Abstract
Mealybugs (Coccoidea: Pseudococcidae) are sap-sucking plant parasites that harbor bacterial endosymbionts within specialized organs. Previous studies have identified two subfamilies, Pseudococcinae and Phenacoccinae, within mealybugs and determined the primary endosymbionts (P-endosymbionts) of the Pseudococcinae to be Betaproteobacteria (“Candidatus Tremblaya princeps”) containing Gammaproteobacteria secondary symbionts. Here, the P-endosymbionts of phenacoccine mealybugs are characterized based on 16S rRNA from the bacteria of 20 species of phenacoccine mealybugs and four outgroup Puto species (Coccoidea: Putoidae) and aligned to more than 100 published 16S rRNA sequences from symbiotic and free-living bacteria. Phylogenetic analyses recovered three separate lineages of bacteria from the Phenacoccinae, and these are considered to be the P-endosymbionts of their respective mealybug hosts, with those from (i) the mealybug genus Rastrococcus belonging to the Bacteroidetes, (ii) the subterranean mealybugs, tribe Rhizoecini, also within Bacteroidetes, in a clade sister to cockroach endosymbionts (Blattabacterium), and (iii) the remaining Phenacoccinae within the Betaproteobacteria, forming a well-supported sister group to “Candidatus Tremblaya princeps.” Names are proposed for two strongly supported lineages: “Candidatus Brownia rhizoecola” for P-endosymbionts of Rhizoecini and “Candidatus Tremblaya phenacola” for P-endosymbionts of Phenacoccinae excluding Rastrococcus and Rhizoecini. Rates of nucleotide substitution among lineages of Tremblaya were inferred to be significantly faster than those of free-living Betaproteobacteria. Analyses also recovered a clade of Gammaproteobacteria, sister to the P-endosymbiont lineage of aphids (“Candidatus Buchnera aphidicola”), containing the endosymbionts of Putoidae, the secondary endosymbionts of pseudococcine mealybugs, and the endosymbionts of several other insect groups.
Many insect species possess specialized cells that contain obligate mutualist bacteria, primary endosymbionts (P-endosymbionts). P-endosymbionts usually show an evolutionary history of codiversification with an insect host and are thought to be essential to the survival of host species (4, 27). Insects with P-endosymbionts are phylogenetically and ecologically heterogeneous but commonly have a nutritionally unbalanced diet. Endosymbiotic bacteria can provide nutritional supplements to their host, permitting exploitation of otherwise inadequate food sources. For example, tsetse fly P-endosymbionts (Wigglesworthia glossinidia) synthesize B vitamins that are lacking in vertebrate blood (1, 3, 32, 33). Several additional examples of blood-feeding insect groups that depend upon P-endosymbionts are known, but the greatest diversity of P-endosymbionts is found in insects that feed on plant sap, a food source poor in amino acids. More than 80% of the approximately 63,000 described insect species with P-endosymbionts feed on plant sap and depend on P-endosymbionts to enrich their diet with amino acids (4, 34, 35, 45, 46).
Insect P-endosymbionts are vertically transmitted (4, 9, 43), and in general, the relationships among the bacteria are highly congruent with those among host taxa due to codiversification. Diverse clades of sap-sucking insects are associated with specific P-endosymbiont lineages. Major sap-sucking groups occur in the hemipteran suborders Auchenorrhycha (11) and Sternorrhyncha (18). Auchenorrhynchan bugs have codiversified with a Bacteroidetes P-endosymbiont lineage, Sulcia muelleri, with just a few exceptions that are interpreted as losses (28). In addition, a few auchenorrhynchan groups, mainly the sharpshooters, house a second P-endosymbiont, Baumannia cicadellinicola (Proteobacteria), in the same bacteriome as S. muelleri (24, 27, 28, 38). Sternorrhynchan insect groups follow the pattern of old and persistent associations with P-endosymbiont lineages but are more heterogeneous, with each of several family level or other higher taxa characterized by unique P-endosymbiont lineages: Aphididae hosts with Buchnera aphidicola P-endosymbionts (23), Diaspididae with Uzinura diaspidicola (16), Pseudococcidae with Tremblaya princeps (5, 13), Psylloidea with Carsonella ruddii (40), and Aleyrodoidea with Portiera aleyrodidarum (39).
