Abstract
A total of 214 rainwater samples from 82 tanks were collected in urban Southeast Queensland (SEQ) in Australia and analyzed for the presence and numbers of zoonotic bacterial and protozoal pathogens using binary PCR and quantitative PCR (qPCR). Quantitative microbial risk assessment (QMRA) analysis was used to quantify the risk of infection associated with the exposure to potential pathogens from roof-harvested rainwater used as potable or nonpotable water. Of the 214 samples tested, 10.7%, 9.8%, 5.6%, and 0.4% were positive for the Salmonella invA, Giardia lamblia β-giardin, Legionella pneumophila mip, and Campylobacter jejuni mapA genes, respectively. Cryptosporidium parvum oocyst wall protein (COWP) could not be detected. The estimated numbers of Salmonella, G. lamblia, and L. pneumophila organisms ranged from 6.5 × 101 to 3.8 × 102 cells, 0.6 × 10° to 3.6 × 10° cysts, and 6.0 × 101 to 1.7 × 102 cells per 1,000 ml of water, respectively. Six risk scenarios were considered for exposure to Salmonella spp., G. lamblia, and L. pneumophila. For Salmonella spp. and G. lamblia, these scenarios were (i) liquid ingestion due to drinking of rainwater on a daily basis, (ii) accidental liquid ingestion due to hosing twice a week, (iii) aerosol ingestion due to showering on a daily basis, and (iv) aerosol ingestion due to hosing twice a week. For L. pneumophila, these scenarios were (i) aerosol inhalation due to showering on a daily basis and (ii) aerosol inhalation due to hosing twice a week. The risk of infection from Salmonella spp., G. lamblia, and L. pneumophila associated with the use of rainwater for showering and garden hosing was calculated to be well below the threshold value of one extra infection per 10,000 persons per year in urban SEQ. However, the risk of infection from ingesting Salmonella spp. and G. lamblia via drinking exceeded this threshold value and indicated that if undisinfected rainwater is ingested by drinking, then the incidences of the gastrointestinal diseases salmonellosis and giardiasis are expected to range from 9.8 × 10° to 5.4 × 101 (with a mean of 1.2 × 101 from Monte Carlo analysis) and from 1.0 × 101 to 6.5 × 101 cases (with a mean of 1.6 × 101 from Monte Carlo analysis) per 10,000 persons per year, respectively, in urban SEQ. Since this health risk seems higher than that expected from the reported incidences of gastroenteritis, the assumptions used to estimate these infection risks are critically examined. Nonetheless, it would seem prudent to disinfect rainwater for use as potable water.
Roof-harvested rainwater has received significant attention as a potential alternative source of potable and nonpotable water in regions where water is scarce (37). To encourage the use of roof-harvested rainwater, governmental bodies of many countries, such as Australia, Denmark, Germany, India, and New Zealand, are providing subsidies to residents to encourage the use of rainwater for domestic purposes. The use of rainwater is quite common in Australia, particularly in rural and remote areas, where reticulated mains or town water is not available. Recent water scarcity in several capital cities prompted the use of rainwater as an alternative source. For instance, the Queensland State Government initiated the “Home Water Wise Rebate Scheme,” which provides subsidies to Southeast Queensland (SEQ) residents who use rainwater as nonpotable water for domestic purposes (49). Over 260,000 householders were granted subsidies up to December 2008, when the scheme was concluded.
There is a general community feeling that roof-harvested rainwater is safe to drink, and this is partially supported by limited epidemiological evidence (26). Some studies have reported that roof-harvested rainwater quality is generally acceptable for use as potable water (13, 29). In contrast, the presence of potential pathogens, such as Aeromonas spp. Campylobacter spp., Campylobacter jejuni, Salmonella spp., Legionella pneumophila, Giardia spp., Giardia lamblia, and Cryptosporidium spp., in roof-harvested rainwater samples has been reported (2, 9, 34, 45, 47, 48). Such pathogens can cause gastrointestinal illness in humans, with nausea, vomiting, and/or diarrhea occurring within 12 to 72 h (Salmonella enterica serovar Typhimurium) to 9 to 15 days (Giardia lamblia) after ingestion of contaminated water. L. pneumophila can cause the respiratory infection pneumonia, and the fatality rate can be 50% in immunocompromised patients (57).
