Abstract
The recent increased detection of acetic acid bacteria (AAB) of the genus Asaia as symbionts of mosquitoes, such as Anopheles spp. and Aedes spp., prompted us to investigate the diversity of these symbionts and their relationships in different mosquito species and populations. Following cultivation-dependent and -independent techniques, we investigated the microbiota associated with four mosquito species, Anopheles stephensi, Anopheles gambiae, Aedes aegypti, and Aedes albopictus, which are important vectors of human and/or animal pathogens. Denaturing gradient gel electrophoresis (DGGE) analysis based on the 16S rRNA gene revealed the presence of several bacterial taxa, among which Asaia sequences were among the dominant in most of the samples. A collection of 281 Asaia isolates in cell-free media was established from individuals belonging to the four species. The isolates were typed by internal transcribed spacer (ITS)-PCR, tRNA-PCR, BOX-PCR, and randomly amplified polymorphic DNA (RAPD)-PCR, revealing that different Asaia strains are present in different mosquito populations, and even in single individuals.
As defined by Anton de Bary (8), symbiosis is the living together of organisms belonging to different species. In the medical entomology area, investigations of the interactions between arthropods and microorganisms focused mainly on the negative interactions, i.e., on pathogenic microorganisms transmitted to vertebrate hosts or on microorganisms pathogenic to the arthropod itself. In recent decades, more interest has been directed toward the study of other forms of arthropod symbiosis, including mutualistic interactions and reproductive parasitism (21). Recently, from the study of mutualistic primary symbionts (e.g., Wigglesworthia in Glossina spp.) and manipulators of reproduction (e.g., Wolbachia), there has been a switch of research toward secondary mutualistic symbionts. Several studies have shown that these secondary symbionts play important roles, even if not vital, in the physiology of their insect hosts (23, 24).
An issue which is still almost unexplored regards the diversity of symbionts belonging to a given taxon within individual hosts. For example, by analyzing the symbionts of different insect hosts, Wolbachia has been shown to encompass a wide molecular diversity (27, 31), but limited information is available for the symbiont diversity in single individual hosts.
Acetic acid bacteria (AAB) are environmental bacteria that can be found in different niches, like the flower of the orchid tree Bauhinia purpurea (34) or are involved in processes like wine fermentation or spoilage (16). In the last decade, several AAB were discovered in association with arthropods, as was Gluconobacter morbifer associated with Drosophila melanogaster (23) or Acetobacter tropicalis associated with the olive fruit fly Bactrocera oleae (17). AAB seem to play a role in the stimulation of the immune system and the protection of the host against pathogens (24). AAB of the genus Asaia (34) have recently been found associated with different insect species (4), like the malaria mosquito vectors Anopheles stephensi (An. stephensi) (11) and Anopheles gambiae (An. gambiae) (7) or the virus vector Aedes aegypti (Ae. aegypti) (4). Asaia has been found as an extracellular bacterium in the gut, the salivary glands, and reproductive organs (11), from which it is transmitted vertically to the progeny (11) by egg smearing (4) and venereally from males to females, and then to the offspring (6). Asaia is also transmitted horizontally, through the cofeeding of mosquitoes on the same food source (11).
The diverse mode of transmission of Asaia spp. among mosquitoes indicates that these insects are potentially exposed to diverse strains, suggesting that differently from symbionts transmitted only (or predominantly) vertically, several strains could coexist in different populations and even within single insect individuals. This aspect of insect symbiosis has rarely been investigated and would be of interest for a better understanding of several biological facets of the symbiosis, like the adaptation of host populations to their specific environment or the efficiency of cross-breeding between populations. We should also consider that Asaia has been proposed as a potential vector of antiplasmodial factors, through an approach known as paratransgenesis (12). Uncovering the level of diversity of Asaia in mosquitoes would be useful also for this potential application, to select the dominant genotype(s) for developing paratransgenetic approaches (2).
Here we present a picture of the diversity of Asaia in mosquitoes, obtained after an investigation on the distribution, isolation, and molecular typing of these symbionts in different populations of four mosquitoes species (An. stephensi, An. gambiae, Ae. aegypti, and Aedes albopictus [Ae. albopictus]), which are implicated in the transmission of infectious diseases to humans and animals.
MATERIALS AND METHODS
Mosquito strains.
For the denaturing gradient gel electrophoresis (DGGE) analysis (see below), the study was carried out with 6 mosquito strains representing 4 species (Table 1). Two strains of An. gambiae, including the G3 strain maintained at the Department of Genetics, University of Stockholm, and a strain maintained at the Centre National de Recherche et de Formation sur le Paludisme, Burkina Faso, were used. The strain Liston of An. stephensi was obtained from a colony reared in the insectary of the Laboratory of Parasitology (University of Camerino, Italy) since 1988. One strain of Ae. aegypti was maintained at the Suisse Tropical Institute (Basel, Switzerland) and reared in the laboratory at the University of Camerino (Italy) since 2006. The other strain of Ae. aegypti came from the rearing facility at the Sezione di Parassitologia, DIPAV University of Milan, Italy. The strain of Ae. albopictus was a wild strain sampled in San Benedetto del Tronto (Italy) and reared in the insectary of the Laboratory of Parasitology (University of Camerino, Italy) since 2008. After DGGE, the following part of the study was carried out with strains listed above, except the An. gambiae G3 strain and the Milan Ae. aegypti strain.