Mealybugs (Pseudococcidae) are the second largest family of scale insects (Hemiptera: Coccoidea), with ca. 2,000 identified species in more than 270 genera (http://www.sel.barc.usda.gov/SCALENET/classif.htm). They have a worldwide distribution and feed on a broad range of hosts but are concentrated in the tropics and favor grasses, legumes, and composites (6, 12). Several mealybug species are serious pests, especially of grapes, figs, apples, citrus, dates, bananas, avocados, and mangos (25). Phylogenetic analyses have identified two primary subgroups within mealybugs, the subfamilies Pseudococcinae and Phenacoccinae (19). Munson et al. (30) and Thao et al. (41) reported monophyly for the mealybug P-endosymbionts, and Downie and Gullan (13) inferred mealybug-P-endosymbiont codiversification, but these studies included samples from only the Pseudococcinae (19). Buchner (8, 9) observed microscopic anatomical differences between the P-endosymbionts of mealybug genera that today are placed in either Pseudococcinae or Phenacoccinae and also considerable variation among phenacoccine species. Hardy et al. (19) hypothesized that a bacteriome containing Tremblaya P-endosymbionts is a synapomorphy (shared feature due to common ancestry) of the Pseudococcinae and that a bacteriome with Tremblaya P-endosymbionts containing Gammaproteobacteria S-endosymbionts (44) is a synapomorphy of the Pseudococcinae, excluding Maconellicoccus and the Ferrisia group. Here, we estimate relationships among 16S rRNA sequences from the bacterial endosymbionts of Phenacoccinae and other insects, as well as some free-living bacteria.
MATERIALS AND METHODS
Nomenclature.
Most symbiotic bacteria of insects cannot be named formally because the difficulty of culturing them outside their host means that characteristics required for description according to the Bacteriological Code 1990 Revision (22) are not known. Instead, they are given names in the category “Candidatus” (22, 27). For example, the P-endosymbiont of Pseudococcinae should be referred to as “Candidatus Tremblaya princeps.” However, most authors drop the “Candidatus” part of the name and italicize the genus and species name (e.g., Tremblaya princeps), which is a practice that we have followed in this paper.
Taxon sampling.
The data set included 140 sequences, of which 27 were bacterial sequences from 23 species of Phenacoccinae and Putoidae. Puto, the only genus in Putoidae, once was included in Pseudococcidae but now is recognized as a distantly related group (10, 17). One host insect (Puto albicans) is represented by three different bacterial sequences. The Phenacoccinae and Putoidae sequences resulted from direct sequencing and cloning of PCR products obtained using general bacterial primers. Voucher specimens of the insects from which the bacterial DNA was extracted are deposited in the Bohart Museum of Entomology, University of California, Davis, and voucher numbers for new data and GenBank (7) accession numbers for all sequences are given in Table S2 at http://bio.bd.psu.edu/gruwell/supplemental/.
The remaining 113 samples (see Table S2 at http://bio.bd.psu.edu/gruwell/supplemental/) were included to help identify bacteria associated with species of Phenacoccinae and Putoidae. These data include 16S rRNA sequences from all known groups of scale insect endosymbionts (15, 16, 41), a diversity of other insect endosymbionts, and various free-living bacteria.
DNA extraction and gene amplification.
All genomic DNA samples of Phenacoccinae and Putoidae were the same as those used by Hardy et al. (19). Sequences of 16S rRNA were amplified with two sets of general PCR primers, 8fBAC-1492BAC and 16SA1-16SB1, which cover almost the entire gene (ca. 1,200 bp). Two PCR protocols were used: a standard three-step PCR protocol with a 52°C annealing temperature, and the EFTD touchdown protocol of Moulton and Wiegmann (29). If initial amplifications did not produce bands that could be sequenced, reamplifications were done using PCR products as the DNA template and the internal primers S30Buch and a1271Diasp (15, 16). Internal primers were used for double coverage in sequencing: TremINTF, TremINTR (Tremblaya-like bacteria), RhizINTF, RhizINTR (bacteria from Rhizoecini), RastINTF, RastINTR (bacteria from Rastrococcus), and S688DIASP (designed from Uzinura diaspidicola, the primary symbiont of armored scale insects). All primer sequences are given in Table 1.