Direct routine monitoring of the microbiological quality of source water for all possible pathogens is not economically, technologically, or practically feasible. Consequently, traditional fecal indicators, such as fecal coliforms, Escherichia coli, and enterococci, have long been used to determine the presence of pathogens. Most studies assess the quality of roof-harvested rainwater based on the numbers of these fecal indicators (13, 30). However, the major limitation in using fecal bacteria as indicators is their poor correlation with the presence of pathogenic microorganisms in water (2, 30). An alternative is the measurement of pathogens using traditional culture-based methods. However, there are several limitations of such methods, including the underestimation of the bacterial number due to the presence of injured or stressed cells (10) and the fact that certain microorganisms in environmental waters can be viable but not culturable (39). Culture-based methods are also generally laborious and costly. Recent advances in molecular techniques such as PCR technology enable rapid, specific, and sensitive detection of many pathogens. Advances in PCR methodology also enable the quantification of potential pathogens in source water that are otherwise difficult and/or laborious to culture using traditional microbiological methods. In view of this, we used binary PCR (presence/absence)- and quantitative PCR (qPCR)-based assays to first detect and then quantify zoonotic pathogens in samples from roof-harvested rainwater in SEQ residential houses.
The aims of the research study were 2-fold: (i) to quantify the number and frequency of occurrence of Salmonella, G. lamblia, and L. pneumophila organisms in a range of domestic water tanks in SEQ by using qPCR-based methods and (ii) to apply quantitative microbial risk assessment (QMRA) analysis in order to estimate the risk of infection from exposure to these pathogens found in roof-harvested rainwater. The uniqueness of this study stems from the fact that instead of measuring fecal indicators, the pathogens that are capable of causing illness were quantified and this information was combined with QMRA to assess the human health risk of using roof-harvested rainwater as potable or nonpotable water.
MATERIALS AND METHODS
Target pathogens.
C. jejuni, L. pneumophila, Salmonella spp., G. lamblia, and Cryptosporidium parvum were selected because these pathogens could be present in the feces of birds, mammals, and reptiles that have access to roofs. Therefore, following rain events, fecal matter could potentially be transported to tanks via roof runoff.
Sampling and analysis.
In all, 214 samples were collected from 82 residential houses in the Brisbane, Gold Coast, and Sunshine Coast regions. The sizes of the sampled tanks ranged between 500 and 20,000 liters (i.e., polyethylene water tanks), and the end use was either (i) outdoor use (65%), including gardening and car washing, and (ii) indoor use (35%), including drinking, showering, and kitchen use. Water samples were collected in sterilized 10-liter containers from the outlet taps located close to the bases of the tanks. Before the tank was sampled, the tap was sterilized with 70% ethanol and allowed to run for 30 to 60 s to flush out water from the tap. Samples were transported to the laboratory on ice and processed within 6 h. Water samples were collected in two phases. In phase one, a total of 100 water samples were collected from 82 tanks and were screened for the presence/absence of C. jejuni, L. pneumophila, Salmonella spp., G. lamblia, and C. parvum using binary PCR assays (3). In phase two, water samples were collected from a subset of tanks (n = 19) sampled in phase one. Tank water samples which were PCR positive for the selected pathogens in phase one were further sampled in order to obtain information on the temporal occurrence of the pathogens. Samples were collected from these 19 tanks every 2 weeks over a period of 3 months (April to June 2009) commencing with a rainfall event and tested using binary PCR. Quantitative PCR methods were then used to quantify these pathogens for all positively identified samples in phases one and two.
DNA extraction.
For binary PCR and qPCR analyses of bacterial pathogens, a 1-liter water sample from each tank was filtered through a 0.45-μm-pore-size membrane (Advantec, Tokyo, Japan). Samples were processed according to a previously published method (2). DNA was extracted using the DNeasy blood and tissue kit (Qiagen, Valencia, CA) and stored at −80°C until use. For binary PCR and qPCR analyses of pathogenic protozoans, a 2.5-liter water sample from each tank was filtered through a 3-μm-pore-size membrane (Advantec). Samples were processed according to a previously published method (22). DNA was extracted directly to the filter using the DNeasy blood and tissue kit (Qiagen).
PCR positive controls.
Strains and purified DNA were purchased from the American Type Culture Collection (ATCC), as follows: Campylobacter jejuni ATCC 33560D (purified DNA), L. pneumophila ATCC 33152 (strain), Salmonella enterica serovar Typhimurium ATCC 14028 (strain), G. lamblia ATCC 30888D (purified DNA of Portland-1 strain), and C. parvum PRA-67D (purified DNA). Bacterial DNA was extracted from the broth cultures of L. pneumophila and S. Typhimurium strains using the DNeasy blood and tissue kit (Qiagen).
Primers, preparation of standard curves, and PCR conditions.
