliquots for preparation of DNA or RNA templates. Genomic DNA used for construction of 16S rRNA and amoA gene clone libraries was extracted using the PowerSoil kit (MoBio, Carlsbad, CA), following the manufacturer's protocol and the method of Griffiths et al. (14), respectively.
Reverse transcription-PCR (RT-PCR) amplification and DGGE analysis of 16S rRNA and amoA genes.
DNA was removed by treating 7 μl of total nucleic acid extract with RQ1 DNase (Promega, Southampton, United Kingdom) before generating cDNA with SuperScript II reverse transcriptase and random hexamer primers (Invitrogen, Paisley, United Kingdom) as described previously (31). PCR amplification and denaturing gradient gel electrophoresis (DGGE) analysis were performed using primers, reagents, and concentrations described previously (31, 50) with primer sets targeting bacterial ammonia oxidizer or thaumarchaeal 16S rRNA genes and bacterial or archaeal amoA genes from both DNA and cDNA templates. Relative (within-lane) intensities of DGGE bands were quantified by densitometry analysis of normalized DGGE profiles using Phoretix 1D gel analysis software (Phoretix International, Newcastle-Upon-Tyne, United Kingdom) as previously described (30). The resulting frequency matrices were used to assess the influence of ammonium addition and incubation time on archaeal community structure. Canoco for Windows 4.0 (49) was used for principal component analysis (PCA) and canonical correspondence analysis (CCA), using 999 permutations to test for significance.
Quantitative PCR analysis of amoA genes and transcripts.
Quantification of bacterial amoA genes was based on the protocol described by Offre et al. (35). Bacterial amoA genes were amplified with primers amoA-1F and amoA-2R (41), and a dilution series (102 to107) of Nitrosospira multiformis genomic DNA was used as a standard. Cycling conditions were 95°C for 5 min, 36 cycles of 95°C for 10 s, 60°C for 30 s, and 81°C for 5 s (followed by collection of fluorescence data), a final extension of 60°C for 10 min, and melting curve analysis (72 to 95°C; holding temperature for 1 s every 0.2°C). Quantification of thaumarchaeal amoA genes was performed using primers crenamoA23F and crenamoA616R (50). Standard curves were generated from known amounts of a PCR product (approximately 1.9 kb in length) amplified from soil fosmid 54d9 (51) and containing amoA and amoB subunit genes. Cycling conditions were 95°C for 5 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, and 82°C for 5 s (followed by collection of fluorescence data), final extension of 72°C for 10 min, and melting curve analysis (72 to 95°C; holding temperature for 1 s every 0.2°C).
Reactions were performed in 25-μl volumes containing 0.2-mg ml−1 bovine serum albumin, 0.8 μM (bacterial amoA assay) or 1 μM (thaumarchaeal amoA assay) each primer, 12.5 μl of QuantiFast qPCR master mix (Qiagen, Crawley, United Kingdom), and 5 μl of nucleic acid template. Amplification was performed in a DNA Engine OPTICON 2 system (GRI Ltd., Braintree, United Kingdom). Melting curves of PCR products were checked at the end of each reaction, and PCR products were confirmed by standard 1% agarose gel electrophoresis. For thaumarchaeal amoA quantitative PCR (qPCR), amplification efficiency was 93 to 97%, and r2 values were approximately 0.99.
Cloning and sequence analysis of 16S rRNA and amoA gene sequences.
Three independent 16S rRNA clone libraries (A, B, and C) were made from three different sampling cores. amoA gene clone libraries were constructed from DNA extracted from soil microcosms taken after incubation for 0 and 20 days. Archaeal 16S rRNA genes were amplified using general archaeal primers Ar109f and Ar915r (11) and thaumarchaeal amoA genes were amplified using primers crenamoA23F and crenamoA616R (50), and PCR products were cloned into the pGEM-T Easy vector (Promega, Southampton, United Kingdom). Selected clones from both 16S rRNA and amoA clone libraries were sequenced using the T7 vector primer. Sequences of chimeric origin were detected by analyzing alignments using the Chimera Check tool in RDPII for 16S rRNA gene sequences (8) and Bellerophon for amoA gene sequences (19). Sequences from short or failed reads were excluded from analysis.
Two databases, one for 16S rRNA gene sequences from all three sample cores and one for amoA predicted amino acid sequences, were built using ARB software (28) and aligned with reference sequences retrieved from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html). The 16S rRNA gene sequence alignment was performed with the SINA aligner tool from the SILVA Project web page (39) and manually corrected with respect to secondary structure. A 50% similarity filter was created for each data set, based on the alignment, leaving 595 nucleotides and 197 amino acids for 16S rRNA and amoA sequence alignments, respectively. Distance, parsimony, and maximum likelihood phylogenetic analyses were performed using neighbor-joining, parsimony, and PHYML tools implemented in ARB, using 1,000, 1,000, and 100 bootstrap values, respectively. The most conservative values for major nodes are represented on the phylogenetic trees.