TABLE 1.
Screening for Asaia in mosquitoes: summary of the results from culture-dependent and -independent investigations
| Mosquito strain/species | Asaia detected by DGGE | Asaia detected by specific PCR | No. of Asaia isolates |
|---|---|---|---|
| Anopheles gambiae strains | |||
| Burkina | Yes | Yes | 73 |
| G3 | Yes | Yes | No isolates |
| Anopheles stephensi | Yes | Yes | 72 |
| Aedes aegypti strains | |||
| Milan | Yes | Yes | No isolates |
| Camerino | Yes | Yes | 71 |
| Aedes albopictus | Yes | Yes | 65 |
| Total | 281 |
Asaia isolation from mosquitoes and DNA extraction.
Asaia strains were isolated from adult mosquitoes using an enrichment medium, named enrichment medium I, as indicated by Lisdiyanti et al. (19). Adults were sterilized in 1% sodium hypochlorite for 2 min, washed three times with 0.9% NaCl, and homogenized by grinding in 200 μl 0.9% NaCl. The homogenate was inoculated into the enrichment medium and allowed to grow at 30°C for 3 days, with shaking. When microbial growth occurred, the microorganisms were streaked on CaCO3 agar plates (pH 6.8) containing 1.0% (wt/vol) d-glucose, 1.0% (wt/vol) glycerol, 1.0% (vol/vol) ethanol, 1.0% (wt/vol) Bacto peptone, 0.5% (wt/vol) yeast extract, 0.7% (wt/vol) CaCO3, and 1.5% (wt/vol) agar. After 3 days growth, circular nonpigmented colonies capable of causing clearing of the CaCO3 were selected and streaked for further studies.
Chromosomal DNA was extracted from both Asaia strains and the mosquito microbiome using the cetyltrimethylammonium bromide (CTAB) method with a prior cell lysis by enzymatic methods and followed by an isopropanol precipitation of the DNA as described by Jara et al. (15).
PCR-DGGE and band sequence analysis.
16S rRNA gene amplification by PCR for DGGE analysis was carried out by using primers 357f (5′-CCTACGGGAGGCAGCAG) and 907r (5′-CCGTCAATTCCTTTRAGTTT) that target a portion of the 16S rRNA gene, including the hypervariable V3 regions, which have been shown to be useful for analyzing bacterial communities (3, 35). Primer 357f incorporated a GC clamp for the first amplification.
DGGE analysis was carried out on each PCR amplicon using a DCode universal mutation detection system (Bio-Rad, Milan, Italy), according to the procedure described previously (9). Electrophoresis was performed in a 0.5-mm polyacrylamide gel (7% [wt/vol] acrylamide-bisacrylamide, 37.5:1) containing a 35 to 60% urea-formamide denaturing gradient (100% corresponds to 7 M urea and 40% [vol/vol] formamide) according to the method of Muyzer et al. (22), increasing in the electrophoretic run direction. The gel was subjected to a constant voltage of 90 V for 15 h at 60°C in 1× Tris-acetate-EDTA (TAE) buffer (50× TAE stock solution consisting of 2 M Tris base, 1 M glacial acetic acid, 50 mM EDTA). After electrophoresis, the DGGE gels were stained in 1× TAE solution containing SYBR green (Molecular Probes, Leiden, Netherlands) for 45 min and photographed under a UV illumination using a GelDoc 2000 apparatus (Bio-Rad). For the sequencing of DGGE bands, bands of interest were excised from the gels with a sterile blade, mixed with 50 μl of sterile water, and incubated overnight at 4°C to allow the DNA of the bands to diffuse out of the polyacrylamide gel blocks. Two microliters of this aqueous solution was used to reamplify the PCR products with the same primers, excluding the GC clamp, according to Sanchez et al. (26).
Nucleotide identity searches were performed in the GenBank database using the basic local alignment search tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi) program to identify the bacterial strains most closely related to isolates and the best matching DNA sequences of the sequenced amplicons. A phylogenetic tree based on the analysis of the 16S rRNA sequences was inferred using the neighbor-joining method (25) with 2,000 replicates as a bootstrap test. The phylogenetic analyses were conducted by using Mega 4 (28).
PCR typing of Asaia isolates.