PCRs of pathogens were performed using previously described primers (3). Standards for qPCR of the L. pneumophila mip, Salmonella invA, and G. lamblia β-giardin genes were prepared from the genomic DNAs of the selected pathogens. The concentration of genomic DNA was determined by measuring the A260 using a Beckman Coulter DU 730 spectrophotometer. The genomic copies were calculated, and a 10-fold dilution ranging from 106 to 10° copies per μl of DNA extract was prepared from the genomic DNA and stored at −20°C until use. For each standard, the concentration was plotted against the cycle number at which the fluorescence signal increased above the threshold value (CT value).
Amplification was performed in 25-μl reaction mixtures using Platinum SYBR green qPCR SuperMix-UDG (Invitrogen, Carlsbad, CA). The PCR mixture contained 12.5 μl of SuperMix, 300 nM each primer, 5.75 μl of DNase- and RNase-free deionized water, and 5 μl of template DNA. For each PCR experiment, corresponding positive DNA and negative controls (i.e., sterile water) were included. The PCRs were performed using the Rotor-Gene 6000 real-time cycler (Corbett Life Sciences).
PCR reproducibility and limit of detection.
The reproducibility of the qPCR was assessed by determining intra-assay repeatability and interassay reproducibility. The coefficient of variation (CV) was calculated using six dilutions (106 to 101 gene copies) of the L. pneumophila, S. Typhimurium, and G. lamblia genomic DNAs. Each dilution was quantified in replicates. The CV for evaluation of intra-assay repeatability was calculated based on the CT value by testing the six dilutions six times in the same experiment. The CV for interassay reproducibility was calculated based on the CT values of six dilutions on six different days. To determine the qPCR limit of detection, known gene copies of L. pneumophila (i.e., 5 × 103 to 5 × 10°), S. Typhimurium (i.e., 5 × 103 to 5 × 10°), and G. lamblia (i.e., 7 × 103 to 7 × 10°) were measured from pure genomic DNA isolated from corresponding control strains and were tested by qPCR. The lowest number of gene copies that was detected consistently in replicate assays was considered the qPCR limit of detection.
Recovery efficiency of the qPCR assays with rainwater samples.
The recovery efficiencies were determined only for Salmonella and G. lamblia qPCR assays. The recovery efficiency for L. pneumophila was assumed to be similar to that for the Salmonella qPCR assay. Deionized water (n = 3) and rainwater (n = 3) samples were spiked with known numbers of S. Typhimurium cells and G. lamblia cysts (obtained from Biotechnology Frontiers, New South Wales, Australia). Initially, samples (n = 3) were collected from several rainwater tanks and were tested for the presence of Salmonella spp. and G. lamblia using binary PCR detection. Water samples from those tanks which showed the absence of Salmonella spp. and G. lamblia were selected for this experiment. The samples were autoclaved to destroy background microbial flora and kept under UV light to minimize any background DNA that could be present. The S. Typhimurium strain was grown overnight in LB broth, and cell numbers were determined using microscopic counts. Ten-fold serial dilutions were made and spiked into 1 liter of deionized water and rainwater samples. Similarly, known numbers of G. lamblia cysts were serially diluted and spiked into 2.5 liters of deionized water and rainwater samples. The samples were filtered through membranes, and DNA extraction was performed according to the method described above. Samples were tested in triplicate for each concentration, and the recovery efficiency (%) was calculated using the following equation: recovery (%) = (number of cells after filtration/number of cells before filtration) × 100. All results were corrected according to their relevant recovery ratios.
Quality control.
To prevent false-positive results for rainwater samples, a method blank was included for each batch (n = 10) of water samples. To prevent false-positive results during DNA extraction, a reagent blank was included in each batch (n = 10) of samples. To separate the specific product from nonspecific products, DNA melting curve analysis was performed for each PCR experiment. Samples were considered to be positive when they were shown to have the same melting temperature as the positive control. To minimize PCR contamination, DNA extraction, PCR setup, and gel electrophoresis were performed in separate laboratories.
QMRA.
We used a four-step quantitative microbial risk assessment (QMRA) process as described by Gerba et al. (20) for estimating the human health risk associated with defined scenarios involving exposure to specified pathogens. The four steps are (i) hazard identification, (ii) exposure assessment, (iii) dose-response assessment, and (iv) risk characterization. The first step of QMRA is hazard identification, which was achieved by collating research literature reporting the presence of specific pathogens such as C. jejuni, L. pneumophila, Salmonella spp., G. lamblia and C. parvum in roof-harvested rainwater tanks (2, 7, 9, 34, 45, 47). The presence of these pathogens (i.e., positive/negative) in a number of water samples was then assessed using binary PCR.
