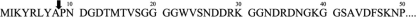Abstract
Two different bacteriocins, carotovoricin and carocin S1, had been found in Pectobacterium carotovorum subsp. carotovorum, which causes soft-rot disease in diverse plants. Previously, we reported that the particular strain Pcc21, producing only one high-molecular-weight bacteriocin, carried a new antibacterial activity against the indicator strain Pcc3. Here, we report that this new antibacterial activity is due to a new bacteriocin produced by strain Pcc21 and named carocin D. Carocin D is encoded by the caroDK gene located in the genomic DNA together with the caroDI gene, which seems to encode an immunity protein. N-terminal amino acid sequences of purified carocin D were determined by Edman degradation. In comparison with the primary translation product of caroDK, it was found that 8 amino acids are missing at the N terminus. This finding proved that carocin D is synthesized as a precursor peptide and that 8 amino acids are removed from its N terminus during maturation. Carocin D has two putative translocation domains; the N-terminal and C-terminal domains are homologous to those of Escherichia coli colicin E3 and Pseudomonas aeruginosa S-type pyocin, respectively. When caroDK and caroDI genes were transformed into carocin D-sensitive bacteria such as Pcc3, the bacteria became resistant to this bacteriocin. Carocin D has one putative DNase domain at the extreme C terminus and showed DNase activity in vitro. This bacteriocin had slight tolerance to heat but not to proteases. The caroDK gene was present in only 5 of 54 strains of P. carotovorum subsp. carotovorum. These results indicate that carocin D is a third bacteriocin found in P. carotovorum subsp. carotovorum, and this bacteriocin can be readily expressed in carocin D-sensitive nonpathogenic bacteria, which may have high potential as a biological control agent in the field.
Pectobacterium carotovorum subsp. carotovorum is a Gram-negative phytopathogen responsible for soft rot, blackleg, or stem rot in various commercially important plants, including Chinese cabbage and potato. Bacterial soft rot is found throughout Korea and causes serious yield loss in the field, in transit, and in storage. Pathogenesis in P. carotovorum subsp. carotovorum is dependent on production of plant cell wall-degrading enzymes that are actively secreted by the bacterium. Various aspects of epidemiology of the disease caused by this phytopathogen are relatively well understood, but no efficient method is available to control the disease (21).
Some bacteria, including plant pathogens, produce one or more antibacterial peptides called bacteriocins. Bacteriocins were originally defined as ribosomally synthesized proteinaceous compounds that killed strains of the same or closely related species (20). They are potent, often highly specific toxins that are usually produced under stressful conditions, causing the rapid elimination of neighboring cells that are not immune or resistant to their effects (14). Elucidation of the ecological significance of inhibitory substances such as bacteriocins produced by plant pathogens is important for understanding factors that affect population dynamics on plant surfaces. Thus, the exploitation of narrow-spectrum bacteriocins is an attractive strategy for targeted attack against bacterial diseases in plant disease control (10).
Among bacteriocins produced by Gram-negative bacteria, colicins and S-type pyocins have been intensively studied. Colicins and S-type pyocins are produced by Escherichia coli and Pseudomonas aeruginosa, respectively. They consist of two proteins, one responsible for antimicrobial activity (the killing protein) and the other for immunity (the immunity protein). The killing proteins are organized in functional domains, with receptor-binding, translocation, and DNase (RNase) activity (9, 15). Their gene promoters, located upstream of the structural genes, include conserved DNA regions, the so-called SOS box of colicins, and the P box of S-type pyocins, and they are inducible by DNA-damaging agents such as mitomycin C (MMC) (1, 19). Colicins and S-type pyocins need to interact with specific membrane receptors on target cells for their activities, and these specific interactions determine the spectrum of target cells, which is generally very narrow. The host strain is protected from its own bacteriocin through interaction with the immunity protein that is coproduced with the bacteriocin. It has been proposed that bacteriocins may play a key role in bacterial population dynamics (16).
Two bacteriocins have been reported in P. carotovorum subsp. carotovorum. One is carotovoricin, a high-molecular-weight bacteriocin, which contains a lysis cassette and a gene cluster for a structural protein and is located in the chromosomal DNA (13, 22). Sequence comparisons showed high homology between carotovoricin and bacteriophage proteins (22). Electron microscopy showed that carotovoricin has an antenna-like structure, with a base plate and tail fibers. Another bacteriocin is a low-molecular-weight bacteriocin, carocin S1, which consists of a killing protein and an immunity protein. Production of carocin S1 is induced by glucose and lactose (4). The carocin S1 gene is homologous to the pyocin S3 and pyocin AP41 genes of P. aeruginosa (4).
Because soft-rot disease in Chinese cabbage is destructive and no efficient control method is known, development of new control methods against the pathogen P. carotovorum subsp. carotovorum is desirable, and any method should be safe for humans and environmentally friendly. The use of bacteriocins may be one of the most feasible methods that satisfies both criteria. Although the rapid occurrence of resistant mutants may limit the efficacy of bacteriocin as a control method, use of combinations of several different bacteriocins will help to overcome this.
In this study, a new low-molecular-weight bacteriocin, carocin D, and its immunity gene were identified and characterized. This new bacteriocin has a rare feature in that it has two translocation domains. Additionally, the domain structure of carocin D suggests that it may have arisen from a chimera of two different bacteriocins: one from colicin E of E. coli and the other from pyocin of P. aeruginosa.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The strains and plasmids used are shown in Table 1. P. carotovorum subsp. carotovorum strains, Pseudomonas aeruginosa, and Escherichia coli were cultured in LB medium (10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl in 1 liter, pH 7.2) at 28°C for Pectobacterium and Pseudomonas and at 37°C for Escherichia coli with shaking, respectively. Rifampin, kanamycin, and ampicillin (all at 50 mg/liter) were added to the LB agar medium where necessary. The P. carotovorum subsp. carotovorum strains were isolated from various host plants and locations in Korea (17).