Distance matrices were exported to calculate rarefaction curves and diversity indices with the DOTUR software (43). Sequences were grouped into operational taxonomic units (OTU) using the furthest-neighbor approach, with an OTU defined as containing sequences that are no more than 1% or 3% different from each other. Diversity and richness were estimated from 16S rRNA gene clone libraries using the Shannon-Weaver diversity index (45) and the Chao1 nonparametric richness estimator (6). Coverage (C) was calculated as C = 1 − (n1/N), where n1 was the number of clones which occurred only once in a library of N clones (13), and relative abundances of major phylogenetic groups were determined.
Statistical analysis.
Nitrification rates and qPCR data from triplicate microcosms were analyzed using a general linear model of analysis of variance using Minitab 15 (Minitab, State College, PA).
Nucleotide sequence accession numbers.
All 16S rRNA gene and amoA sequences have been deposited in the GenBank database with accession numbers HQ233247 to HQ233489.
RESULTS
Nitrification dynamics.
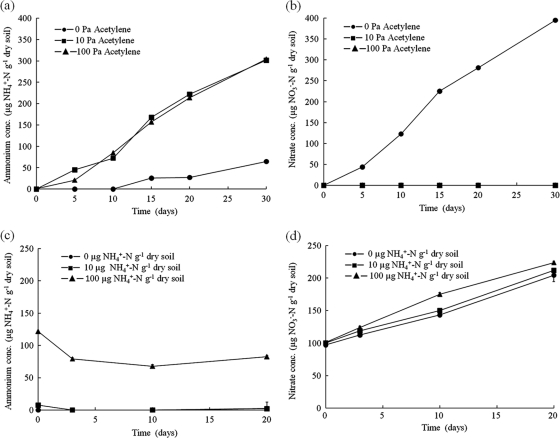
Net nitrification rate was determined as changes in nitrate concentration, assuming linear kinetics, during incubation of microcosms in the presence and absence of acetylene, an inhibitor of autotrophic nitrification. The conditions employed minimize the denitrification rate, and net and gross nitrification rates, measured in terms of nitrate production, are likely to be similar. In the absence of acetylene (Fig. 1a) the nitrification rate was 13.8 (SE, 0.8) μg NO3−-N g of dry soil−1 day−1, and nitrate concentration increased from below the detection limit to 395 (SE, 2) μg NO3−-N g of dry soil−1 after incubation for 30 days. There was no evidence of inhibition of nitrification through, for example, a possible decrease in soil pH. The ammonium concentration was low in comparison, increasing from below the detection limit for the first 10 days of incubation to 65 (SE, 7) μg NH4+-N g of dry soil−1 after incubation for 30 days (Fig. 1a). Nitrification was inhibited completely in microcosms with 10 and 100 Pa C2H2 headspace concentration (Fig. 1b), and ammonium concentration increased continuously during incubation through mineralization of organic nitrogen, at rates of 10.8 (SE, 0.6) and 10.6 (SE, 0.8) μg g of dry soil−1 day−1, assuming linear kinetics. The ammonium concentration increased to 303 (SE, 2.7) μg NH4+-N g dry soil−1 after incubation for 30 days, while the nitrate concentration was below the detection limit throughout the experiment. Mineralization of organic nitrogen therefore generated significant amounts of ammonia, supporting the high rates of nitrification observed in control microcosms.
FIG. 1.
The influence of acetylene and ammonium amendment on nitrification in peat soil microcosms. (a and b) Changes in ammonium (a) and nitrate (b) concentrations in soil microcosms incubated at 28°C for 30 days containing 0, 10, or 100 Pa acetylene headspace concentration. The y axes for these two graphs show the ammonium concentration (NH4+-N g dry soil−1) and nitrate concentration (NO3−-N g dry soil−1 day−1), respectively. (c and d) Changes in ammonium (c) and nitrate (d) in soil microcosms amended with 0, 10, or 100 μg NH4+-N g dry soil−1 and incubated at 22°C for 20 days. The y axes for these two graphs show the ammonium concentration (NH4+-N g dry soil−1) and nitrate concentration (NO3−-N g dry soil−1 day−1), respectively. For both experiments, triplicate microcosms were destructively sampled at each time point and mean values are plotted. Error bars represent standard errors but were usually smaller than the plotted symbols. Symbols overlap in panels b and c where values are close to zero.