TABLE 1.
Primers designed for 16S rRNA gene sequences from bacteriaa
| Primer name | Description | Sequence |
|---|---|---|
| 8FBAC | Universal bacteria | AGAGTTTGATCCTGGCTCAG |
| 1492RBac | Universal bacteria | GGTTACCTTGTTACGACTT |
| S30 Buch16S | Universal bacteria | GGCGGCAAGCCTAACACATGCAAGTCG |
| A1446 Buch16S | Universal bacteria | CTCCCATGGTGTGACGGGCGGTGTG |
| s688DIASP | Universal bacteria | GGAATGTATGGTGTAGCGGTGAAATGC |
| TremintF | Tremblaya internal | CTACTGCCAGCAGCAGCCGCGG |
| TremintR | Tremblaya internal | CCGCGGCTGCTGCTGGCAGTAG |
| RHIZINTF | Brownia internal | GCAAACAGGATTAGATACCCTGG |
| RHIZINTR | Brownia internal | CCAGGGTATCTAATCCTGTTTGC |
| RastintF | Rastrococcus internal | GCCGTAAACGAATGATCACTAG |
| RastintR | Rastrococcus internal | CTAGTGATCATTCGTTTACGGC |
Universal primers were used for amplification, and internal primers were used for sequencing.
Each PCR was run with a total volume of 25 μl made up of 13 μl double-distilled water (ddH2O), 2 μl 50 mM MgCl2, 3 μl 10× buffer, 2.5 μl of 10 mM deoxynucleoside triphosphate (dNTP), 2.5 μl loading dye, 1 μl each primer (10 pmol/μl), 0.1 μl Taq polymerase, and 1 μl of genomic DNA. Each PCR was run with negative controls. Reactions were confirmed as successful using agarose gel electrophoresis and sent to the High Throughput Genomics Unit (Seattle, WA) for cleaning and DNA sequencing.
The samples PG48, NH12, NH59, PG96, and NH57 were cloned using a TOPO One Shot cloning kit (Invitrogen Corp.) and sequenced with M13 forward and reverse primers. The samples were chosen for cloning to obtain coverage across the Phenacoccinae and Putoidae. Only PG48 returned multiple distinct sequences during cloning.
Sequences and alignment.
Sequences were edited, assembled, and proofread in Sequencher 4.2 (GeneCodes Corp., 2003). Sequences were aligned using MAFFT (version 6), with default parameter values under the E-INS-i strategy that is optimized for sequences with multiple conserved domains and long gaps. The alignment was edited by eye, and ambiguous regions were excluded using MacClade (version 4.08; Sinauer Associates).
Phylogenetic analysis.
Phylogenies were estimated using maximum likelihood (ML), maximum parsimony, and Bayesian methods via the Cipres Portal (http://www.phylo.org/). Bayesian analyses were run in MrBayes 3.04b (21) using a general time-reversible (GTR) model with gamma-distributed among-site rate variation, as selected by MrModeltest (version 2; distributed by J. A. A. Nylander, Evolutionary Biology Centre, Uppsala University). Two runs of 10,000,000 generations were completed. Burn-in (250,000 generations) was determined by visual inspection of a graph of likelihood values. Trees sampled before burn-in were discarded using Mesquite (version 2.6; http://mesquiteproject.org). RAxML (37) ML searches consisted of 1,500 bootstrap pseudoreplicates under GTR with CAT approximation of gamma-distributed among-site rate variation. Every fifth bootstrap tree was used as a starting tree for ML inferences on the original alignment under GTR + γ. Parsimony runs were completed under Fitch optimization using heuristic searches implemented with a ratchet algorithm (31) in PAUP* (version 4.0b10; Sinauer Associates, Sunderland, MA) as a batch file in PAUPRAT (version 1; distributed by D. S. Sikes and P. O. Lewis, Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, CT). For each of 2,500 iterations, 20% of characters were reweighted. Trees were filtered for length and duplication.