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or oligonucleotide | Relevant features or sequence | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| EPI300-T1R | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ−rpsL nupG trfA dhfr | Epicentre |
| EC100 pir+ | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ−rpsL nupG trfA dhfr | Epicentre |
| DH5α | F− φ80dlacZΔM15 endA41 recAl hsdR17(rK− mK−) supE44 thi-1 gyrA96 | BRL |
| BL21 | hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV-T7 gene 1) | Novagen |
| P. carotovorum subsp. carotovorum | ||
| Pcc3 | Carocin D−, sensitive to carocin D, Rifr | 17 |
| Pcc21 | Carocin D+, Rifr | 17 |
| Pcc21DK1,2,6,7,9 | Pcc21; caroDK::Tn5, carocin D−, Rifr Kanr | This study |
| Plasmids | ||
| pBluescript II KS(+) | AmprlacZ, cloning vector | Stratagene |
| pGEM-T Easy | TA-cloning vector; Ampr | Promega |
| pLUG-multi | TA-cloning vector; Ampr | Intron |
| pGFPuv | Ampr, lac promoter | Clontech |
| pDSK519 | Cloning vector; Kanr | 8 |
| pCC1FOS | Fosmid vector; Chlr | Epicentre |
| pRF6-A4 | Fosmid library, pCC1FOS with 35-kb genomic DNA fragment | This study |
| pRB41 | pBluescript II KS(+) with ClaI/BamHI 4.1-kb fragment | This study |
| pRG3431 | pGEM-T Easy with 221 to ∼3,651 bp of ClaI/BamHI 4.1-kb fragment | This study |
| pRG2878 | pGEM-T Easy with 385 to ∼3,263 bp of ClaI/BamHI 4.1-kb fragment | This study |
| pRG2808 | pGEM-T Easy with 221 to ∼3,028 bp of ClaI/BamHI 4.1-kb fragment | This study |
| pRL1208 | pLUG-multi with 1,821 to ∼3,028 bp of ClaI/BamHI 4.1-kb fragment | This study |
| pRG369 | pGEM-T Easy with 2,895 to ∼3,262 bp of ClaI/BamHI 4.1-kb fragment | This study |
| pRP369 | pGFPuv with HindIII/EcoRI 0.4-kb fragment of pRG369 | This study |
| pRD2808 | pDSK519 with SalI/BamHI 2.9-kb fragment of pRG2808 | This study |
| Oligonucleotides | ||
| caro-F-1762 | 5′-GAA AAC GCA GGC GGA AAC AGG TC-3′ | This study |
| caro-F-1926 | 5′-TTT AGC CAG ATC AGA GAA GTC-3′ | This study |
| caro-F-3362 | 5′-TTG CCG AAG AAA AAG AGA TAA-3′ | This study |
| caro-F-4436 | 5′-AAC CGT TGT ACA CTG ATG CT-3′ | This study |
| caro-R-3263 | 5′-AGC GCG TAT TAA TTA AGA ACA CA-3′ | This study |
| caro-R-4569 | 5′-GCA AAA ACG GTA AGC ACA TCT-3′ | This study |
| caro-R-4803 | 5′-GCG CGT ATT AAT TAA GAA CAC A-3′ | This study |
| caro-R-5192 | 5′-CGG CGT CAG CGT GGT GGT ATC-3′ | This study |
| DK3751 | 5′-GTA GCG GGC AGT TAT GAC AAA AAT-3′ | This study |
| DK4121 | 5′-ATC GGA CCG CCT GCC TTA TC-3′ | This study |
| DI4578 | 5′-CAG ATT ATA AAC AAT TGG ATA GAC-3′ | This study |
| DI4723 | 5′-CAC CCA ATT AGT CCC TTT TAC-3′ | This study |
| DKI3744 | 5′-CAG AGG TGT AGC GGG CAG TTA TGA-3′ | This study |
| DKI4730 | 5′-GCC ACC CAA TTA GTC CCT TTT ACA-3′ | This study |
Bacteriocin assay.
Antibacterial activity was detected by the spot-on-lawn method for screening of inhibitory activity against the P. carotovorum subsp. carotovorum Pcc3 strain. A spot was made on solid LB medium with the bacterial strain to be tested as a producer. The spotted producer strains were incubated for 12 h at 28°C and exposed to 700 μl chloroform vapor for 10 min. After aeration for 20 min to remove traces of chloroform, the surface of the solid medium was subsequently overlaid with 7 ml of 0.7% soft agar containing 10 μl of an overnight culture of the indicator organism. The production of antibacterial substance was detected as growth-free inhibition zones (clear zones) around the spotted area after overnight incubation at 28°C (12). Induction of bacteriocin production by UV or mitomycin C (MMC) treatment was conducted as described previously (17).
Transposon mutagenesis and cloning of mutated genes.
P. carotovorum subsp. carotovorum strain Pcc21 was subjected to transposon mutagenesis using the EZ-Tn5<R6Kγori/KAN-2>Tnp transposome kit (Epicentre Biotechnologies, Madison, WI) according to the manufacturer's protocol. Briefly, the transposome complex was electroporated into P. carotovorum subsp. carotovorum Pcc21, and the cell suspension was diluted and plated on LB agar, supplemented with kanamycin and rifampin. Approximately 5,000 colonies were screened, from which 12 mutants were selected for further characterization. Genomic DNA was prepared from these 12 strains by using the G-spin kit for bacterial genomic DNA extraction (Intron Biotechnology, Seongnam, South Korea), digested with EcoRI, and then immediately ligated. Ligation reaction mixtures were electroporated into strain E. coli EC100 pir+ (Epicentre Biotechnologies), and kanamycin-resistant transformants were selected. Plasmid DNAs were purified from these strains and sequenced with primers KAN-2 FP-1 and R6KAN-2 RP-1 (Epicentre Biotechnologies). Sequence data were analyzed with BLAST to identify sites of transposon insertion, based on the fact that transposon EZ-Tn5 generates a 9-bp duplication at its insertion site.
Construction of a fosmid library.