Net nitrification kinetics were not affected by addition of ammonium, with rates of 6.18 (SE, 0.34), 5.73 (0.19), and 5.55 (0.37) μg NO3−-N g dry soil−1 day−1 in microcosms amended with 100, 10, and 0 μg NH4+-N g dry soil−1, respectively (Fig. 1d). These rates were lower than those reported for the acetylene experiment, due to the lower incubation temperature of 22°C, compared to 28°C. In addition, the final nitrate concentration was not affected significantly by ammonium amendment with 10 μg NH4+-N g dry soil−1 (P = 0.688) or 100 μg NH4+-N g dry soil−1 (P = 0.278). Ammonium concentration was always below the detection limit in control microcosms until day 20, when the mineralization rate exceeded the nitrification rate. Ammonium became undetectable after amendment with 10 μg NH4+-N g dry soil−1 but increased after incubation for 20 days. After addition of 100 μg NH4+-N g dry soil−1, ammonium concentration decreased to 68.7 (SE, 1.95) μg NH4+-N g dry soil−1 and increased to 86.4 (SE, 2.6) μg NH4+-N g dry soil−1 at day 20 (Fig. 1c). Soil pH did not change significantly during incubation, despite high rates of nitrification.
Community structure of archaea and ammonia oxidizers.
Total (dormant and active) thaumarchaeal and ammonia oxidizer communities were characterized by DGGE analysis of 16S rRNA and amoA genes amplified from DNA extracted from soil incubated for 0, 4, 10, and 20 days at 22°C, following addition of 0, 10, and 100 μg NH4+-N g dry soil−1. The composition of active communities was determined by DGGE analysis of 16S rRNA and amoA gene transcripts following RT-PCR amplification of extracted RNA.
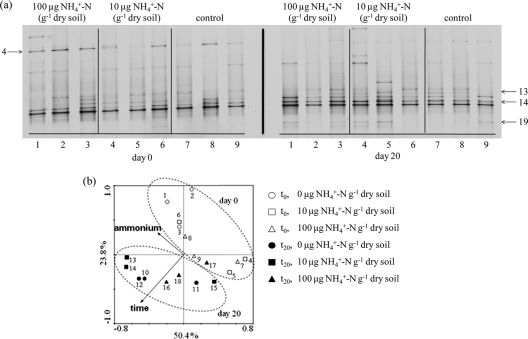
16S rRNA and amoA genes of ammonia-oxidizing bacteria were not detected, despite using a range of PCR primers and conditions (data not shown) that have been used to successfully amplify these genes from other soils. In contrast, thaumarchaeal 16S rRNA and amoA genes and gene transcripts were amplified from soil samples taken from all microcosms at all sampling points. DGGE profiles of archaeal amoA genes were not affected by amendment with ammonium or length of incubation, indicating little differential growth or shift within the community. In contrast, small but reproducible changes were observed in DGGE profiles of archaeal amoA gene transcripts after incubation for 20 days (Fig. 2a), but not for 4 or 10 days (data not shown). These changes were not influenced by amendment with ammonium. Principal component analysis (Fig. 2b) and canonical correspondence analysis of archaeal amoA transcript DGGE bands confirmed the effect of incubation time on community structure (P = 0.002) and the lack of effect of amendment with ammonium (P > 0.05).
FIG. 2.
DGGE analysis of archaeal amoA gene transcripts from soil incubated with 0, 10, or 100 μg NH4+-N g dry soil−1. (a) DGGE profiles of mRNA transcripts. Each lane represents a profile derived from an individual microcosm. The numbers of bands highlighted by arrows were derived using Phoretix 1-D (Phoretix International, Newcastle-Upon-Tyne, United Kingdom), with highlighted bands showing a decrease (band 4) or increase (bands 13, 14, and 19) in relative intensity during incubation. (b) PCA of the different archaeal amoA gene transcript communities based on the relative intensities of DGGE bands. Each symbol represents one community derived from an individual microcosm.
Abundance of amoA genes and gene transcripts.
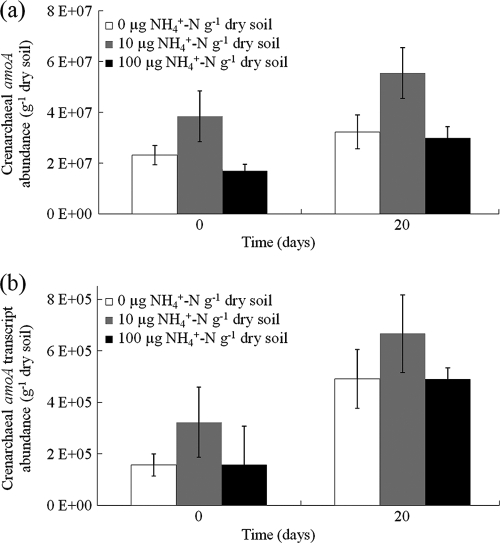
Abundance and transcriptional activity of ammonia oxidizers were assessed by quantification of amoA genes and gene transcripts, respectively, using qPCR assays with primers specific for bacterial and thaumarchaeal amoA genes. Neither bacterial amoA genes nor gene transcripts were detected in qPCR analysis of nucleic acids extracted from any of the microcosms. Thaumarchaeal amoA gene abundance increased significantly (1.5-fold; P = 0.022) during incubation for 20 days, from (2.62 [SE, 0.095] to 3.92 [SE, 0.45]) × 107 g dry soil−1 (Fig. 3a), but increases were not affected by ammonium amendment (P > 0.05). Archaeal amoA transcript abundance also increased significantly (2.5-fold; P < 0.005), from (2.12 [SE, 0.31] to 5.49 [SE, 0.12]) × 105 g dry soil−1 (Fig. 3b). Again, no effect of ammonium amendment was detected (P > 0.05), and transcript abundance was approximately 2 orders of magnitude lower than gene abundance.