The 16S rRNA gene was amplified with universal bacterial 16S rRNA gene primers 27F (5′-TCGACATCGTTTACGGCGTG) and 1492R (5′-CTACGGCTACCTTGTTACGA). Internal transcribed spacer (ITS)-PCR was performed, with primer ITSF (5′-GCCAAGGCATCCAAC) and ITSR (5′-GTCGTAACAAGGTAGCCGTA), according to Daffonchio et al. (5). tRNA-PCR was performed, with tRNA primer T3A (5′-GGGGGTTCGAATTCCCGCCGGCCCCA) according to Borin et al. (1). BOX-PCR was performed, with BOX-A1 primer (5′-CTACGGCAAGGCGACGCTGAC), according to Urzì et al. (29). Randomly amplified polymorphic DNA (RAPD)-PCRs were performed, using RAPD OPA4 (5′-AATCGGGCTG), OPA7 (5′-GAAACGGGTC), and OPA10 (5′-GTGATCGCAG) primers according to Daffonchio et al. (5).
PCR fingerprintings were resolved by agarose gel electrophoresis under UV light. Band profiles were analyzed using the Quantity One software (version 4.6.5). MVSP software (version 3.13n) was used to estimate levels of similarity between strains based on the merged profiles obtained from ITS-PCR, tRNA-PCR, BOX-PCR, and RAPD-PCR and to generate unweighted pair group method with arithmetic average (UPGMA) trees.
Nucleotide accession number.
The 16S rRNA gene sequences were deposited under accession numbers FN814275 to FN814281, FN814283 to FN814297, FN814299 to FN814301, and FN814303.
RESULTS
PCR-DGGE from the whole insect body.
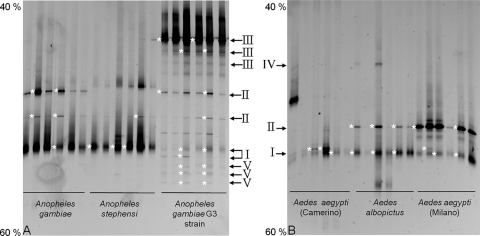
Bacterial PCR-DGGE fingerprints were obtained from the whole body of Anopheles and Aedes mosquitoes by the use of insects reared in laboratories but having different origins (Table 1). For each population, six individuals were tested independently. Whole-insect-body samples from the same origin had similar DGGE profiles (Fig. 1). All of the profiles from all of the individuals showed some common bands and some bands specific to certain populations. Other bands were found only in individuals having the same origin. Some other profiles were specific to a genus and absent from the other. Sequences of selected bands representative of most of the different migration classes in the gels showed that a major band in the middle of the gel and common to all of the profiles matched with Asaia spp. We emphasize that banding patterns were well conserved within mosquito species, even between individuals of different sex, with the exception of a few minor bands (Fig. 1). Considering only the Anopheles genus (Fig. 1A), in addition to Asaia spp., another band matching Burkholderia sp. was found in all the samples. A second band identified as Asaia sp. was present in An. gambiae. The individuals from the G3 strain of An. gambiae also presented bands that were identified after sequencing as Elizabethkingia sp. In addition, further bands were identified as deriving from Burkholderia sp. It is worth noticing that sequences identified as belonging to the genus Bacillus were also found in the G3 strain (lowest faint bands). For the profiles of individuals of the Aedes genus (Fig. 1B), the already mentioned band common to all of the specimens was identified as deriving from Asaia. In these mosquitoes, however, this band was intense for some individuals but faint for some others. The bands in the middle of the gel were identified as belonging to Burkholderia sp. This bacterium was mostly associated with the strain of Ae. aegypti from Milan and with Ae. albopictus. Another band was observed in the upper part of the gel, but it produced unreadable sequences, possibly due to the presence of comigrating amplicons.
FIG. 1.
DGGE profiles of 16S rRNA gene fragments amplified by PCR using general primers (357f and 907r) from whole-mosquito DNA templates. (A) DGGE profiles from mosquitoes of the genus Anopheles. Bands showing sequence identity with Asaia, Burkholderia, Elizabethkingia, and Bacillus spp. are indicated with I, II, III, and V, respectively. (B) DGGE profiles from mosquitoes of the genus Aedes. Bands with sequence identity with Asaia and Burkholderia spp. are indicated with I and II, respectively. IV indicates a band that was not identified, due to unreadable sequences. The numbers (40% and 60%) indicate the percentage of denaturating condition at the respective gel positions. Anopheles gambiae strain G3 indicates a laboratory strain reared in Stockholm, Sweden. Aedes aegypti (Camerino) and Aedes aegypti (Milano) indicate individuals reared in the laboratories in Camerino and Milan. Asterisks on the left of the bands indicate the bands that were cut and sequenced.
Asaia isolation and typing.
Two hundred eighty-one isolates of Asaia spp. were obtained from individuals belonging to all four mosquito species. Even if Asaia was detected in all of the strains by DGGE, it was not possible to get it in culture from the Ae. aegypti strain reared in Milan (Table 1). Along with Asaia, seven isolates identified as Gluconacetobacter liquefaciens were also obtained.