Estimated branch lengths among samples of Tremblaya were conspicuously greater than those among closely related free-living lineages of Betaproteobacteria. To test if the rate of 16S rRNA evolution was significantly different between Tremblaya and its free-living sister group, we compared two molecular clock models: a null 1-rate model and an alternative 2-rate model in which Tremblaya and its sister group were each assigned a unique rate (as in reference 36). A rooted ML topology was inferred from an alignment of Betaproteobacteria 16S rRNA sequences with RAxML (detailed phylogenetic tree available; see Fig. S1 posted at http://bio.bd.psu.edu/gruwell/supplemental) (37). The RAxML analysis consisted of 500 bootstrap replicates under GTR + CAT, with every fifth bootstrap tree used as a starting tree for more thorough optimization under GTR + γ. We used the baseml module of PAML 4.2b (47) to estimate branch lengths under HKY + γ and to calculate the likelihoods of the null and alternative clock models. We evaluated the difference between the likelihoods of our clock models with a likelihood ratio test (14).
Assigning P-endosymbiont status.
It was unclear that the amplified 16S rRNA sequences were from bacteria occurring within the host bacteriome. Direct assessment of endosymbiotic properties of bacteria was not undertaken. Endosymbiont status was allocated to bacterial sequences on the basis of three criteria: (i) relationships among P-endosymbiont sequences should reflect those of their hosts, (ii) P-endosymbiont lineages should be related to other known P-endosymbiont lineages, and (iii) it should be possible to amplify P-endosymbiont sequences consistently from closely related specimens. These premises assume that (i) P-endosymbionts are vertically transmitted, (ii) the likelihood of a host's successful acquisition of an endosymbiont increases if the candidate bacterium is already an endosymbiont, and (iii) P-endosymbionts are always present in the host bacteriome.
RESULTS
P-symbiont identities.
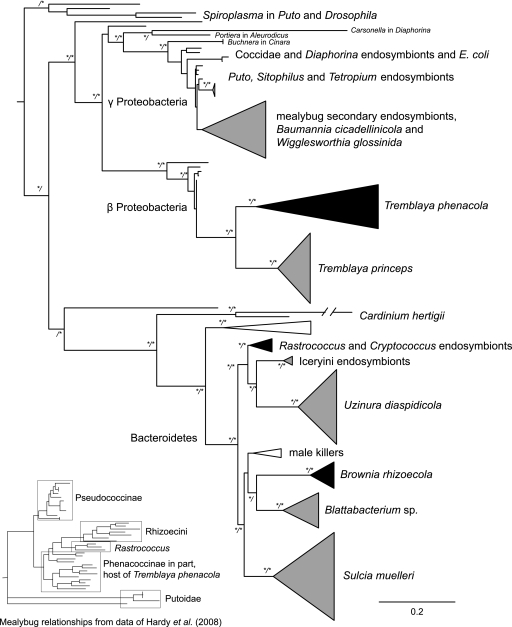
Three groups of bacteria were recovered from phenacoccine mealybugs (Fig. 1), as detailed in the next three paragraphs.
FIG. 1.
Salient features of the maximum likelihood estimate of relationships among 16S rRNA sequences from free-living and endosymbiotic bacteria. Nodes having posterior probabilities (PP) ≥95% and/or bootstrap support (BS) ≥80% are denoted with asterisks (PP/BS). Free-living bacteria lineages are not named. Endosymbiont clades of the Phenacoccinae are collapsed and in black; the bacterial groups Tremblaya phenacola and Brownia rhizoecola are designated in this paper. Endosymbiont lineages of other insect groups are in gray.