FIG. 3.
Archaeal amoA gene abundance (a) and transcript abundance (b) in soil microcosms amended with 0, 10, or 100 μg NH4+-N g dry soil−1 after incubation for 0 or 20 days. Values plotted are means and standard errors from triplicate microcosms. Values on the y axes are the gene or transcript abundance per g of dry soil.
Phylogenetic analysis.
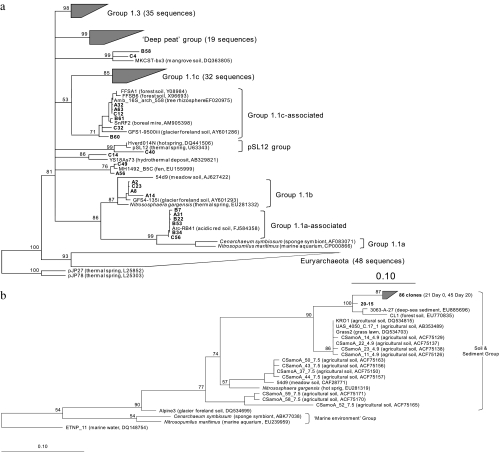
Of 156 sequences from the three archaeal 16S rRNA gene clone libraries, 108 fell within the Thaumarchaeota and 48 within the Euryarchaeota (Fig. 4a). Most thaumarchaeal sequences were placed within lineages that have not yet been implicated in ammonia oxidation, with only 8% placed within thaumarchaeal lineages 1.1b and 1.1a-associated. Two dominant clades (each containing 29% of thaumarchaeal sequences) belonging to Thaumarchaeota group 1.1c and 1.3 were observed, and 18% of thaumarchaeal sequences were associated with a recently identified “deep peat” lineage (40). Archaeal amoA clone libraries generated from microcosm samples taken after incubation for 0 and 20 days generated 89 clones (Fig. 4b). All sequences fell within the major lineage dominated by soil and sediment amoA sequences and are thought to be associated with group 1.1b thaumarchaea (38).
FIG. 4.
Maximum likelihood phylogenetic analysis of thaumarchaeal 16S rRNA genes from three clone libraries (A, B, and C) from bog soil (a) and translated amoA gene sequences from soil microcosms (b) sampled after incubation for 0 and 20 days, with reference sequences of environmental clones and cultivated organisms described as follows: clone name (environmental source, accession number). For panel a, names of lineages are given according to the methods described by Prosser and Nicol (38), except for the deep peat (DP) group (40). The majority of sequences fell within three clades, highlighted by gray blocks (group 1.1c, deep peat, and group 1.3) and are presented fully expanded in Fig. S1 of the supplemental material. Multifurcation indicates where the relative branching order of major lineages could not be determined in the majority of bootstrap replicates. For panel b, 86 of 87 sequences formed one specific clade within the soil (assumed group 1.1b) clade. In both trees, the length of each branch represents the maximum branch length obtained. Bars, an estimated 0.1 change per nucleotide (a) or amino acid (b) position.
OTU were formed at a ≤3% genetic distance for thaumarchaeal 16S rRNA genes, and rarefaction curves were inferred using the DOTUR software (43). Rarefaction curves from two (A and B) of the three clone libraries approached an asymptote (see Fig. S2 in the supplemental material). These differences are reflected in greater coverage values for clone libraries A and B, compared to C, and lower estimated richness (Table 1). Shannon diversity values were similar in the three libraries. Libraries were dominated by three groups, group 1.1c (33% [SE, 3]), group 1.3 (17% [SE, 3]), and the DP group (13% [SE, 3]) (Table 1).
TABLE 1.