The second step is exposure assessment, where the pathogen number in the source water (i.e., tank rainwater) and the volume ingested/inhaled by a person are estimated. For the estimation of pathogen number, number of the genomic copies (determined by qPCR) of each pathogen was converted to bacterial cells or protozoan cysts. L. pneumophila mip (14) and Salmonella invA (17) are single-copy genes and therefore allow the estimation of cells (i.e., one gene copy of L. pneumophila or Salmonella = one cell of L. pneumophila or Salmonella). The G. lamblia β-giardin gene is expressed as a single-copy gene within the nucleus of each trophozoite (28). Cysts of Giardia contain two trophozoites that have undergone multiple steps of nuclear division, resulting in 16 copies of total genetic information within each cyst (6). Therefore, there are 16 copies of the β-giardin gene per Giardia cyst (22). However, only a proportion of the PCR-detected/quantified cells and cysts may be viable and infectious (51). In the present study, because of a lack of information regarding the proportion of PCR-detected cells and cysts that are viable, it was assumed all the PCR-detected cells and cysts were viable and capable of causing infections. It is acknowledged that this assumption is likely to overestimate the risk of infection by the QMRA analysis. Nonetheless, a highly conservative “worst-case” approach was chosen to determine the risk of infection from exposure to pathogens in roof-harvested rainwater.
To estimate the possible pathogen dose received by an individual, the likely infection routes appropriate to each pathogen must be considered. Infection may occur by ingesting (accidentally during hosing or deliberately via drinking) water containing Salmonella spp. or G. lamblia. Another possible route is to inhale and swallow aerosols containing these pathogens. For L. pneumophila to cause infection, cells must be inhaled deep into the lungs. Given these possible routes, the infection risk associated with each of a total of six scenarios was estimated.
For salmonellosis and giardiasis risk, the scenarios were (i) liquid ingestion due to drinking of rainwater on a daily basis, (ii) accidental liquid ingestion due to garden hosing twice a week, (iii) aerosol ingestion due to showering on a daily basis, and (iv) aerosol ingestion due to hosing twice a week. For legionellosis risk, the scenarios were (i) aerosol inhalation due to showering on a daily basis and (ii) aerosol inhalation due to hosing twice a week. For liquid ingestion, volumes were assumed to be 1,000 ml per day due to drinking (54) and 1 ml per event for accidental liquid ingestion due to hosing (50). For aerosol inhalation during showering, an estimate of the volume of shower water that is deposited in the alveoli of adults is required. This information requires knowledge of the aerosol size distribution at the receptor and the proportion of inhaled aerosols that is deposited in the alveoli of the receptor. This information is quite difficult to obtain. However, several studies which have estimated the aerosol size distributions measured next to a shower were identified (33, 40, 58). Schlesinger (46) provided estimates of alveolar depositional efficiencies across different aerosol size classes. This information was used to adjust the data of O'Toole et al. (40) and Keating and McKone (33) to estimate total deposition in the alveoli. Zhou et al. (58) estimated deposition using a lung model. Based on these estimates, the volume of shower water inhaled was calculated for an adult breathing 20 liters per min, as is consistent with a previous study for “light activity” (27) during a 7-min hot shower. The volume of shower water inhaled, represented by the 0.3- to 6.0-μm (respirable) aerosol size class, was calculated to range from 0.02 μl to 0.84 μl for a 7-min hot shower, across the range of different experimental conditions, shower heads, and flow rates used in the studies. For the scenario of aerosol inhalation by showering, the worst-case volume of 0.84 μl was chosen for subsequent infection risk calculations.
For aerosol inhalation during hosing, it was also assumed that exposure would occur only during that portion of time that the recipient was actually downwind of the hose nozzle. Hence, exposure to aerosols during hosing was assumed to take place for 7 min with the user breathing at 20 liters per min, as for showering. The volume of hosing water that would be deposited in the alveoli of the user was calculated from the aerosol size distributions measured for a hose spraying against a car door. O'Toole et al. (40) adjusted for differences in alveolar depositional efficiencies across aerosol size classes as described by Schlesinger (46). The volume of hose water represented by the 0.3- to 6.0-μm aerosol size class was calculated to be 0.008 to 0.04 μl for a high-pressure hose under spray and jet settings and 0.09 to 0.5 μl for a garden hose with a trigger nozzle using spray or jet settings. For the scenario of aerosol inhalation by hosing, the worst-case volume of 0.5 μl was chosen for subsequent infection risk calculations. It has been suggested that aerosols above 6 μm tend to be deposited in the upper respiratory tract, where they would be swallowed (40). Accordingly, the volume of shower or hose water ingested represented by the >6.0-μm aerosol size class was calculated to range from 58 μl to 1.9 ml for showering and from 0.002 μl to 1.9 μl for hosing, for the same time of exposure and inhalation rate described above. For the scenarios of aerosol inhalation by showering and hosing, the worst-case volumes of 1.9 ml and 1.9 μl, respectively, were chosen for subsequent infection risk calculations.