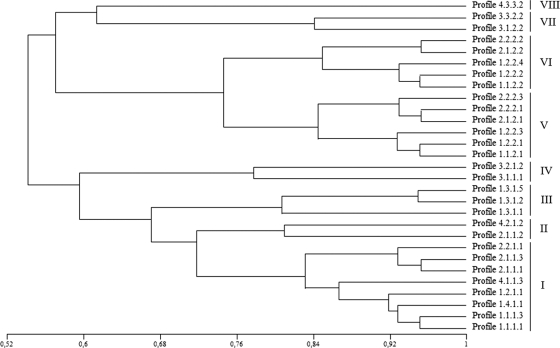
The diversity of the Asaia symbionts was assayed with different fingerprinting approaches: ITS-PCR, tRNA-PCR, BOX-PCR, and RAPD-PCR (5). The 281 Asaia isolates from the mosquitoes exhibited the same ITS-PCR and tRNA-PCR fingerprinting profiles, presenting single bands of about 850 and 1,000 bp, respectively (data not shown). BOX-PCR and RAPD-PCR were able to distinguish several profiles among the strains examined. BOX-PCR discriminated four band pattern types, with more than 10 bands in the range between 200 and 3,000 bp. The RAPD-PCR using the primers OPA4, OPA7, and OPA10 unraveled, respectively, four, three, and five genotypes which were characterized by complex band patterns between 400 and 4,000 bp. The merging of the results obtained from all the six PCRs allowed us to distinguish 29 genotype patterns. The dominant profile (profile 1.1.1.1), represented by 65 strains (Table 2), was present in all of the mosquito species except Ae. aegypti. Some rarer profiles (profiles 1.2.2.4, 2.2.1.1, and 2.1.1.3) were present in only one species, while other patterns (profiles 1.1.2.2, 2.1.1.1, and 4.1.1.3) were present in more than one species. The analysis of these profiles showed that they were very similar, sharing more than 50% similarity (Fig. 2). When a threshold of 75% similarity is adopted, only eight groups can be distinguished, the first group (group I) is composed of eight profiles and includes 156 strains representing 55.51% of the isolates. Different Asaia types were found to be simultaneously associated with single mosquito individuals, indicating that multiple infections can occur during the mosquito life span. A similar situation has been recorded for Wolbachia, for which individual insect specimens have been found to harbor more than one strain of this bacterium (27, 30).
TABLE 2.
Number of strains of Asaia for each profile pattern and their distribution among the mosquito populations
| Profile | No. of strains | Mosquito species harboring the Asaia profilea |
|---|---|---|
| 1.1.1.1 | 65 | Ae. albopictus (34), An. gambiae (22), An. stephensi (9) |
| 1.1.1.3 | 29 | An. stephensi (26), Ae. aegypti (3) |
| 1.1.2.1 | 10 | Ae. albopictus (10) |
| 1.1.2.2 | 51 | Ae. aegypti (48), An. gambiae (3) |
| 1.2.1.1 | 9 | An. gambiae (7), Ae. albopictus (2) |
| 1.2.2.1 | 8 | An. gambiae (6), Ae. albopictus (2) |
| 1.2.2.2 | 5 | Ae. albopictus (3), An. gambiae (1), Ae. aegypti (1) |
| 1.2.2.3 | 4 | An. stephensi (4) |
| 1.2.2.4 | 11 | Ae. aegypti (11) |
| 1.3.1.1 | 2 | Ae. albopictus (2) |
| 1.3.1.2 | 1 | Ae. albopictus (1) |
| 1.3.1.5 | 1 | Ae. aegypti (1) |
| 1.4.1.1 | 2 | Ae. albopictus (2) |
| 2.1.1.1 | 9 | An. gambiae (6), Ae. albopictus (3) |
| 2.1.1.2 | 4 | An. gambiae (3), Ae. albopictus (1) |
| 2.1.1.3 | 8 | An. stephensi (8) |
| 2.1.2.1 | 6 | An. gambiae (6) |
| 2.1.2.2 | 4 | An. gambiae (3), Ae. aegypti (1) |
| 2.2.1.1 | 13 | An. gambiae (13) |
| 2.2.2.1 | 3 | An. stephensi (3) |
| 2.2.2.2 | 1 | An. gambiae (1) |
| 2.2.2.3 | 1 | Ae. aegypti (1) |
| 3.1.1.1 | 3 | An. stephensi (3) |
| 3.1.2.2 | 1 | Ae. aegypti (1) |
| 3.2.1.2 | 1 | Ae. albopictus (1) |
| 3.3.2.2 | 3 | An. gambiae (3) |
| 4.1.1.3 | 21 | An. stephensi (19), Ae. albopictus (2) |
| 4.2.1.2 | 2 | An. gambiae (2) |
| 4.3.3.2 | 3 | Ae. albopictus (3) |
The numbers in parentheses indicate the number of strains obtained for each profile from the various mosquito species.
FIG. 2.
Similarity tree between the various profile patterns of Asaia spp. isolated from the mosquitoes. The tree, generated with the UPGMA methods using the Jaccard coefficient, allowed us to cluster the different profiles in a tree, presenting a similarity higher than 50% among all of the patterns. We can distinguish 8 groups (from I to VIII) that share a similarity higher than 75%.