The genomic DNA of P. carotovorum subsp. carotovorum Pcc21 was obtained with a genomic DNA isolation kit (Promega, Madison, WI) and size fractionated in a 0.5% low-melting-point agarose gel. DNA fragments of >30 kb were collected for library construction. The construction of the genomic library was performed according to the manufacturer's protocol. Sheared and end-repaired DNA fragments were ligated into fosmid vector pCC1FOS (Epicentre Biotechnologies), and the ligated DNA was packaged using MaxPlax lambda packaging extracts (Epicentre Biotechnologies). E. coli EPI300 (Epicentre Biotechnologies) cells were infected using the packaged DNA and plated on LB agar medium supplemented with chloramphenicol. The presence of recombinant plasmids and the polymorphism of the insert DNA were examined by agarose gel electrophoresis after digestion of plasmid DNA purified from randomly selected E. coli transformants with BamHI. In total, 1,920 fosmid library clones were stored in a 96-well plate at −70°C.
Genetic engineering techniques.
Total DNA was isolated by the method of Sambrook et al. (18). Plasmid DNA was isolated with the DNA-spin plasmid DNA isolation kit (Intron Biotechnology). DNA electrophoresis, restriction digestion, ligation, and transformation procedures were done as described by Sambrook et al. (18). DNA primers were synthesized by Genotech, Inc. (Daejeon, South Korea). PCR amplification of serial deletion constructs of carocin D and the immunity gene was performed with the specific primers listed in Table 1. PCR was performed on a PTC-100 DNA thermal cycler (MJ Research, Inc., Watertown, MA), and Maxime PCR PreMix i-MAX (Intron Biotechnology) was used in a final volume of 20 μl containing 50 nM (each) primer and 1 μg to 50 ng of plasmid DNA. The program used was 95°C for 5 min for the first cycle and then 95°C for 1 min, 55°C for 1 min, and 72°C for 1 to 3 min for the next 30 cycles and 72°C for 5 min for the final cycle.
RT-PCR.
Total RNA was extracted from the Pcc21 strain using the RNeasy minikit (Qiagen) according to the manufacturer's procedure and purified by incubation with an RNase-free DNase set (Qiagen) at 37°C for 1 h to eliminate contaminating DNA, followed by phenol extraction and ethanol precipitation. Reverse transcription (RT) was carried out at 42°C for 1 h in a reaction mixture (20 μl) consisting of 5 μg of the total RNA, 20 μM random 6-mer, 10 mM deoxynucleoside triphosphate (dNTP), 20 U of RNase inhibitor, 10 U of PrimeScript RTase, and RT buffer (Takara). The reaction was terminated by heating the reaction mixture at 70°C for 15 min; the reaction mixture was used as a template to amplify cDNA by PCR (30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min) in the presence of each specific primer pair. PCR products were analyzed on 1% agarose gels.
Purification and N-terminal sequencing.
E. coli BL21 Codon plus (DE3) carrying the carocin DK open reading frame (ORF) in pET41b and carocin DI ORF in pCDFDuet (Novagen) was expressed. His tag was fused only on caroDK. To express caroDK and caroDI, transformed cells were grown at 37°C and then induced by the additional of 1 mM isopropyl-β-d-thiogalactopyranoside. The induced cells were grown for 16 h at 20°C, harvested by centrifugation, and sonicated in buffer A (50 mM HEPES, pH 7.6, 200 mM NaCl). CaroDK and CaroDI proteins were then copurified from the cell lysate using immobilized metal affinity chromatography with buffer B (buffer A containing 250 mM imidazole). The pooled fractions containing CaroDK and CaroDI were concentrated and loaded on a Superdex 200 gel filtration column (GE Health) as a final purification step.
The purified proteins were run on an SDS-PAGE gel and electroblotted onto a polyvinylidene difluoride (PVDF) membrane in transfer buffer (25 mM Tris-HCl, pH 8.2, 19.2 mM glycine, 10% [vol/vol] ethanol) at 50 V for 2 h. Purified carocin D was subjected to Edman degradation and sequenced in a Procise 491 protein sequencer (Korea Basic Science Institute [KBSI]).
Sensitivity of bacteriocin to heat and enzyme treatment.
To lyse the cell, chloroform (5%) was added to the overnight culture of P. carotovorum subsp. carotovorum or E. coli containing plasmids. Cell debris and genomic DNA were removed by centrifugation (10,000 × g, 30 min). The supernatant was filtered through an 0.22-μm membrane. Filtered cell crude extract was then treated with each of the following enzymes: lipase (0.1 mg/ml, 120 min, 37°C), trypsin (2.5 mg/ml, 60 min, 25°C), pronase E (0.2 mg/ml, 60 min, 37°C), proteinase K (0.2 mg/ml, 60 min, 37°C), DNase (0.2 mg/ml, 60 min, 37°C), and RNase A (0.2 mg/ml, 60 min, 37°C) (7). Bacteriocin thermostability was investigated by heating the mixture for 10 min at 60, 70, and 80°C and then quickly chilling it in ice water. Residual bacteriocin activity was determined as described previously (6).
Bacteriocin assay for DNase activity.
To confirm DNase activity, 1 μg genomic DNA from P. carotovorum subsp. carotovorum was mixed with 10 μl of the cell crude extract containing carocin D or purified carocin D (3 μg) and incubated at 37°C for 3 h. After incubation, the samples were analyzed by 1.0% agarose gel electrophoresis in Tris-acetate-EDTA buffer.
Analysis of sequence data.
The nucleotide sequence and the deduced amino acid sequence of carocin D were compared using the BLAST programs of the National Center for Biotechnology Information server. Sequence data were compiled using the DNAstar software (DNAstar, Inc., Madison, WI).
Nucleotide sequence accession number.
The nucleotide sequence was submitted to GenBank under accession number GU354220.
RESULTS
Transposon mutagenesis and isolation of bacteriocin-defective mutants.