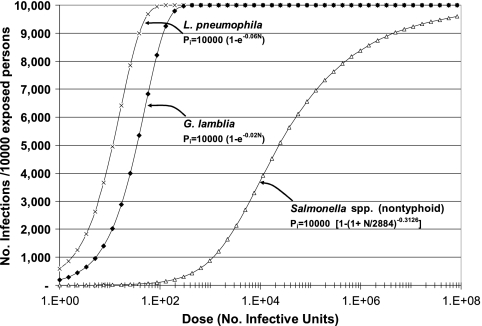
The third step is dose-response assessment, which describes the relationship between the administered dose and the probability of infection in the exposed population. The dose-response relationships used for this study were obtained from the literature. For L. pneumophila (4) and G. lamblia (43), an exponential dose-response model was used, while for Salmonella (23), a beta-Poisson dose-response relationship was used (Fig. 1).
FIG. 1.
Dose-response relationships for a single event. Exponential dose-response relationships were used for Giardia lamblia (43) and Legionella pneumophila (4), and a beta-Poisson dose-response relationship for nontyphoid Salmonella was used for Salmonella serovar Typhimurium) (23). These dose-response relationships relate N, the number of infective units ingested, to Pi, the expected number of infections per 10,000 exposed persons. A second beta-Poisson dose-response relationship for nontyphoid Salmonella relates N to Pill, the number of expected illnesses per 10,000 exposed persons.
The fourth and final step of QMRA is risk characterization, where exposure and dose-response assessment are combined to estimate the probability of infection (expressed as likely numbers of infections per 10,000 persons per year) for the urban SEQ community, and comparison with an arbitrary but commonly accepted risk level of one extra infection per 10,000 persons per year (53). The number of infections caused by a specific pathogen per 10,000 persons in SEQ was determined as the number of infections per 10,000 exposed persons × the proportion of persons in SEQ who were exposed to specific pathogens through drinking, showering, or hosing of rainwater. To estimate the latter, market survey data were used to establish the number of households in Brisbane that use roof-harvested rainwater as potable water (19). From the total number of households in urban SEQ (807,555), the survey estimated that 208,110 had rainwater tanks retrofitted to existing dwellings and that 28,295 were in new dwellings with mandated tanks with internal connections. Within each of these groups, 22% and 19%, respectively, frequently use the rainwater for cooking and drinking purposes (19). This suggests that almost 30% of urban SEQ households possess a rainwater tank and that 6.3% of urban SEQ households use the rainwater for potable water and therefore are at risk of exposure to each pathogen identified in the tank water samples. It was conservatively assumed that all urban SEQ households with rainwater tanks also use the water for their gardens. It was also assumed that only those households using rainwater for potable water used the tank water for showering.
Assuming that the pathogen distribution measured in the sampled roof-harvested rainwater tanks was representative of that in all the tanks in urban SEQ, the percentage of the urban SEQ population exposed to each pathogen could be estimated for each risk scenario as follows: % of rainwater tanks in which the pathogen was detected by binary PCR × % of urban SEQ households that use the rainwater as specified in the risk scenario. The probability of infection per single exposure was converted to the probability of infection per year using the equation for multiple exposures given by Haas et al. (23): number of infections per 10,000 persons in urban SEQ per year = 1 − (1 − Pi)E, where E = number of exposure events per year and Pi = number of infections per 10,000 persons in urban SEQ from a single exposure. The number of exposure events per year was determined as the number of events per year adjusted by the proportion of the year that pathogens were present in the tanks. The latter was estimated from the sampling conducted every 2 weeks in phase two of the study described in “Sampling and analysis” above.
Monte Carlo analysis of scenarios showing a significant health risk.
For those scenarios that indicated a significant health risk (greater than one infection per 10,000 persons in urban SEQ per year) based on “worst-case” assumptions, Monte Carlo analysis was performed using input parameter distributions based on our data and the literature (see Table S1 in the supplemental material) and performing 500,000 runs to determine a more robust probability of obtaining greater than one extra illness per 10,000 persons in urban SEQ per year.