Diversity estimators calculated for thaumarchaeal 16S rRNA clone libraries from three replicate soil samplesa
| Clone library | No. of clones | No. of phylotypes | Shannon indexb | Chao1 OTUc | % coverage | % in group 1.1c | % in group 1.3 | % in DP group |
|---|---|---|---|---|---|---|---|---|
| A | 35 | 10 | 1.99 (1.71-2.26) | 13 (10.4-33.0) | 86 | 30 | 10 | 10 |
| B | 34 | 10 | 2.09 (1.86-2.33) | 11.5 (10.2-25.1) | 91 | 30 | 20 | 20 |
| C | 39 | 15 | 2.148 (1.77-2.51) | 30 (18.5-79.2) | 74 | 40 | 20 | 10 |
| Mean (SE) | 36 (1) | 12 (1) | 2.07 (1.78-2.37) | 18.2 (13.0-45.8) | 84 (4) | 33 (3) | 17 (3) | 13 (3) |
Clone libraries from three soil samples(A, B, and C) were evaluated; summary means and standard errors for each estimator are also shown. Enumeration of thaumarchaeal phylotypes and calculation of diversity indices were completed at a 97% identity threshold between thaumarchaeal 16S rRNA gene sequences.
Shannon-Weaver diversity index; values in parentheses are 95% confidence intervals. The overall SE(not shown in the graph) was 0.045(95% confidence interval, 0.042-0.075).
Number of OTU in the original sample, obtained using the Chao1 estimator; values in parentheses are 95% confidence intervals. The overall SE(not shown in the graph) was 5.9(95% confidence interval, 2.75-16.9).
DISCUSSION
There is evidence that both archaea and bacteria contribute to ammonia oxidation in soil, but the factors determining their relative contributions are not clear (38). Martens-Habbena et al. (29) showed recently that the marine archaeon N. maritimus has the greatest substrate affinity determined for any autotrophic ammonia oxidizer, with nitrification kinetics matching those measured in the low-nutrient open ocean. Although bulk soil ammonia concentrations are generally higher than in oceans, localized concentrations may be low, and physical heterogeneity will restrict transport of ammonia through the soil. Availability of ammonia will also be significantly reduced in acid soils, due to ionization of ammonia to ammonium. Ammonium has a pKa of 9.24 at 25°C. The highest ammonium amendment in this study was designed to give a total ammonium concentration of 100 μg N g soil−1, equivalent to 7.2 mM. At pH 4, the ammonia concentration of such a solution will be 31 nM, which is of similar magnitude to the reported Km value for N. maritimus but 2 to 3 orders of magnitude lower than values for AOB (29). The two major sources of ammonia in soil are release during mineralization of organic matter and, in managed soils, input at high concentration of animal urine and manure or inorganic fertilizer. Mineralization is likely to lead to low, localized concentrations, particularly if ammonia oxidizers and heterotrophs assimilate ammonia from colonized degrading particulate material. Archaeal ammonia oxidizers might therefore be expected to be of greater importance in soils with high organic matter and little history of fertilization with inorganic nitrogen or acid soils.
This was tested by investigation of nitrification and ammonia oxidizer communities in an acidic peat soil with high organic matter content. Net nitrification rates were high, reaching 13.8 μg NO3−-N g dry soil−1 day−1, which is in the upper range of in situ gross nitrification rates analyzed in a metastudy of approximately 100 different soils (4). Nitrification was inhibited by acetylene at both concentrations used (10 and 100 Pa), as found in previous studies (2, 35, 53). The rate of ammonia production in acetylene-treated microcosms was slightly lower than the nitrification rate, suggesting possible inhibition of mineralization by either acetylene or ammonia that accumulated during mineralization or stimulation of immobilization.
Domination of ammonia oxidation by archaea.
Archaea appear to play the major role in ammonia oxidation in this soil. Despite use of a range of PCR primers and attempts to increase sensitivity, it was not possible to detect bacterial ammonia oxidizer 16S rRNA or amoA genes. This may have resulted from incomplete coverage of primers or specific problems associated with extraction of nucleic acids from organic soils. However, it is most likely that bacterial ammonia oxidizer abundance was low and that bacteria did not contribute significantly to nitrification in this soil. Bacterial ammonia oxidizer abundance decreases with soil pH (31, 34) and in acid soils can be below detection limits of both molecular-based (44) and cultivation-based (23) enumeration methods. In contrast, thaumarchaeal 16S rRNA and amoA genes and gene transcripts were readily amplified from all microcosms. amoA genes and gene transcripts increased significantly during nitrification, providing strong evidence for growth of a transcriptionally active, archaeal ammonia oxidizer community. amoA gene abundance increased to 3.92 × 107 g dry soil−1 after incubation for 20 days, which is similar to abundances found in other soils (27, 31), where amoA abundances of approximately 4 × 106 g soil−1 gave a net nitrification rate that was approximately 25% of the level determined in this study, suggesting that thaumarchaeal ammonia oxidizers could support the relatively high rates observed. amoA transcript abundance was approximately 100-fold lower than gene abundance, reaching 5.49 × 105 g dry soil−1. This difference in abundance has been observed in other studies of ammonia oxidizers (27, 31) and other functional groups (12) and may be due to inefficient conversion of mRNA to cDNA, a high rate of mRNA turnover, and/or a large proportion of amoA-containing archaea not actively oxidizing ammonia.