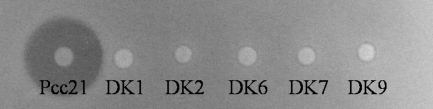
In previous work, the presence of a new bacteriocin(s) in P. carotovorum subsp. carotovorum isolated in Korea was suggested (17). To find the gene(s) responsible for the new antibacterial activity, P. carotovorum subsp. carotovorum strain Pcc21 was chosen among several candidate strains because this strain secreted an antibacterial substance(s) other than a low-molecular-weight bacteriocin, carocin S1, and a high-molecular-weight bacteriocin, carotovoricin (17). In total, 5,184 mutants were generated by Tn5-based mutagenesis and screened for the loss of antibacterial activity due to a new low-molecular-weight substance(s). Among the 5,184 mutants, five mutants lost their antibacterial activities against P. carotovorum subsp. carotovorum Pcc3, which is sensitive to this antibacterial activity (Fig. 1). Because the wild-type P. carotovorum subsp. carotovorum Pcc21 produces carotovoricin, a high-molecular-weight bacteriocin, mutants still showed a clear zone that was restricted to the immediate surroundings of the colony, but the clear zone was lost farther away from the colony. Tn5-inserted DNA regions of five mutants that lost antibacterial activity were cloned and sequenced. Interestingly, all five mutants had Tn5 insertions at different positions within the same open reading frame (ORF) (Fig. 2B).
FIG. 1.
Antibacterial activity of Tn5 insertion mutants of P. carotovorum subsp. carotovorum Pcc21 against the indicator strain Pcc3. Pcc21, P. carotovorum subsp. carotovorum Pcc21 wild-type strain; DK1, -2, -6, -7, and -9, P. carotovorum subsp. carotovorum Pcc21 mutant strains (loss of low-molecular-weight bacteriocin activity). The photo was taken 24 h after the indicator strain had been overlaid.
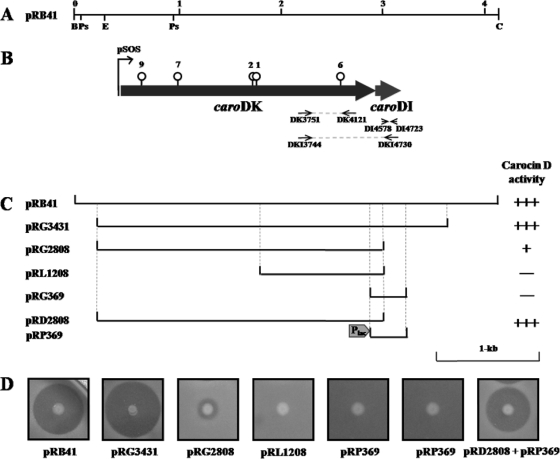
FIG. 2.
(A) Physical maps of the BamHI-ClaI 4.1-kb region of pRB41. Abbreviations are for the endonuclease recognition sites: C, ClaI; B, BamHI; Ps, PstI; E, EcoRI. The numbers indicate kb. (B) Thick horizontal arrows indicate the predicted ORFs and the direction of ORF transcription. The thin arrow indicates the potential promoter region. The vertical lines with circular heads on the ORF map show the points of transposon insertion. The circular heads indicate Tn5, and the numbers (1, 2, 6, 7, and 9) are the same as those of Tn5 mutants shown in Fig. 1. (C) The horizontal lines under the map represent plasmids derived from the cloned fosmid library DNA fragments listed on the left. The dotted vertical lines represent the ends of the amplified PCR fragment of pRB41 used to clone the bacteriocin determinant. Carocin D activity is shown, based on results in panel D. +++, full activity; +, low activity; −, no activity. (D) Antibacterial activity expressed in E. coli DH5α for each subclone.
Isolation of clones responsible for an antibacterial activity.
To isolate the full gene(s) responsible for the antibacterial activity, a fosmid library carrying about 40-kb fragments of P. carotovorum subsp. carotovorum Pcc21 genomic DNA was screened. Because Pectobacterium belongs to the Enterobacteriaceae family, like E. coli, many genes isolated from Pectobacterium are expressed well in E. coli. When 1,920 fosmid library clones were screened with an indicator strain, P. carotovorum subsp. carotovorum Pcc3, 16 clones showed antibacterial activity. Based on the DNA sequence of the Tn5 insertion region mentioned above, PCR primers caro-F-1929 and caro-R-4804 were generated (Table 1) and they amplified those regions to confirm the existence of the gene. All 16 clones showed an amplified DNA band at the “correct” location (data not shown), and a clone carrying pRF6-A4 was selected for further study. The pRF6-A4 plasmid contained 35-kb genomic DNA of P. carotovorum subsp. carotovorum Pcc21 in the pCC1FOS vector. Serial subclones were generated to narrow down the DNA region responsible for the production of the antibacterial substance against P. carotovorum subsp. carotovorum Pcc3 (data not shown). Among the subclones, the plasmid pRB41 showed full antibacterial activity in E. coli, as did P. carotovorum subsp. carotovorum Pcc21 (Fig. 2C and D). This plasmid contained a 4.1-kb BamHI-ClaI DNA fragment and was chosen for further in-depth analysis.
Analysis of a genetic structure in a 4.1-kb DNA region.