For comparison with epidemiological studies, the probability of infection was converted to the probability of illness by multiplying by the fraction of persons with infections who show symptoms of the illness. For giardiasis, the illness-to-infection (cyst excretion) ratio is highly variable (43), and we assumed that 50% of infections were symptomatic, as reported by Veazie et al. (55) during a giardiasis epidemic. For salmonellosis, FAO/WHO (15) provided a dose-response relationship for illness based on epidemiological data. However, at low doses the fitted relationship indicated illness rates that were higher than those indicated for infection rates using the relationship described by Haas et al. (23). For consistency of approach across all tested pathogens, we chose to use the relationship described by Haas et al. (23) and assumed that 100% of Salmonella infections were symptomatic.
RESULTS
Quantitative PCR standards, reproducibility, and limit of detection.
DNAs from 10-fold dilutions of quantified L. pneumophila, S. Typhimurium, and G. lamblia strains were analyzed in order to determine the reaction efficiencies. The standard curves had a linear range of quantification from 106 to 101 genomic copies per μl of DNA extracts. The amplification efficiencies were >95% for all qPCR, assays and the correlation coefficient (r2) was >0.98 for all three assays. The reproducibility of each qPCR assay was determined by assessing intra-assay and interassay coefficients of variation (CVs) of the standards. The mean intra-assay and interassay CV values and standard deviations, respectively, were 3.4% ± 0.8% and 1.9% ± 1.1% (for L. pneumophila mip), 1.9% ± 0.8% and 1.9% ± 1.3% (for Salmonella invA), and 3.2% ± 1.2% and 4.5% ± 2.1% (for the G. lamblia β-giardin gene), indicating high reproducibility. The qPCR limit of detection was as low as five gene copies for the L. pneumophila mip and Salmonella invA genes. For the G. lamblia β-giardin gene, the limit of detection was seven gene copies.
Recovery efficiency.
The estimated recovery efficiency in autoclaved distilled water samples ranged between 93% and 48% (for Salmonella) and 43% and 23% (for G. lamblia), with the greatest variability occurring at lower cell and cyst counts. The mean recovery efficiencies were 72% ± 16% (for Salmonella) and 35% ± 11% (for G. lamblia). The estimated recovery efficiency in autoclaved rainwater samples ranged between 91% and 45% (for Salmonella) and 41% and 19% (for G. lamblia), with the greatest variability occurring at lower cell and cyst counts. The mean recovery efficiencies were 66% ± 17% (for Salmonella) and 33% ± 12% (for G. lamblia).
Number of pathogens in roof-harvested rainwater.
Of the 214 samples tested during phases one and two, the Salmonella invA, G. lamblia β-giardin, and L. pneumophila mip genes were detected in 23 (10.7%), 21 (9.8%), and 12 (5.6%) rainwater samples, respectively, using binary PCR. However, certain samples were nonquantifiable (Table 1). The C. jejuni mapA gene was detected in one sample by binary PCR but was nonquantifiable. None of the samples were positive for C. parvum oocyst wall protein (COWP) genes. The numbers of Salmonella invA, G. lamblia β-giardin, and L. pneumophila mip genes in quantifiable samples ranged from 6.5 × 101 to 3.8 × 102, 0.9 × 101 to 5.7 × 101, and 6.0 × 101 to 1.7 × 102 genomic copies per 1,000 ml of water, respectively (Table 2). After conversion of number of genomic copies to number of cells, the numbers of Salmonella, G. lamblia, and L. pneumophila organisms in water samples ranged from 6.5 × 101 to 3.8 × 102 cells (for Salmonella spp.), 0.6 × 10° to 3.6 × 10° cysts (for G. lamblia), and 6.0 × 101 to 1.7 × 102 cells (for L. pneumophila) per 1,000 ml of water. The ranges of viable and infective cells and cysts, assuming that 100% of the quantified pathogens are viable and infective, are shown in Table 3.
TABLE 1.
Binary PCR and qPCR results for potential pathogens
| Target pathogen (gene) | No. of binary PCR-positive samples/no. of samples tested (% of positive samples) | No. of qPCR-quantifiable samples/no. of samples tested (% of quantifiable samples) |
|---|---|---|
| C. jejuni (mapA) | 1/214 (0.4) | 0/214 (0) |
| C. parvum (COWP gene) | 0/214 (0) | 0/214 (0) |
| Salmonella (invA) | 23/214 (10.7) | 14/214 (6.5) |
| G. lamblia (β-giardin gene) | 21/214 (9.8) | 17/214 (7.9) |
| L. pneumophila (mip) | 12/214 (5.6) | 9/214 (4.2) |
TABLE 2.