There was also some evidence of changes in archaeal ammonia oxidizer communities during nitrification, through DGGE analysis of archaeal amoA gene transcripts, but not genes, after incubation for 20 days. Transcriptional activity may represent the most sensitive measure of change in relative activities within a functional group, but the results suggest that nitrification did not lead to significant selection of archaeal ammonia oxidizer phylotypes. Although laboratory conditions will unavoidably differ from those in the natural environment, nitrification of ammonia released through mineralization represents a much smaller disturbance than addition of high concentrations of inorganic nitrogen, and it is therefore less likely to lead to changes in community structure.
Ammonium addition did not influence nitrification rate or ammonia oxidizers.
Addition of ammonium to soil microcosms as ammonium sulfate in solution at final concentrations of 10 and 100 μg NH4+-N g dry soil−1 had no detectable effect on nitrification rate and did not affect increases in ammonia oxidizer abundance or ammonia oxidizer community structure. This suggests that (i) ammonia oxidation was limited by factors other than ammonia concentration, (ii) ammonia did not reach active ammonia oxidizers, and/or (iii) active ammonia oxidizers were adapted to growth at ammonia concentrations lower than those in added solutions. Initial reductions in ammonium concentration following amendment are therefore likely due to immobilization by heterotrophs or adsorption of ammonium by clay minerals and organic matter, rather than ammonia oxidation, as nitrification rates and nitrate yields were not affected. Ammonium increased in all microcosms between 10 and 20 days, suggesting that the mineralization rate exceeded the nitrification rate. The inability of ammonia oxidizers to completely oxidize ammonia may have been due to a local decrease in soil pH, although bulk soil pH in microcosms did not decrease during incubation. There was no evidence for a decrease in nitrification rate, and the increase in ammonium was therefore likely due to an increased mineralization rate or decreased assimilation of ammonia into microbial biomass.
Although the influence of amendment of soil with ammonium on the relative activities of archaeal and bacterial ammonia oxidizers has been poorly studied, our results are consistent with several other studies. Höfferle et al. (18) found that a high ammonia concentration influences bacterial more than archaeal ammonia oxidizers, and Chen et al. (7) found little effect of fertilizer addition on archaea, which were influenced more by root exudates, suggesting mixotrophic or heterotrophic metabolism. Similarly, Di et al. (10) observed bacterial but not archaeal ammonia oxidizer growth in soil microcosms after addition of urine.
Community composition.
Phylogenetic analysis of 16S rRNA gene sequences indicated dominance of thaumarchaeal communities by groups 1.1c, 1.3, and deep peat lineages, which are not known to possess amo gene homologues. The high abundance of these groups was previously reported in other studies. For example, group 1.1c thaumarchaea are found predominantly in acidic soils (3, 21, 22, 26, 36), and thaumarchaeal 16S rRNA gene sequences dominating in a Finnish acidic peat soil fell within the lineages dominating in this study (i.e., groups 1.1c and 1.3 and the deep peat lineage) (40). Ammonia oxidation may therefore have been due to the less-abundant thaumarchaeal populations, such as group 1.1b, which are known to possess amo homologues, and links between 16S rRNA gene-derived community data and ammonia oxidation are less likely.
Phylogenetic analysis from soil microcosms placed all 89 amoA sequences within group 1.1b (31). Sequences from low-pH environments grouped together in a single clade (31), within which amoA sequences from this study formed a tight and separate cluster with low diversity. No amoA sequence grouping in the 1.1a clade could be retrieved from the amoA clone libraries, suggesting that 1.1b organisms are dominant and presumably more active than 1.1a-associated crenarchaea.
In conclusion, acetylene-sensitive ammonia oxidation in an acidic, organic, peat soil was dominated by thaumarchaea, with no evidence for activity or growth of bacterial ammonia oxidizers. Thaumarchaea utilized and grew on ammonia released during mineralization of organic matter, and nitrification rates and community changes were not influenced by addition of inorganic ammonium, suggesting that thaumarchaea preferentially utilized ammonia produced at low rates through mineralization. Further research is required to determine whether this feast-or-famine growth strategy results from preference for low ammonia concentration or other physiological characteristics. However, if widespread among AOA, there are implications for application of ammonia-based fertilizers to soils in which ammonia oxidizers are dominated by archaea.
Supplementary Material
Acknowledgments
This work was supported by the Slovenian Ministry of Higher Education (grant P4-0116; N.S.), award of an NERC Advanced Fellowship (NE/D010195/1; G.W.N.), and an NERC Research Grant (NE/F021909/1; C.G.-R.).