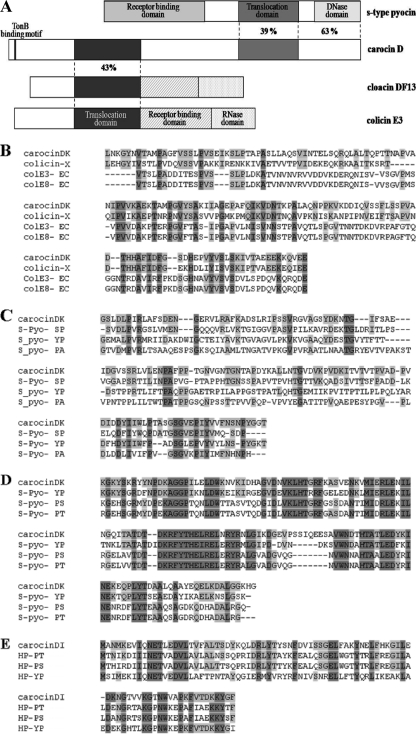
A 4.1-kb chromosomal DNA region of P. carotovorum subsp. carotovorum Pcc21 carrying an antibacterial activity was sequenced. Computer analysis revealed two open reading frames (ORF1 and ORF2) in the same direction (Fig. 2B). The predicted amino acid sequences of ORF1 and ORF2 were compared with amino acid sequences deposited in the SwissProt protein sequence database. The predicted molecular masses were 91 kDa and 9.8 kDa for ORF1 and ORF2, respectively, and similar sizes of bands were confirmed with SDS-PAGE (data not shown). Significant similarities were found between the amino acid sequence of ORF1 in P. carotovorum subsp. carotovorum Pcc21 and those of E-group colicins from E. coli, such as cloacin DF13 or colicin E3, and S-type pyocin of P. aeruginosa (Fig. 3A), indicating that orf1 encoded a new bacteriocin. The ORF1 of 836 amino acids contained two characteristic translocation domains at amino acids 156 to 310 and 547 to 688. The first region showed 30% identity with the cloacin translocation domain of E. coli colicin E3 (Fig. 3B) and 27% identity with the S-type pyocin translocation domain of Pseudomonas pyosin (Fig. 3C). The C-terminal sequence of ORF1 was very similar to those of DNase-type bacteriocins. It showed 55% identity with the DNase activity domain of Pseudomonas pyocin (Fig. 3D). At 13 amino acids from the beginning of the ORF1 protein, a TonB-binding motif (DTMTV) was found. The amino acid sequences of ORF2 in P. carotovorum subsp. carotovorum Pcc21 had no similarity with that of the immunity protein of S-type pyocin in P. aeruginosa but had 65% homology with those of hypothetical immunity proteins from Pseudomonas syringae, Yersinia pestis, and Yersinia pseudotuberculosis (Fig. 3E). To date, only three bacteriocins, colicin D, klebicin D, and colicin D-157, have been shown to carry two different translocation domains. Thus, the bacteriocin produced by ORF1 was designated carocin D, and genes encoding the bacteriocin killing protein and the immunity protein were named caroDK and caroDI, respectively.
FIG. 3.
Alignment of amino acid sequences of carocin D and the organization of functional domains. (A) Regions similar to S-type pyocin and to cloacin DF13 and colicin E3 are shown; the location of the TonB-binding motif, the percentages of identical residues, and the positions of functional domains are given. (B) Peptide sequence alignment of the N-terminal translocation domain of carocin D (positions 156 to 310, amino acid sequences). (C) Peptide sequence alignment of the C-terminal translocation domain of carocin D (positions 547 to 688, amino acid sequences). (D) Peptide sequence alignment of the DNase activity domain of carocin D (positions 689 to 688, amino acid sequences). (E) Alignment of carocin D immunity protein and homologs from Pseudomonas syringae pv. syringae B728a, Yersinia pestis CO92, and Yersinia pseudotuberculosis IP 32953. Amino acid residues conserved in the majority of sequences are highlighted in light gray, whereas those conserved in all sequences are shown in dark gray. Abbreviations for bacteriocins are as follows: colE3, colicin E3; colE8, colicin E8; S-Pyo, S-type pyocin; HP, hypothetical protein. Abbreviations for bacteria are as follows: X, Xenorhabdus nematophila; EC, Escherichia coli; SP, Serratia proteamaculans; YP, Yersinia pseudotuberculosis; PA, Pseudomonas aeruginosa; PS, Pseudomonas syringae pv. syringae; PT, Pseudomonas syringae pv. tabaci.
In order to confirm whether the ORF1 is an active ORF carrying antibacterial activity, N-terminal amino acid sequences were determined by Edman degradation. The sequences of five N-terminal amino acid residues of ORF1 purified from E. coli BL21 Codon plus (DE3) with antibacterial activity were proline (P), asparagine (N), aspartic acid (D), glycine (G), and aspartic acid (D). In comparison with the primary translation product of ORF1, it was found that 8 amino acids are missing at the N terminus. This finding proved that the carocin D is synthesized as a precursor peptide and that 8 amino acids are removed from its N terminus during maturation (Fig. 4).
FIG. 4.
Deduced amino acid sequence of the N-terminal region of carocin D. The amino acid sequence of purified carocin D was determined by Edman degradation. The bacteriocin processing site is indicated by the vertical arrow.
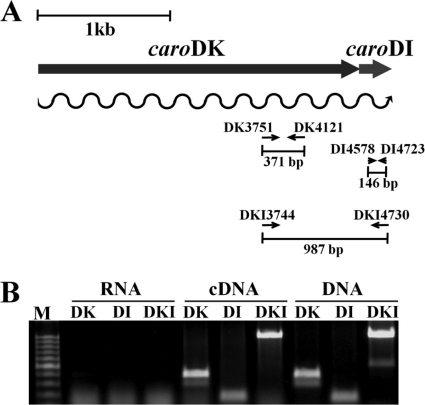
Analysis of the caroDK gene promoter region showed a potential Shine-Dalgarno sequence (GGAGAA) at the −11-bp position from the putative ATG start codon and a possible SOS box, CTGTAT(N)4ATACAG, at positions −44 to −28 bp, like those in the genes encoding colicins (Fig. 5). Additionally, an RdgB box for Mor binding [TAA(N)6TTA] was found 107 bp upstream of the start codon (data not shown). Similarly to caroDK, caroDI has a putative Shine-Dalgarno sequence (GGAGGG) located at the 3′ end of the caroDK gene. This sequence was 18 nucleotides from the stop codon, TAA, of caroDK and 11 nucleotides from the initiation codon of the caroDI gene (data not shown). However, no obvious promoter for the caroDI gene was identified, suggesting that caroDK and caroDI comprise a single operon (Fig. 6). An operon organization of the genes caroDK and caroDI as a single transcriptional unit was further confirmed by RT-PCR. A cDNA with a 987-bp nucleotide sequence covering parts of both caroDK and caroDI sequences as well as the intergenic sequence was produced. Under the same reaction conditions, two cDNAs carrying partial sequences of the genes caroDK and caroDI, respectively, also were produced (Fig. 6).
FIG. 5.