Numbers of genomic copies of pathogens in roof-harvested rainwater samples
| Tank no. (sampling occasion)c | No. of genomic copies per 1,000 ml of water |
||
|---|---|---|---|
| Salmonella invA | G. lamblia β-giardin gene | L. pneumophila mip | |
| 1 (1)a | 7.5 × 101 | 1.6 × 101 | |
| 1 (4)b | 1.1 × 102 | ||
| 2 (1)a | 1.5 × 102 | ||
| 3 (1)a | 1.4 × 102 | ||
| 3 (2)a | 6.5 × 101 | ||
| 3 (3)a | 1.7 × 102 | ||
| 3 (4)b | 1.8 × 102 | 1.1 × 102 | |
| 3 (5)b | 1.0 × 102 | 1.4 × 101 | |
| 7 (1)a | 1.8 × 102 | 5.7 × 101 | 1.0 × 102 |
| 8 (1)a | 1.5 × 102 | ||
| 11 (1)a | 2.7 × 102 | ||
| 11 (3)b | 9.0 × 101 | ||
| 11 (4)b | 1.4 × 102 | ||
| 14 (1)a | 1.4 × 101 | ||
| 15 (1)a | 6.0 × 101 | ||
| 15 (2)b | 2.8 × 101 | ||
| 15 (3)b | 1.3 × 101 | ||
| 18 (1)a | 1.9 × 101 | ||
| 18 (2)b | 2.1 × 102 | 1.1 × 101 | |
| 18 (3)b | 1.1 × 102 | ||
| 20 (1)a | 5.1 × 101 | ||
| 28 (1)a | 7.0 × 101 | ||
| 32 (1)a | 2.1 × 101 | ||
| 32 (2)a | 1.8 × 101 | 8.0 × 101 | |
| 38 (1)a | 3.8 × 102 | 0.9 × 101 | |
| 38 (2)a | 1.6 × 101 | ||
| 39 (1)a | 3.0 × 102 | 4.8 × 101 | |
| 40 (1)a | 3.6 × 101 | ||
| 44 (1)a | 3.3 × 102 | 5.6 × 101 | |
| 45 (1)a | 2.1 × 101 | ||
| Minimum | 6.5 × 101 | 0.9 × 101 | 6.0 × 101 |
| Maximum | 3.8 × 102 | 5.7 × 101 | 1.7 × 102 |
Phase one.
Phase two.
Samples showing no detectable pathogens are not listed.
TABLE 3.
Numbers of viable and infective pathogens in roof-harvested rainwater samples
| Pathogen(s) | Range per 1,000 ml of water |
|
|---|---|---|
| Genomic copies | Cells and cystsa | |
| Salmonella spp. | 6.5 × 101-3.8 × 102 | 6.5 × 101-3.8 × 102 |
| G. lamblia | 9.0 × 10°-5.7 × 101 | 0.6 × 10°-3.6 × 10° |
| L. pneumophila | 6.0 × 101-1.7 × 102 | 6.0 × 101-1.7 × 102 |
Genomic copies were converted to cells and cysts. It is assumed that 100% of the cells and cysts are viable and infective.
Occurrence of pathogens in roof-harvested rainwater.
During phase two of the sampling program, 114 rainwater samples were collected from 19 tanks (i.e., a subset of 82 total tanks) to determine the occurrence of pathogens over time. Pathogens were found to be present in the tanks between 0 and 32% of the time during the 3 months, with averages of 4.4% of the time for Salmonella spp., 5.3% of the time for G. lamblia, and 3.5% of the time for L. pneumophila (Table 4). The overall results suggest that the pathogens are present approximately 5% of the time.
TABLE 4.