Bacterial sequences from the Phenacoccinae, excluding Rastrococcus and the Rhizoecini, belonged within the Betaproteobacteria, as sister to Tremblaya princeps, the pseudococcine endosymbiont lineage (bootstrap support [BS] = 100; posterior probabilities [PP] = 100). Relationships inferred among these bacterial sequences reflected those inferred among their hosts (19). We propose the designation “Candidatus Tremblaya phenacola” for the P-endosymbiont of mealybug species in the Phenacoccinae, excluding Rastrococcus and Rhizoecini. This lineage has unique sequences at the following 16S rRNA sites that distinguish it from Tremblaya princeps (homologous to the Escherichia coli positions): positions 466 to 469 are AATT (ACCA in T. princeps), 817 is G (C in T. princeps), 831 is A (G in T. princeps), 848 and 849 are TT (AA in T. princeps), and 1257 to 1261 are AGTT (GGTC in T. princeps).
Bacterial sequences from the mealybug tribe Rhizoecini were recovered as a clade within Bacteroidetes. A sister relationship with cockroach endosymbionts (Blattabacterium) was weakly supported. We propose the designation “Candidatus Brownia rhizoecola” for the P-endosymbiont of mealybug species in the tribe Rhizoecini. The genus name is in honor of Spencer W. Brown, who was a pioneer of scale insect cytogenetics, and the species name is in reference to the host taxon. This bacterial lineage has sequences that differ from its sister taxon Blattabacterium at the following loci, which correlate to the E. coli positions of 16S rRNA: positions 729 to 736 are GGCAAGCT (AGCAGGT in Blattabacterium), 1037 to 1048 are AACGCAAG (AGTACAAG in Blattabacterium), 1117 to 1121 are GTCG (ATTG in Blattabacterium), and 1138 to 1140 are GCT (GTC in Blattabacterium).
Bacterial sequences from the genus Rastrococcus were recovered within Bacteroidetes, in a clade containing a bacterial sequence from Cryptococcus ulmi (Eriococcidae) (15). Strong support (BS = 76; PP = 100) was discovered for a sister relationship between this clade and a group of symbiont sequences from two other scale insect families (Monophlebidae and Diaspididae). In this case, relationships among the bacteria do not mirror those among hosts. Therefore, we do not propose a name for this putative P-endosymbiont of Rastrococcus.
Most sequences from species of Putoidae were recovered from within the Gammaproteobacteria, in a weakly supported group comprised of symbiont sequences from Tetropium castaneum (Cerambycidae) (S. Gruenwald, unpublished data), Sitophilus oryzae (Dryophthoridae) (20), Glossina austeni (the symbiont Wigglesworthia glossinidia) (2), Paromenia isabellina (the symbiont Baumannia cicadellinicola) (38), and Pseudococcinae mealybugs (secondary symbionts) (41). Sequences from a psyllid secondary symbiont (Arsenophonus) (40, 42) and an uncharacterized soft scale (Coccidae) symbiont were sisters to this group with weak support. Monophyly was not identified for the putoid-associated bacterial sequences within this group. One sequence from Puto albicans was quite distantly related; it was recovered as sister to a Spiroplasma (Mollicutes) lineage from Drosophila with an unknown function.
Rate differences.
A significant difference (P < 0.0001) in the rate of 16S rRNA evolution between Tremblaya lineages and free-living Betaproteobacteria was inferred from comparison of the null 1-rate clock model (log likelihood score [lnL] = −10,007.11) to an alternative 2-rate model (lnL = −9,942.20) in which the rates of Tremblaya and its sister group were independent. The estimated local clock rate for Tremblaya was more than 7-fold faster than that of the included free-living Betaproteobacteria.
DISCUSSION
Sap-sucking insect lineages have had relationships with P-endosymbionts characterized by persistence and stability. Nearly all of Auchenorrhyncha share a P-endosymbiont in Sulcia muelleri, and three of four Sternorrhyncha superfamilies are each thought to have a single lineage of P-endosymbiont. In contrast, among scale insects (Coccoidea), mealybugs alone have three distantly related P-endosymbiont lineages.