Promoter region of the caroDK gene. Cloacin DF13, the colicin family (E3 and E6), and carocin D all have −10 boxes and SOS boxes (bold and underlined) in the promoter regions. The “ATG” at the right end is the start codon of the structural gene.
FIG. 6.
Detection of the caroDK/caroDI transcripts by RT-PCR. (A) Schematic representation of the RT-PCR experiment. The small arrows indicate the positions and directions of the gene-specific primers used in RT-PCR with lengths of the expected PCR product, and the wavy arrow indicates transcription. (B) Agarose gel electrophoresis of RT-PCR products. The reactions were carried out in the presence of a reverse transcriptase (cDNA) or with the genomic DNA as a template (DNA). DK indicates the caroDK gene-specific product, DI indicates the caroDI-specific product, and DKI indicates the caroDK and caroDI product.
Functional complementation and deletion analysis.
To verify that carocin D was responsible for the antibacterial activity of P. carotovorum subsp. carotovorum Pcc21, plasmid pRB41, carrying both caroDK and caroDI genes, was transformed into P. carotovorum subsp. carotovorum Pcc21DK1, which had a mutation in caroDK (Fig. 2B) and had lost antibacterial activity against P. carotovorum subsp. carotovorum Pcc3 (Fig. 1). Full antibacterial activity of P. carotovorum subsp. carotovorum Pcc21DK1(pRB41) against the indicator strain Pcc3 was restored, as it was in the wild-type P. carotovorum subsp. carotovorum Pcc21 (Table 2). Indicator strain P. carotovorum subsp. carotovorum Pcc3 became toxic to its own Pcc3 strain after receiving the plasmid pRB41. The production of antibacterial substances was also induced by UV and MMC treatment in both Pcc21DK1(pRB41) and Pcc3(pRB41) strains, just like carocin D of the Pcc21 wild-type strain (Table 2). These results indicated that carocin D, encoded by the caroDK gene, together with the caroDI-encoded protein, was responsible for the antibacterial activity of P. carotovorum subsp. carotovorum Pcc21 against indicator strains tested in this study.
TABLE 2.
Induction test
| Producer strain | Activity against P. carotovorum subsp. carotovorum Pcc3c |
||
|---|---|---|---|
| LB | UVa | MMCb | |
| P. carotovorum subsp. carotovorum | |||
| Pcc21 | + | ++ | +++ |
| Pcc21DK1 | − | − | − |
| Pcc21DK1(pRB41) | ++ | +++ | ++++ |
| Pcc3 | − | − | − |
| Pcc3(pRB41) | ++ | +++ | ++++ |
| E. coli | |||
| DH5α(pRB41) | ++ | ++ | ++ |
| BL21(pRB41) | ++ | +++ | ++++ |
The tested strain was spotted onto a plate and exposed to UV light (320 nm) for 15 min at a distance of 25 cm.
The MMC experiments were conducted in LB medium containing 0.5 μg/ml mitomycin C.
The data represent the averages of three independent experiments. Symbols: +, 0- to 0.5-cm-diameter inhibition zone; ++, 0.5- to 1-cm-diameter inhibition zone; +++, 1- to 1.5-cm-diameter inhibition zone; ++++, 1.5- to 2-cm-diameter inhibition zone; −, no inhibition.
When the plasmid pRB41 was introduced into E. coli DH5α, which is recA negative, antibacterial activity of carocin D was not induced, but it was induced in E. coli BL21, which is recA+ (Table 2). This indicates that for full induction of carocin D activity, the recA gene is required. These data confirm that the SOS box in the promoter region is important in the regulation of carocin D expression (Fig. 5).
To identify the immunity function of caroDI, a 196-bp DNA fragment of orf2 was deleted from plasmid pRG3431, and a new plasmid, pRG2808, was constructed (Fig. 2C). Although the plasmid pRG2808 contained a deleted ORF2, responsible for the immunity function, E. coli DH5α(pRG2808) was able to produce carocin D. However, the size of the clear zone produced by the E. coli DH5α carrying plasmid pRG2808 was much smaller than that produced by E. coli DH5α carrying plasmid pRG3431 (Fig. 2D). Because E. coli DH5α(pRG2808) did not contain an immunity gene for its own protection, it was hypothesized that the highest number of E. coli DH5α(pRG2808) cells were killed by the antibacterial activity from the caroDK gene in plasmid pRG2808. To test this, the immunity gene constructed under a constitutively expressed UV promoter was introduced into E. coli DH5α(pRG2808). After the strain received the plasmid pRP369, full carocin D activity and immunity were recovered in E. coli DH5α(pRD2808). Immunity activity was also confirmed by the introduction of plasmid pRP369 into the bacteriocin-susceptible indicator strain P. carotovorum subsp. carotovorum Pcc3. The Pcc3 transformant carrying pRP369 was not killed by strains carrying a fully functional carocin D gene. However, Pcc3 containing the caroDK gene without caroDI was susceptible to strains carrying a functional carocin D gene (Table 3). Thus, the immunity gene under the UV promoter was active and fully functional, and this gene product inhibited the antibacterial activity of P. carotovorum subsp. carotovorum Pcc21.
TABLE 3.
Complementation test
| Producer strain of P. carotovorum subsp. carotovorum | Activity against indicator straina: |
||||
|---|---|---|---|---|---|
| Pcc3 | Pcc3(pRB41) | Pcc3(pRG369) | Pcc3(pRP369) | Pcc3(pRG2808) | |
| Pcc3 | − | − | − | − | − |
| Pcc3(pRB41) | +++ | − | +++ | − | +++ |
| Pcc21 | +++ | − | +++ | − | +++ |
| Pcc21DK1 | − | − | − | − | − |
| Pcc21DK1(pRB41) | +++ | − | +++ | − | +++ |
The data represent the averages of three independent experiments. Symbols: +, 0- to 0.5-cm-diameter inhibition zone; ++, 0.5- to 1-cm-diameter inhibition zone; +++, 1- to 1.5-cm-diameter inhibition zone; −, no inhibition.
Characterization of other carocin D features.