Phylogenetic analysis.
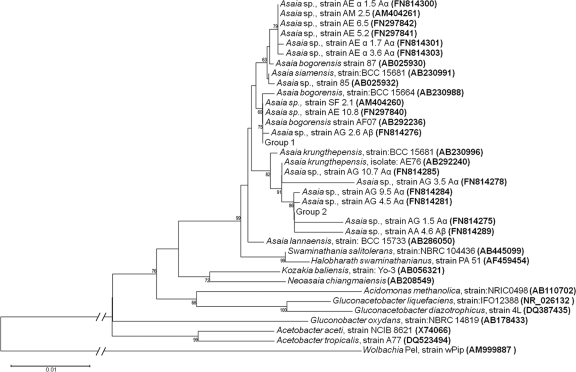
Almost the entire 16S rRNA gene was sequenced for 26 Asaia isolates. The sequences obtained were compared to the databases using BLAST (Table 3) and used for phylogenetic analysis. Phylogenetic analysis included AAB sequences retrieved from the databases, which in most cases derive from strains not associated with insects (hereafter indicated as environmental strains). The phylogenetic tree that was generated groups AAB strains of environmental origin with the strains isolated from mosquitoes. Indeed, mosquito strains clustered into two groups within the Asaia genus, one clustering with Asaia krungthepensis and one clustering with Asaia bogorensis, together with isolates from environmental niches (Fig. 3), and this was not dependent on the origin of the strain. It is interesting to note that the clustering of Asaia strains from mosquitoes in the 16S rRNA-based phylogenetic tree is not congruent with the clustering in the UPGMA tree based on the PCR banding profiles (Fig. 2).
TABLE 3.
Identification of strains based on the 16S rRNA gene sequences
| Isolate | Closest relativea | % Similarity | Accession no. of the closest relative | Profile |
|---|---|---|---|---|
| AG 1.5 α | Asaia krungthepensis | 100 | AB292240 | 1.1.1.1 |
| AG 2.6 β | Asaia sp. SF2.1* | 100 | AM404260 | 3.3.2.2 |
| AG 2.7 β | Asaia sp. SF2.1* | 100 | AM404260 | 3.3.2.2 |
| AG 3.5 α | Asaia krungthepensis | 99 | AB292240 | 2.2.1.1 |
| AG 3.7 α | Asaia krungthepensis | 100 | AB292240 | 2.2.1.1 |
| AG 4.5 α | Asaia krungthepensis | 99 | AB292240 | 1.2.1.1 |
| AG 6.6 α | Asaia krungthepensis | 100 | AB292240 | 1.1.1.1 |
| AG 7.6 α | Asaia krungthepensis | 99 | AB292240 | 1.2.1.1 |
| AG 9.5 α | Asaia krungthepensis | 99 | AB292240 | 2.1.2.1 |
| AG 10.7 α | Asaia krungthepensis | 100 | AB292240 | 4.2.1.1 |
| AA 1.5 β | Asaia krungthepensis | 100 | AB292240 | 1.1.1.1 |
| AA 1.6 β | Asaia krungthepensis | 100 | AB292240 | 1.1.1.1 |
| AA 3.7 α | Asaia krungthepensis | 100 | AB292240 | 1.1.1.1 |
| AA 4.6 β | Asaia krungthepensis | 99 | AB292240 | 1.4.1.1 |
| AA 5.6 α | Asaia krungthepensis | 100 | AB292240 | 1.1.2.1 |
| AA 6.7 α | Asaia krungthepensis | 99 | AB292240 | 1.1.2.1 |
| AA 7.5 α | Asaia sp. SF2.1* | 100 | AM404260 | 4.4.3.2 |
| AA 8.6 β | Asaia krungthepensis | 100 | AB292240 | 1.1.1.1 |
| AA 8.7 β | Asaia krungthepensis | 100 | AB292240 | 1.1.1.1 |
| AA 9.6 β | Asaia krungthepensis | 100 | AB292240 | 2.1.1.1 |
| AS 1.7 α | Asaia sp. SF2.1* | 100 | AM404260 | 1.1.1.3 |
| AS 2.6 α | Asaia sp. SF2.1* | 100 | AM404260 | 4.1.1.3 |
| AS 3.6 α | Asaia sp. SF2.1* | 100 | AM404260 | 1.1.1.3 |
| AE α 1.5 α | Asaia sp. SM2.5* | 100 | AM404261 | 1.1.2.2 |
| AE α 1.7 α | Asaia sp. SM2.5* | 99 | AM404261 | 1.1.2.4 |
| AE α 3.6 α | Asaia sp. SM2.5* | 99 | AM404261 | 1.1.2.2 |
*, strains clustering along with Asaia bogorensis.
FIG. 3.