Contrary to the hypothesis of Hardy et al. (19) that Tremblaya P-endosymbionts were a synapomorphy of the Pseudococcinae, many species in the Phenacoccinae also possess Tremblaya P-endosymbionts. It is the specific lineage of Tremblaya princeps that is synapomorphic of the Pseudococcinae, with Tremblaya phenacola occurring in all Phenacoccinae mealybugs other than Rastrococcus and the Rhizoecini. This suggests an endosymbiotic relationship between Tremblaya and mealybugs that evolved in conjunction with or predates the mealybug radiation.
The drastically elevated rates of 16S rRNA evolution observed among Tremblaya samples in comparison to rates among closely rated Betaproteobacteria lineages are consistent with the results of other empirical studies (e.g., references 26 and 42) and with theoretical predictions centered on small populations sizes and a lack of lateral gene transfer in endosymbiont lineages (reviewed in reference 45).
This work further establishes the mealybugs and in particular the Phenacoccinae as a system in which to study comparative endosymbiosis. Obligate relationships with P-endosymbionts have been more fundamental in the evolutionary history of mealybugs than historical biogeography or host associations (19). Therefore, mealybugs should be an ideal model for researchers wishing to isolate the history of endosymbiosis from other historical and ecological processes.
Acknowledgments
We thank the generous support from the scale insect community in obtaining specimens. We gratefully acknowledge the laboratory support provided by undergraduates Doug Brummell and Emily DiBlasi (SUNY at Buffalo). P.J.G. thanks Paul Baumann for initiating her interest in mealybug endosymbionts. M.E.G. thanks Benjamin B. Normark for inspiring interest in scale insects and endosymbionts and for continued support and direction during the development of this project. This paper benefited from the critical comments of two anonymous reviewers.
This research was supported by funds from the State University of New York at Buffalo, Pennsylvania State University at Erie, Behrend College, and Hatch funding from the California Agricultural Experiment Station.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shiba, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402-407. [DOI] [PubMed] [Google Scholar]
- 2.Aksoy, S. 1995. Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. Int. J. Syst. Bacteriol. 45:848-851. [DOI] [PubMed] [Google Scholar]
- 3.Aksoy, S. 2000. Tsetse—a haven for microorganisms. Parasitol. Today 16:114-118. [DOI] [PubMed] [Google Scholar]
- 4.Baumann, P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59:155-189. [DOI] [PubMed] [Google Scholar]
- 5.Baumann, L., and P. Baumann. 2005. Cospeciation between the primary endosymbionts of mealybugs and their hosts. Curr. Microbiol. 50:84-87. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Dov, Y. 1994. A systematic catalogue of the mealybugs of the world (Insecta: Homoptera: Coccoidea: Pseudococcidae and Putoidae) with data on geographical distribution, host plants, biology and economic importance. Intercept Ltd., Andover, Hants, United Kingdom.
- 7.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2004. GenBank: update. Nucleic Acids Res. 32:D23-D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchner, P. 1957. Endosymbiosestudien an schildläusen. 5. Die gattung Rastrococcus Ferris (Ceroputo Sulc). Z. Morphol. Ökol. Tiere. 46:111-148. [Google Scholar]
- 9.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Inc., New York, NY.
- 10.Cook, L. G., P. J. Gullan, and H. E. Trueman. 2002. A preliminary phylogeny of the scale insects (Hemiptera: Sternorrhyncha: Coccoidea) based on nuclear small subunit ribosomal DNA. Mol. Phylogenet. Evol. 25:43-52. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich, C. H. 2009. Auchenorrhyncha (cicadas, spittlebugs, leafhoppers, treehoppers, and planthoppers), p. 56-64. In V. H. Resh and R. T. Cardé (ed.), Encyclopedia of insects, 2nd ed. Elsevier, San Diego, CA.