How temperature affected the inhibitory activity of carocin D of strain Pcc21 was determined. Cell crude extract containing carocin D completely lost antibacterial activity when it was held at 70°C and 80°C for 10 min. However, when the extract was held at 60°C for 10 min, approximately 50% of the antibacterial activity was retained. As no activity change occurred after 5 min at 60°C, this bacteriocin was relatively resistant to heat (data not shown). The antimicrobial activity was retained with treatment with lipase, DNase, and RNase, but treatment with protease K, trypsin, or pronase E inactivated the antimicrobial activity, indicating that the inhibitory compound was proteinaceous (data not shown). When we tested the antibacterial activity of purified carocin D, up to 5 ng/ml of carocin D showed antibacterial activity against P. carotovorum subsp. carotovorum Pcc3.
Because a ClustalW alignment showed maximum similarity between the C-terminal region of carocin D and the DNase domain of S-type pyocin, the DNase activity of carocin D was tested using total DNA isolated from P. carotovorum subsp. carotovorum Pcc21 cells. Carocin D was able to degrade genomic DNA of P. carotovorum subsp. carotovorum Pcc21. UV or MMC treatment of P. carotovorum subsp. carotovorum Pcc21 increased the DNase activity on genomic DNA (data not shown). The DNase activity was not specific to Pcc21 because carocin D was also able to degrade plasmid DNA and genomic DNAs from other bacteria, including Xanthomonas and Pseudomonas species. Since mutants were screened by the loss of antibacterial activity, no transposon-insertional mutants showed DNase activity (data not shown).
Distribution of carocin D in other P. carotovorum subsp. carotovorum strains in Korea.
The distribution of carocin D was investigated using PCR amplification with primers specific for the caroDK gene. A 2.8-kb product was amplified with the primer pair caro-F-1926 and caro-R-4804 from P. carotovorum subsp. carotovorum Pcc21. Among 54 other strains of P. carotovorum subsp. carotovorum isolated from various hosts (Chinese cabbage, potato, lettuce, tobacco, tomato, carrot, and radish) and diverse geographic regions in Korea tested (17), only five strains of P. carotovorum subsp. carotovorum showed a 2.8-kb product (data not shown).
DISCUSSION
This report shows that a new antibacterial activity of P. carotovorum subsp. carotovorum Pcc21 was due to a new bacteriocin named carocin D. The genetic determinant of carocin D consists of two structural genes, caroDK and caroDI, which have homology to the S-type pyocin and E-group colicin genes that encode the killer protein and immunity protein of P. aeruginosa strains and E. coli, respectively. Expression of these two genes in non-bacteriocin-producing strains P. carotovorum subsp. carotovorum Pcc3, E. coli, and P. carotovorum subsp. carotovorum Pcc21M-15 resulted in the production of a bacteriocin that was released into the growth medium.
Analysis of the caroDK and caroDI genes showed that both genes carried a potential Shine-Dalgarno sequence (GGAGAA) upstream of the ATG start codons. However, a promoter sequence was present only upstream of the caroDK gene, suggesting that the two genes are controlled by one promoter and consist of an operon. In this promoter region were two distinct sequence motifs, a possible SOS box [CTGTAT(N)4ATACAG] and an RdgB box for Mor binding [TAA(N)6TTA]. The SOS box is a binding site for LexA, the repressor of DNA damage-inducible genes. This may explain why expression of carocin D was recA dependent and was not induced in E. coli DH5α but was successfully induced in P. carotovorum subsp. carotovorum and E. coli BL21. The RdgB box was found slightly farther away from the main promoter region (107 bp upstream of the start codon). This box is needed for binding of a DNA damage-responsive transcriptional activator, RdgB (23). This is correlated with induction of the caroDK gene by DNA damage, such as that by UV treatment. It will be interesting to test whether both boxes are important for the regulation of caroDK and caroDI genes under particular environmental conditions.
Based on homology searches, the protein sequence of the caroDK gene product likely has two translocation domains; the N-terminal and C-terminal ones are homologous to the cloacin domain of cloacin and E-group colicins and to the pyocin domain of S-type pyocin, respectively. When tested, none of 5 E. coli strains (DH5α, BL21, EPI300-T1R, EC100 pir+, and JM109) or 5 Pseudomonas aeruginosa strains (ATCC 9027, 10145, 17503, 25619, and 27853) was sensitive to carocin D. To date, only three bacteriocins, klebicin D, colicin D, and colicin D-157, have been shown to have two translocation domains. However, the function of each translocation domain has not yet been clearly determined (3, 5). In the case of colicin D, each translocation domain is correlated with its DNase activity and RNase activity. Carocin D has DNase activity, probably due to the domain at the C-terminal region that is homologous to the DNase domain of S-type pyocin. This may indicate that the C-terminal translocation domain of carocin D is correlated with DNase activity. In contrast, carocin D had no RNase activity (data not shown). Interestingly, carocin D bacteriocin has a putative TonB binding motif (DTMTV) at the N terminus. Some bacteriocins have been shown to enter target cells in a TonB-dependent manner (2, 11), although the exact mechanism is unclear. This may indicate that the N-terminal translocation domain is correlated with binding to receptors for promoting entrance into target cells. It will be interesting to determine whether both translocation domains are required for full bacteriocin activity of carocin D.
Generally, bacteriocins can be divided into three parts: a receptor-binding domain, a translocation domain, and an activity domain. Carocin D seems to have translocation domains and a domain with DNase activity, but it shows no strong homology with any known receptor-binding domain. Because OmpF may be the receptor for carocin D (E. Roh et al., unpublished data), in-depth analysis for a putative receptor-binding domain with other bacteriocins for which the receptor is OmpF was carried out (data not shown). From this analysis, we found weak homology between the receptor-binding domain of colicin E3 and the two translocation domains of carocin D, but more study is necessary to further characterize this.
Among 54 strains of P. carotovorum subsp. carotovorum that we tested, only five strains carried the caroDK gene. This suggests that most of the strains in the field may be sensitive to carocin D. Because E. coli strains carrying caroDK and caroDI genes were resistant to carocin D, carocin D can be readily expressed by transferring these two genes into other bacterial strains that are nonphytopathogenic but can survive the harsh environmental conditions in the field. These engineered strains could then be applied to potentially control the pathogen causing soft-rot disease. The search for new bacteriocins that are effective against plant pathogens that cause serious diseases in important crops will be helpful in developing new and environmentally friendly methods for disease control in the field.