Phylogenetic positions of the strains of Asaia indicated in Table 3, based on 16S rRNA gene sequences (neighbor-joining method; Kimura correction; all positions containing gaps and missing data were eliminated from the data set). Numbers at each node represent the bootstrap percentages of replications calculated from 2,000 replicated trees. The scale bar represents sequence divergence. Group 1 includes in the same cluster Asaia strains AG2.7 Aβ (FN814277), AA7.5 Aα (FN814292), AS1.7 Aα (FN814296), AS2.6 Aα (FN814297), and AS3.6Aα (FN814299). Group 2 includes in the same cluster Asaia strains: AG3.7 Aα (FN814279), AG6.6 Aα (FN814281), AG7.6 Aα (FN814283), AA1.5 Aβ (FN814286), AA1.6 Aβ (FN814287), AA3.7 Aα (FN814288), AA5.6 Aα (FN814290), AA 6.7 Aα (FN814291), AA 8.6Aβ (FN814293), AA8.7 Aβ (FN814294), and AA9.6 Aβ (FN814295).
DISCUSSION
The analysis of the 16S rRNA gene sequences of the DGGE bands indicates that the microbial community associated with mosquitoes is characterized by a limited bacterial diversity. This observation is comparable to similar observations made for Drosophila by Ryu et al. (24), in which seven bacterial species were found to be associated with a laboratory strain. This is in agreement with experimental evidence showing that the transfer of wild insects to laboratory insectaries leads to a reduction of the diversity of the associated microflora, likely due to an adaptation to the rearing conditions (18).
Culture methods allowed us to identify another species from the AAB associated with mosquitoes (G. liquefaciens), which was not detected by DGGE. This could be due to the detection limits of the PCR used to amplify the total DNA extracted from the insect and to the enrichment steps that may have led to the selection of some species at the expense of others. The fact that Asaia was detected with both culture-independent and -dependent techniques in all of the mosquito species examined suggests that this bacterium is a truly constant component of the microbial community associated with these insects. This also agrees with the experimental evidence that Asaia sp. is able to cross-colonize different species of mosquitoes (4).
Asaia was detected in the Milan Ae. aegypti population by culture-independent tools, but all attempts at isolation failed. This situation is similar to the case described by Crotti et al. (4) in which they were able to detect Asaia in the leafhopper Scaphoideus titanus but not to isolate it in culture. This is possibly due to the fact that some AAB can be found in a viable but not culturable state, as was described by Millet and Lonvaud-Funel (20). This state is characterized by the ability of the cells to perform most metabolic functions but not to grow.
The present study and other studies (4) demonstrate that Asaia is capable of colonizing rather diverse insect species. Here, different strains of Asaia, possibly representing different species, were shown to be able to colonize the same insect species. Such an occurrence is similar to what was observed with D. melanogaster, which can be colonized by two species of Gluconacetobacter, Gluconacetobacter intermedius and Gluconacetobacter rhaeticus (24). These diverse species could be selected by the capability to colonize slightly different microniches in the host gut.
Despite Asaia typing experiments indicating a rather high heterogeneity associated with mosquitoes, the 16S rRNA gene-based phylogenetic analysis indicated that all of the strains fall inside two of the validly described species, A. bogorensis and A. krungthepensis. Such an apparent incoherence can be explained by considering that the phylogenetic analysis of Asaia was based on a 16S rRNA gene which is a highly conserved gene (33), while the typing is based mostly on randomly distributed regions (14, 32) that may be subject to a higher rate of substitution. However, typing results showed a rather high similarity of the profiles (over 50%).
It is interesting to note that a single mosquito individual can harbor up to five strains of Asaia. For example, we recorded individuals harboring strains with profiles 1.1.1.1, 1.2.1.1, 1.3.1.2, 2.1.1.1, and 4.4.3.2. It is generally thought that symbionts evolve toward reduced virulence and even to mutualistic interaction with the hosts where the symbiont transmission is mainly vertical and the symbiotic population is made up of closely related individuals, a condition that might promote virulence reduction through kin selection (10, 13). In the case of mosquitoes, Asaia does not apparently exert any pathogenic effect on the host, but rather, considering its widespread presence in mosquitoes, it could be beneficial indeed. Considering that Asaia can follow different ways for horizontal transmission (4, 6, 11), as indirectly evidenced by the results reported here showing the clustering of symbiotic and environmental isolates, vertical transmission does not appear to be the main mechanism for the maintenance of this bacterium in the insect hosts. Indeed, horizontal transmission tends to lead single hosts to be infected by multiple strains, a case reported here for several mosquito individuals. Despite the above evidence for frequent horizontal transmission and multiple infections of single hosts, Asaia is likely useful to the mosquito, otherwise it would be difficult to explain its 100% prevalence and its preservation in laboratory conditions. Current models to explain the evolution of virulence in host-symbiont interactions are thus probably not adequate to interpret the Asaia-mosquito symbiotic system. While primary symbionts in insects are generally vertically transmitted to the progeny, and likely form clones, it is perhaps more common for secondary symbionts to experience horizontal transmission. We would thus suggest that investigations of the diversity of symbionts are extended to those secondary symbionts that are subjected to horizontal transmission.