- 12.Downie, D. A., and P. J. Gullan. 2004. Phylogenetic analysis of mealybugs (Hemiptera: Coccoidea: Pseudococcidae) based on DNA sequences from three nuclear genes, and a review of the higher classification. Syst. Entomol. 29:238-259. [Google Scholar]
- 13.Downie, D. A., and P. J. Gullan. 2005. Phylogenetic congruence of mealybugs and their primary endosymbionts. J. Evol. Biol. 18:315-324. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 15.Gruwell, M. E., C. D. von Dohlen, K. Patch, and B. B. Normark. 2005. Preliminary PCR survey of bacteria associated with scale insects (Hemiptera: Coccoidea), p. 101-115. In L. Erkiliç and M. B. Kaydan (ed.), Proceedings of the X International Symposium on Scale Insect Studies (2004). Adana Zirai Muscadele Arastirma Enstitusu, Adana, Turkey.
- 16.Gruwell, M. E., G. E. Morse, and B. B. Normark. 2007. Phylogenetic congruence of armored scale insects (Hemiptera: Diaspididae) and their primary endosymbionts from the phylum Bacteroidetes. Mol. Phylogen. Evol. 44:267-280. [DOI] [PubMed] [Google Scholar]
- 17.Gullan, P. J., and L. G. Cook. 2007. Phylogeny and higher classification of the scale insects (Hemiptera: Sternorrhyncha: Coccoidea). In Z. Q. Zhang and W. A. Shear (ed.), Linnaeus tercentenary: progress in invertebrate taxonomy. Zootaxa 1668:413-425. [Google Scholar]
- 18.Gullan, P. J., and J. H. Martin. 2009. Sternorrhyncha (jumping plant-lice, whiteflies, aphids, and scale insects), p. 957-967. In V. H. Resh and R. T. Cardé (ed.), Encyclopedia of insects, 2nd ed. Elsevier, San Diego, CA.
- 19.Hardy, N. B., P. J. Gullan, and C. J. Hodgson. 2008. A subfamily-level classification of mealybugs (Hemiptera: Pseudococcidae) based on integrated molecular and morphological data. Syst. Entomol. 33:51-71. [Google Scholar]
- 20.Heddi, A., C. Hubert, C. Khatchadourian, G. Bonnot, and P. Nardon. 1998. Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: a peculiar G + C content of an endocytobiotic DNA. J. Mol. Evol. 47:52-61. [DOI] [PubMed] [Google Scholar]
- 21.Huelsenbeck, J. P., and F. Ronquist. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 22.Lapage, S. P., P. H. A. Sneath, E. F. Lessel, V. B. D. Skerman, H. P. R. Seeliger, and W. A. Clark. 1990. International code of nomenclature of bacteria: bacteriological code 1990 revision. International Union of Microbiological Societies, ASM Press, Washington, DC. [PubMed]
- 23.Martinez-Torrez, D., C. Buades, A. Lattore, and A. Moya. 2001. Molecular systematics of aphids and their primary endosymbionts. Mol. Phylogenet. Evol. 20:437-449. [DOI] [PubMed] [Google Scholar]
- 24.McCutcheon, J. P., and N. A. Moran. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. U. S. A. 104:19392-19397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, D. R., G. L. Miller, and G. W. Watson. 2002. Invasive species of mealybugs (Hemiptera: Pseudococcidae) and their threat to U.S. agriculture. Proc. Entomol. Soc. Washington 104:825-836. [Google Scholar]
- 26.Moran, N. A. 1996. Accelerated evolution and Muller's ratchet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. U. S. A. 93:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran, N. A., J. P. McCutcheon, and A. Nakabachi. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165-190. [DOI] [PubMed] [Google Scholar]
- 28.Moran, N. A., P. Tran, and N. M. Gerardo. 2005. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterium phylum Bacteroidetes. Appl. Environ. Microbiol. 71:8802-8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moulton, J. K., and B. M. Weigmann. 2004. Evolution and phylogenetic utility of CAD (rudimentary) among Mesozoic-aged Eremoneuran Diptera (Insecta). Mol. Phylogenet. Evol. 31:363-378. [DOI] [PubMed] [Google Scholar]