Footnotes
Published ahead of print on 1 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ausec, L., B. Kraigher, and I. Mandić-Mulec. 2009. Differences in the activity and bacterial community structure of drained grassland and forest peat soils. Soil Biol. Biochem. 41:1874-1881. [Google Scholar]
- 2.Berg, P., L. Klemedtsson, and T. Rosswall. 1982. Inhibitory effect of low partial pressures of acetylene on nitrification. Soil Biol. Biochem. 14:301-303. [Google Scholar]
- 3.Bomberg, M., and S. Timonen. 2007. Distribution of cren- and euryarchaeota in Scots pine mycorrhizospheres and boreal forest humus. Microb. Ecol. 54:406-416. [DOI] [PubMed] [Google Scholar]
- 4.Booth, M. S., J. M. Stark, and E. Rastetter. 2005. Controls on nitrogen cycling in terrestrial ecosystems: a synthetic analysis of literature data. Ecol. Monogr. 75:139-157. [Google Scholar]
- 5.Brochier-Armanet, C., B. Boussau, S. Gribaldo, and P. Forterre. 2008. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6:245-252. [DOI] [PubMed] [Google Scholar]
- 6.Chao, A. 1987. Estimating the population-size for capture recapture data with unequal catchability. Biometrics 43:783-791. [PubMed] [Google Scholar]
- 7.Chen, X. P., Y. G. Zhu, Y. Xia, J. P. Shen, and J. Z. He. 2008. Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ. Microbiol. 10:1978-1987. [DOI] [PubMed] [Google Scholar]
- 8.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Torre, J. R., C. B. Walker, A. E. Ingalls, M. Könneke, and D. A. Stahl. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10:810-818. [DOI] [PubMed] [Google Scholar]
- 10.Di, H. J., K. C. Cameron, J. P. Shen, C. S. Winefield, M. O'Callaghan, S. Bowatte, and J. Z. He. 2009. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat. Geosci. 2:621-624. [Google Scholar]
- 11.Egert, M., S. Marhan, B. Wagner, S. Scheu, and M. W. Friedrich. 2004. Molecular profiling of 16S rRNA genes reveals diet-related differences of microbial communities in soil, gut, and casts of Lumbricus terrestris L. (Oligochaeta: Lumbricidae). FEMS Microbiol. Ecol. 48:187-197. [DOI] [PubMed] [Google Scholar]
- 12.Freitag, T. E., and J. I. Prosser. 2009. Correlation of methane production and functional gene transcriptional activity in a peat soil. Appl. Environ. Microbiol. 75:6679-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Good, I. J. 1953. The population frequencies of species and the estimation of the population parameters. Biometrika 40:237-264. [Google Scholar]
- 14.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatzenpichler, R., E. V. Lebedeva, E. Spieck, K. Stoecker, A. Richter, H. Daims, and M. Wagner. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105:2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, J., J. Shen, L. Zhang, Y. Zhu, Y. Zheng, M. Xu, and H. J. Di. 2007. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ. Microbiol. 9:2364-2374. [DOI] [PubMed] [Google Scholar]
- 17.Herndl, G. J., T. Reinthaler, E. Teira, H. van Aken, C. Veth, A. Pernthaler, and J. Pernthaler. 2005. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71:2303-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Höfferle, Š., G. W. Nicol, P. Levin, J. Hacin, J. I. Prosser, and I. Mandić-Mulec. 2010. Ammonium supply rate influences archaeal and bacterial ammonia oxidisers in a wetland soil vertical profile. FEMS Microbiol. Ecol. 74:302-315. [DOI] [PubMed] [Google Scholar]
- 19.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 20.Jia, Z. J., and R. Conrad. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11:1658-1671. [DOI] [PubMed] [Google Scholar]
- 21.Jurgens, G., K. Lindstrom, and A. Saano. 1997. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl. Environ. Microbiol. 63:803-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemnitz, D., S. Kolb, and R. Conrad. 2007. High abundance of Crenarchaeota in a temperate acidic forest soil. FEMS Microbiol. Ecol. 60:442-448. [DOI] [PubMed] [Google Scholar]
- 23.Klemedtsson, L., Q. Jiang, A. K. Klemedtsson, and L. Bakken. 1999. Autotrophic ammonium-oxidising bacteria in Swedish mor humus. Soil Biol. Biochem. 31:839-847. [Google Scholar]
- 24.Könneke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 25.Koops, H. P., U. Purkhold, A. Pommerening-Röser, G. Timmermann, and M. Wagner. 2006. The lithoautotrophic ammonia-oxidizing bacteria, p. 778-811. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, vol. 5. Springer, New York, NY. [Google Scholar]
- 26.Lehtovirta, L. E., J. I. Prosser, and G. W. Nicol. 2009. Soil pH regulates the abundance and diversity of group 1.1c Crenarchaeota. FEMS Microbiol. Ecol. 70:367-376. [DOI] [PubMed] [Google Scholar]