Acknowledgments
This work was supported by grant MIUR-PRIN 2007 to A.A., D.D., and G.F.
We thank Davide Sassera and Lorenzo Brusetti for their helpful suggestions. B.C. and C.B. thank Massimo Pajoro for driving them into the area of applied entomology.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Borin, S., D. Daffonchio, and C. Sorlini. 1997. Single strand conformation polymorphism analysis of PCR-tDNA fingerprinting to address the identification of Bacillus species. FEMS Microbiol. Lett. 157:87-93. [DOI] [PubMed] [Google Scholar]
- 2.Bourtzis, K. 2008. Wolbachia-based technologies for insect pest population control. Adv. Exp. Med. Biol. 627:104-113. [DOI] [PubMed] [Google Scholar]
- 3.Chakravorty, S., D. Helb, M. Burday, N. Connell, and D. Alland. 2007. A detailed analysis of 16S ribosomal gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69:330-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crotti, E., C. Damiani, M. Pajoro, E. Gonella, A. Rizzi, I. Ricci, I. Negri, P. Scuppa, P. Rossi, P. Ballarini, N. Raddadi, M. Marzorati, L. Sacchi, E. Clementi, M. Genchi, M. Mandrioli, C. Bandi, G. Favia, A. Alma, and D. Daffonchio. 2009. Asaia, a versatile acetic acid bacterial symbiont, capable of cross-colonizing insects of phylogenetically distant genera and orders. Environ. Microbiol. 11:3252-3264. [DOI] [PubMed] [Google Scholar]
- 5.Daffonchio, D., S. Borin, G. Frova, P. L. Manachini, and C. Sorlini. 1998. PCR fingerprinting of the whole genome, the spacers between the 16S and 23S rRNA genes and of the intergenic tRNA gene regions reveal a different intraspecific genomic variability of Bacillus cereus and Bacillus licheniformis. Int. J. Syst. Bacteriol. 48:107-116. [DOI] [PubMed] [Google Scholar]
- 6.Damiani, C., I. Ricci, E. Crotti, P. Rossi, A. Rizzi, P. Scuppa, F. Esposito, C. Bandi, D. Daffonchio, and G. Favia. 2008. Paternal transmission of symbiotic bacteria in malaria vectors. Curr. Biol. 18:1087-1088. [DOI] [PubMed] [Google Scholar]
- 7.Damiani, C., I. Ricci, E. Crotti, P. Rossi, A. Rizzi, P. Scuppa, A. Capone, U. Ulissi, S. Epis, M. Genchi, N. Sagnon, I. Faye, A. Kang, B. Chouaia, C. Whitehorn, G. W. Moussa, M. Mandrioli, F. Esposito, L. Sacchi, C. Bandi, D. Daffonchio, and G. Favia. 23 June 2010. Mosquito-bacteria symbiosis: the case of Anopheles gambiae and Asaia. Microb. Ecol. doi: 10.1007/s00248-010-9704-8. [DOI] [PubMed]
- 8.de Bary, A. 1879. The phenomenon of symbiosis. Karl J. Trubner, Strasbourg, Germany.
- 9.De Vero, L., E. Gala, M. Gullo, L. Solieri, S. Landi, and P. Giudici. 2006. Application of denaturing gradient gel electrophoresis (DGGE) analysis to evaluate acetic acid bacteria in traditional balsamic vinegar. Food Microbiol. 23:809-813. [DOI] [PubMed] [Google Scholar]
- 10.Ebert, D., and E. A. Herre. 1996. The evolution of parasitic diseases. Parasitol. Today 12:96-101. [DOI] [PubMed] [Google Scholar]
- 11.Favia, G., I. Ricci, C. Damiani, N. Raddadi, E. Crotti, M. Marzorati, A. Rizzi, R. Urso, L. Brusetti, S. Borin, D. Mora, P. Scuppa, L. Pasqualini, E. Clementi, M. Genchi, S. Corona, I. Negri, G. Grandi, A. Alma, L. Kramer, F. Esposito, C. Bandi, L. Sacchi, and D. Daffonchio. 2007. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc. Natl. Acad. Sci. U. S. A. 104:9047-9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Favia, G., I. Ricci, M. Marzorati, I. Negri, A. Alma, L. Sacchi, C. Bandi, and D. Daffonchio. 2008. Bacteria of the genus Asaia: a potential paratransgenic weapon against malaria. Adv. Exp. Med. Biol. 627:49-59. [DOI] [PubMed] [Google Scholar]
- 13.Griffin, A. S., S. A. West, and A. Buckling. 2004. Cooperation and competition in pathogenic bacteria. Nature 430:1024-1027. [DOI] [PubMed] [Google Scholar]
- 14.Hadrys, H., M. Balick, and B. Schierwater. 1992. Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol. Ecol. 1:55-63. [DOI] [PubMed] [Google Scholar]
- 15.Jara, C., E. Mateo, J. M. Guillamón, M. J. Torija, and A. Mas. 2008. Analysis of several methods for the extraction of high quality DNA from acetic acid bacteria in wine and vinegar for characterization by PCR-based methods. Int. J. Food Microbiol. 128:336-341. [DOI] [PubMed] [Google Scholar]