- 30.Munson, M. A., P. Baumann, and N. A. Moran. 1992. Phylogenetic relationships of the endosymbionts of mealybugs (Homoptera: Pseudococcidae) based on 165 rRNA sequences. Mol. Phylogenet. Evol. 1:26-30. [DOI] [PubMed] [Google Scholar]
- 31.Nixon, K. C. 1999. The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics 15:407-414. [DOI] [PubMed] [Google Scholar]
- 32.Nogge, G. 1981. Significance of symbionts for the maintenance of an optimal nutritional state for successful reproduction in hematophagous arthropods. Parasitology 82:299-304. [Google Scholar]
- 33.Pais, R., C. Lohs, Y. Wu, J. Wang, and S. Aksoy. 2008. The obligate mutualist Wigglesworthia glossinida influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 74:5965-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramsey, J. S., S. J. MacDonald, G. Jander, A. Nakabach, G. H. Thomas, and A. E. Douglas. 2010. Genomic evidence for complementary purine metabolism in the pea aphid, Acyrthosiphon pisum, and its symbiotic bacterium Buchnera aphidicola. Insect Mol. Biol. 19(Suppl. 2):241-248. [DOI] [PubMed] [Google Scholar]
- 35.Sandstrom, J., A. Telang, and N. A. Moran. 2000. Nutritional enhancement of host plants by aphids—a comparison of three aphid species on grasses. J. Insect Physiol. 46:33-40. [DOI] [PubMed] [Google Scholar]
- 36.Schuettpelz, E., and K. M. Pryer. 2006. Reconciling extreme branch length differences: decoupling time and rate through the evolutionary history of filmy ferns. Syst. Biol. 55:485-502. [DOI] [PubMed] [Google Scholar]
- 37.Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688-2690. [DOI] [PubMed] [Google Scholar]
- 38.Takiya, D. M., P. L. Tran, C. H. Dietrich, and N. A. Moran. 2006. Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol. Ecol. 15:4175-4191. [DOI] [PubMed] [Google Scholar]
- 39.Thao, M. L., and P. Baumann. 2004. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl. Environ. Microbiol. 70:3401-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thao, M. L., M. A. Clark, D. H. Burckhardt, N. A. Moran, and P. Baumann. 2001. Phylogenetic analysis of vertically transmitted psyllid endosymbionts (Candidatus Carsonella ruddii) based on atpAGD and rpoC: comparisons with 16S-23S rDNA-derived phylogeny. Curr. Microbiol. 42:419-421. [DOI] [PubMed] [Google Scholar]
- 41.Thao, M. L., P. J. Gullan, and P. Baumann. 2002. Secondary (Gammaproteobacteria) endosymbionts infect the primary (Betaproteobacteria) endosymbiont of mealybugs multiple times and coevolve with their hosts. Appl. Environ. Microbiol. 68:3190-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thao, M. L., N. A. Moran, P. Abbot, E. B. Brennan, D. H. Burckhardt, and P. Baumann. 2000. Cospeciation of psyllids and their prokaryotic endosymbionts. Appl. Environ. Microbiol. 66:2898-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tremblay, E. 1989. Coccoidea endocytobiosis, p. 145-173. In W. Schwemmler and G. Gassner (ed.), Insect endocytobiosis: morphology, physiology, genetics, evolution. CRC Press Inc., Boca Raton, FL.
- 44.von Dohlen, C. D., S. Kohler, S. T. Alsop, and W. R. McManus. 2001. Mealybug β-proteobacterial endosymbionts contain γ-proteobacterial symbionts. Nature 412:433-436. [DOI] [PubMed] [Google Scholar]
- 45.Wernegreen, J. J. 2002. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3:850-861. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, A. C. C., P. D. Ashton, F. Calevro, H. Charles, S. Colella, G. Febvay, G. Jander, P. F. Kushlan, S. J. Macdonald, J. F. Schwartz, G. H. Thomas, and A. E. Douglas. 2010. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol. Biol. 19(Suppl. 2):249-258. [DOI] [PubMed] [Google Scholar]
- 47.Yang, Z. 2007. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586-1591. [DOI] [PubMed] [Google Scholar]