- 27.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 28.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, K. Yadhu, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martens-Habbena, W., P. M. Berube, H. Urakawa, J. R. de la Torre, and D. A. Stahl. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976-979. [DOI] [PubMed] [Google Scholar]
- 30.McCaig, A. E., L. A. Glover, and J. I. Prosser. 2001. Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl. Environ. Microbiol. 67:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicol, G. W., S. Leininger, C. Schleper, and J. I. Prosser. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966-2978. [DOI] [PubMed] [Google Scholar]
- 32.Nicol, G. W., and C. Schleper. 2006. Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol. 14:207-212. [DOI] [PubMed] [Google Scholar]
- 33.Nicol, G. W., D. Tscherko, T. M. Embley, and J. I. Prosser. 2005. Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ. Microbiol. 7:337-347. [DOI] [PubMed] [Google Scholar]
- 34.Nugroho, R. A., W. F. M. Röling, A. M. Laverman, and H. A. Verhoef. 2007. Low nitrification rates in acid scots pine forest soils are due to pH-related factors. Microb. Ecol. 53:89-97. [DOI] [PubMed] [Google Scholar]
- 35.Offre, P., J. I. Prosser, and G. W. Nicol. 2009. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microb. Ecol. 70:99-108. [DOI] [PubMed] [Google Scholar]
- 36.Oline, D. K., S. K. Schmidt, and M. C. Grant. 2006. Biogeography and landscape-scale diversity of the dominant crenarchaeota of soil. Microb. Ecol. 52:480-490. [DOI] [PubMed] [Google Scholar]
- 37.Ouverney, C. C., and J. A. Fuhrman. 2000. Marine planktonic Archaea take up amino acids. Appl. Environ. Microbiol. 66:4829-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prosser, J. I., and G. W. Nicol. 2008. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ. Microbiol. 10:2931-2941. [DOI] [PubMed] [Google Scholar]
- 39.Pruesse, E., C. Quast, K. Knittel, B. M. Fuchs, W. Ludwig, J. Peplies, and F. O. Glockner. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Putkinen, A., H. Juottonen, S. Juutinen, E. S. Tuittila, H. Fritze, and K. Yrjälä. 2009. Archaeal rRNA diversity and methane production in deep boreal peat. FEMS Microbiol. Ecol. 70:87-98. [DOI] [PubMed] [Google Scholar]
- 41.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schleper, C., G. Jurgens, and M. Jonuscheit. 2005. Genomic studies of uncultivated archaea. Nat. Rev. Microbiol. 3:479-488. [DOI] [PubMed] [Google Scholar]
- 43.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt, C. S., K. A. Hultman, D. Robinson, K. Killham, and J. I. Prosser. 2007. PCR profiling of ammonia-oxidizer communities in acidic soils subjected to nitrogen and sulphur deposition. FEMS Microbiol. Ecol. 61:305-316. [DOI] [PubMed] [Google Scholar]
- 45.Shannon, C. E., and W. Weaver. 1963. The mathematical theory of communication. University of Illinois Press, Urbana, IL.
- 46.Spang, A., R. Hatzenpichler, C. Brochier-Armanet, T. Rattei, P. Tischler, E. Spieck, W. Streit, D. A. Stahl, M. Wagner, and C. Schleper. 2010. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 18:331-340. [DOI] [PubMed] [Google Scholar]
- 47.Stephen, J. R., A. E. McCaig, Z. Smith, J. I. Prosser, and T. M. Embley. 1996. Molecular diversity of soil and marine 16S rRNA gene sequences related to beta-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 62:4147-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevenson, F. J., and M. A. Cole. 1999. Cycles of soil: carbon, nitrogen, phosphorus, sulfur, micronutrients, 2nd ed. John Wiley and Sons, New York, NY.
- 49.ter Braak, C. J. F., and P. Smilauer. 2002. CANOCO reference manual and CanoDraw for Windows user's guide: software for canonical community coordination (version 4.5). Microcomputer Power, Ithaca, NY.
- 50.Tourna, M., T. E. Freitag, G. W. Nicol, and J. I. Prosser. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10:1357-1364. [DOI] [PubMed] [Google Scholar]
- 51.Treusch, A. H., S. Leininger, A. Kletzin, S. C. Schuster, H. P. Klenk, and C. Schleper. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985-1995. [DOI] [PubMed] [Google Scholar]
- 52.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Y. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 53.Walter, H. M., D. R. Keeney, and I. R. Fillery. 1979. Inhibition of nitrification by acetylene. Soil Sci. Soc. Am. J. 43:195-196. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.