Acknowledgments
This work was supported by a grant (code 20070301034035) from the BioGreen 21 Program, Rural Development Administration, Republic of Korea.
Footnotes
Published ahead of print on 24 September 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Akutsu, A., H. Masaki, and T. Ohta. 1989. Molecular structure and immunity specificity of colicin E6, an evolutionary intermediate between E-group colicins and cloacin DF13. J. Bacteriol. 171:6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cascales, E., S. K. Buchanan, D. Duche, C. Kleanthous, R. Lloubes, K. Postle, M. Riley, S. Slatin, and D. Cavard. 2007. Colicin biology. Microbiol. Mol. Biol. Rev. 71:158-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chavan, M., H. Rafi, J. Wertz, C. Goldstone, and M. A. Riley. 2005. Phage associated bacteriocins reveal a novel mechanism for bacteriocin diversification in Klebsiella. J. Mol. Evol. 60:546-556. [DOI] [PubMed] [Google Scholar]
- 4.Chuang, D. Y., Y. C. Chien, and H. P. Wu. 2007. Cloning and expression of the Erwinia carotovora subsp. carotovora gene encoding the low-molecular-weight bacteriocin carocin S1. J. Bacteriol. 189:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Zamaroczy, M., and R. H. Buckingham. 2002. Importation of nuclease colicins into E. coli cells: endoproteolytic cleavage and its prevention by the immunity protein. Biochimie 84:423-432. [DOI] [PubMed] [Google Scholar]
- 6.Jabrane, A., A. Sabri, P. Compere, P. Jacques, I. Vandenberghe, J. Van Beeumen, and P. Thonart. 2002. Characterization of serracin P, a phage-tail-like bacteriocin, and its activity against Erwinia amylovora, the fire blight pathogen. Appl. Environ. Microbiol. 68:5704-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawamoto, S., J. Shima, R. Sato, T. Eguchi, S. Ohmomo, J. Shibato, N. Horikoshi, K. Takeshita, and T. Sameshima. 2002. Biochemical and genetic characterization of mundticin KS, an antilisterial peptide produced by Enterococcus mundtii NFRI 7393. Appl. Environ. Microbiol. 68:3830-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 9.Michel-Briand, Y., and C. Baysse. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499-510. [DOI] [PubMed] [Google Scholar]
- 10.Montesinos, E. 2007. Antimicrobial peptides and plant disease control. FEMS Microbiol. Lett. 270:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Mora, L., M. Klepsch, R. H. Buckingham, V. Heurgue-Hamard, S. Kervestin, and M. de Zamaroczy. 2008. Dual roles of the central domain of colicin D tRNase in TonB-mediated import and in immunity. J. Biol. Chem. 283:4993-5003. [DOI] [PubMed] [Google Scholar]
- 12.Morgan, S., R. P. Ross, and C. Hill. 1995. Bacteriolytic activity caused by the presence of a novel lactococcal plasmid encoding lactococcins A, B, and M. Appl. Environ. Microbiol. 61:2995-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen, A. H., T. Tomita, M. Hirota, T. Sato, and Y. Kamio. 1999. A simple purification method and morphology and component analyses for carotovoricin Er, a phage-tail-like bacteriocin from the plant pathogen Erwinia carotovora Er. Biosci. Biotechnol. Biochem. 63:1360-1369. [DOI] [PubMed] [Google Scholar]
- 14.Pugsley, A. P. 1984. The ins and outs of colicins. Part I: production, and translocation across membranes. Microbiol. Sci. 1:168-175. [PubMed] [Google Scholar]
- 15.Riley, M. A. 1998. Molecular mechanisms of bacteriocin evolution. Annu. Rev. Genet. 32:255-278. [DOI] [PubMed] [Google Scholar]
- 16.Riley, M. A., and J. E. Wertz. 2002. Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 56:117-137. [DOI] [PubMed] [Google Scholar]
- 17.Roh, E., S. Lee, Y. Lee, D. Ra, J. Choi, E. Moon, and S. Heu. 2009. Diverse antibacterial activity of Pectobacterium carotovorum subsp. carotovorum isolated in Korea. J. Microbiol. Biotechnol. 19:42-50. [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Sano, Y., H. Matsui, M. Kobayashi, and M. Kageyama. 1993. Molecular structures and functions of pyocins S1 and S2 in Pseudomonas aeruginosa. J. Bacteriol. 175:2907-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tagg, J. R., A. S. Dajani, and L. W. Wannamaker. 1976. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 40:722-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitehead, N. A., J. T. Byers, P. Commander, M. J. Corbett, S. J. Coulthurst, L. Everson, A. K. Harris, C. L. Pemberton, N. J. Simpson, H. Slater, D. S. Smith, M. Welch, N. Williamson, and G. P. Salmond. 2002. The regulation of virulence in phytopathogenic Erwinia species: quorum sensing, antibiotics and ecological considerations. Antonie Van Leeuwenhoek 81:223-231. [DOI] [PubMed] [Google Scholar]
- 22.Yamada, K., M. Hirota, Y. Niimi, H. A. Nguyen, Y. Takahara, Y. Kamio, and J. Kaneko. 2006. Nucleotide sequences and organization of the genes for carotovoricin (Ctv) from Erwinia carotovora indicate that Ctv evolved from the same ancestor as Salmonella typhi prophage. Biosci. Biotechnol. Biochem. 70:2236-2247. [DOI] [PubMed] [Google Scholar]
- 23.Yamada, K., J. Kaneko, Y. Kamio, and Y. Itoh. 2008. Binding sequences for RdgB, a DNA damage-responsive transcriptional activator, and temperature-dependent expression of bacteriocin and pectin lyase genes in Pectobacterium carotovorum subsp. carotovorum. Appl. Environ. Microbiol. 74:6017-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]