- 16.Kersters, K., P. Lisdiyanti, K. Komagata, and J. Swings. 2006. The family Acetobacteraceae: the genera Acetobacter, Acidomonas, Asaia, Gluconacetobacter, Gluconobacter, and Kozakia, p. 163-200. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 5. Springer, New York, NY. [Google Scholar]
- 17.Kounatidis, I., E. Crotti, P. Sapountzis, L. Sacchi, A. Rizzi, B. Chouaia, C. Bandi, A. Alma, D. Daffonchio, P. Mavragani-Tsipidou, and K. Bourtzis. 2009. Acetobacter tropicalis is a major symbiont of the olive fruit fly (Bactrocera oleae). Appl. Environ. Microbiol. 75:3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehman, R. M., J. G. Lundgren, and L. M. Petzke. 2009. Bacterial communities associated with the digestive tract of the predatory ground beetle, Poecilus chalcites, and their modification by laboratory rearing and antibiotic treatment. Microb. Ecol. 57:349-358. [DOI] [PubMed] [Google Scholar]
- 19.Lisdiyanti, P., H. Kawasaki, T. Seki, Y. Yamada, T. Uchimura, and K. Komagata. 2001. Identification of Acetobacter strains isolated from Indonesian sources, and proposals of Acetobacter syzygii sp. nov., Acetobacter cibinongensis sp. nov., and Acetobacter orientalis sp. nov. J. Gen. Appl. Microbiol. 47:119-131. [DOI] [PubMed] [Google Scholar]
- 20.Millet, V., and A. Lonvaud-Funel. 2000. The viable but non-culturable state of wine micro-organisms during storage. Lett. Appl. Microbiol. 30:136-141. [DOI] [PubMed] [Google Scholar]
- 21.Moran, N. A. 2006. Symbiosis. Curr. Biol. 24:R866-R871. [DOI] [PubMed] [Google Scholar]
- 22.Muyzer, G., T. Brinkhoff, U. Nubel, C. Santegoeds, H. Schafer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 3.4.4/1-3.4.4/27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 23.Roh, S. W., Y. D. Nam, H. W. Chang, K. H. Kim, M. S. Kim, J. H. Ryu, S. H. Kim, W. J. Lee, and J. W. Bae. 2008. Phylogenetic characterization of two novel commensal bacteria involved with innate immune homeostasis in Drosophila melanogaster. Appl. Environ. Microbiol. 74:6171-6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryu, J. H., S. H. Kim, H. Y. Lee, J. Y. Bai, Y. D. Nam, J. W. Bae, D. G. Lee, S. C. Shin, E. M. Ha, and W. J. Lee. 2008. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319:777-782. [DOI] [PubMed] [Google Scholar]
- 25.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 26.Sánchez, O., J. M. Gasol, R. Massana, J. Mas, and C. Pedros-Alio. 2007. Comparison of different denaturing gradient gel electrophoresis primer sets for the study of marine bacterioplankton communities. Appl. Environ. Microbiol. 73:5962-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinkins, S. P., T. Walker, A. R. Lynd, A. R. Steven, B. L. Makepeace, H. C. Godfray, and J. Parkhill. 2005. Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature 436:257-260. [DOI] [PubMed] [Google Scholar]
- 28.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 29.Urzì, C., L. Brusetti, P. Salamone, C. Sorlini, E. Stackebrandt, and D. Daffonchio. 2001. Biodiversity of Geodermatophilaceae isolated from altered stones and monuments in the Mediterranean basin. Environ. Microbiol. 3:471-479. [DOI] [PubMed] [Google Scholar]
- 30.Werren, J. H., D. Windsor, and L. R. Guo. 1995. Distribution of Wolbachia among neotropical arthropods. Proc. R. Soc. Lond. B 262:197-204. [Google Scholar]
- 31.Werren, J. H., L. Baldo, and M. E. Clark. 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6:741-751. [DOI] [PubMed] [Google Scholar]
- 32.Williams, J. G., A. R. Kubelik, K. J. Livak, J. A. Rafalski, and S. V. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woese, C. R., G. E. Fox, L. Zablen, T. Uchida, L. Bonen, K. Pechman, B. J. Lewis, and D. Stahl. 1975. Conservation of primary structure in 16S ribosomal RNA. Nature 254:83-86. [DOI] [PubMed] [Google Scholar]
- 34.Yamada, Y., K. Katsura, H. Kawasaki, Y. Widyastuti, S. Saono, T. Seki, T. Uchimura, and K. Komagata. 2000. Asaia bogorensis gen. nov., sp. nov., an unusual acetic acid bacterium in the alpha-Proteobacteria. Int. J. Syst. Evol. Microbiol. 50:823-829. [DOI] [PubMed] [Google Scholar]
- 35.Yu, Z., and M. Morrison. 2004. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:4800-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]