Occurrence of pathogens in selected rainwater tanks sampled at every 2 weeks over 3 months
| Tank no. |
Salmonella invA gene |
G. lamblia β-giardin gene |
L. pneumophila mip gene |
|||
|---|---|---|---|---|---|---|
| No. of binary PCR-positive samplings/no. of sampling occasions | Occurrence (%) | No. of binary PCR-positive samplings/no. of sampling occasions | Occurrence (%) | No. of binary PCR-positive samplings/no. of sampling occasions | Occurrence (%) | |
| 1 | 1/6 | 17 | 0/6 | 0 | 0/6 | 0 |
| 2 | 0/6 | 0 | 0/6 | 0 | 0/6 | 0 |
| 3 | 2/6a | 33 | 1/6 | 17 | 1/6 | 17 |
| 7 | 0/6 | 0 | 0/6 | 0 | 0/6 | 0 |
| 8 | 0/6 | 0 | 0/6 | 0 | 0/6 | 0 |
| 11 | 0/6 | 0 | 0/6 | 0 | 2/6a | 33 |
| 12 | 0/6 | 0 | 0/6 | 0 | 0/6 | 0 |
| 14 | 0/6 | 0 | 0/6 | 0 | 0/6 | 0 |
| 15 | 0/6 | 0 | 2/6a | 33 | 0/6 | 0 |
| 18 | 2/6a | 33 | 1/6 | 17 | 0/6 | 0 |
| 20 | 0/6 | 0 | 0/6 | 0 | 0/6 | 0 |
| 28 | 0/6 | 0 | 1/6 | 17 | 0/6 | 0 |
| 32 | 0/6 | 0 | 0/6 | 0 | 1/6 | 17 |
| 33 | 0/6 | 0 | 1/6 | 17 | 0/6 | 0 |
| 38 | 0/6 | 0 | 0/6 | 0 | 0/6 | 0 |
| 39 | 0/6 | 0 | 0/6 | 0 | 0/6 | 0 |
| 40 | 0/6 | 0 | 0/6 | 0 | 0/6 | 0 |
| 44 | 0/6 | 0 | 0/6 | 0 | 0/6 | 0 |
| 45 | 0/6 | 0 | 0/6 | 0 | 0/6 | 0 |
| Avg % occurrence | 4.4 | 5.3 | 3.5 | |||
PCR-positive samples were obtained on two consecutive occasions.
Likely dose received by exposed persons.
Estimates of the ingestion dose (i.e., range) of each pathogen per person exposed according to the six scenarios are shown in Table 5. Due to the 100-fold-higher number of Salmonella than G. lamblia organisms in the roof-collected rainwater samples, the Salmonella dose was 2 orders of magnitude higher than that of G. lamblia. The liquid ingestion dose via drinking was several orders of magnitude greater than the dose received by the other scenarios due to the greater volume of water ingested. For liquid ingestion via drinking, 6.5 × 101 to 3.8 × 102 Salmonella cells and 5.6 × 10−1 to 3.6 × 10° G. lamblia cysts may be ingested.
TABLE 5.
Exposure pathways, ingested/inhaled volumes, and calculated ingested/inhaled pathogen doses for individuals exposed to tank water containing pathogens
| Pathogen exposure and risk scenario | Vol per day or event | Dose range (cells or cysts) |
|---|---|---|
| Salmonella spp. | ||
| Liquid ingestion via drinking | 1,000 ml | 6.5 × 101-3.8 × 102 |
| Liquid ingestion via hosing | 1 ml | 6.5 × 10−2-3.8 × 10−1 |
| Aerosol ingestion via showering | 1.9 ml | 1.2 × 10−1-7.2 × 10−1 |
| Aerosol ingestion via hosing | 1.9 μl | 1.2 × 10−4-7.2 × 10−4 |
| G. lamblia | ||
| Liquid ingestion via drinking | 1,000 ml | 5.6 × 10−1-3.6 × 10° |
| Liquid ingestion via hosing | 1 ml | 5.6 × 10−4-3.6 × 10−3 |
| Aerosol ingestion via showering | 1.9 ml | 1.0 × 10−3-6.8 × 10−3 |
| Aerosol ingestion via hosing | 1.9 μl | 1.1 × 10−6-6.8 × 10−6 |
| L. pneumophila | ||
| Aerosol inhalation via showering | 0.84 μl | 5.2 × 10−5-1.4 × 10−4 |
| Aerosol inhalation via hosing | 0.5 μl | 3.0 × 10−5-8.4 × 10−5 |
Infection risk for exposed persons.
Because G. lamblia is more infectious than Salmonella spp. according to the dose-response relationships used, the infection risks per 10,000 exposed persons per exposure event for Salmonella spp. and G. lamblia were similar for each scenario (Table 6). L. pneumophila is indicated to be the most infectious of the three pathogens (Fig. 1), but due to the low doses received by the aerosol inhalation infection route, the infection risk per 10,000 exposed persons per event remained low (up to 8.6 × 10−2) compared to those calculated for Salmonella spp. and G. lamblia for the routes of liquid ingestion via drinking (up to 6.8 × 102), liquid ingestion via hosing (up to 7.1 × 10−1), and aerosol ingestion via showering (up to 1.3 × 10°). Very low risks of infection were calculated for the route of aerosol ingestion via hosing for Salmonella spp. and G. lamblia (up to 1.3 × 10−3) due to the extremely low volumes ingested (Table 6).
TABLE 6.
Infection risks for individuals exposed to contaminated tank water for six risk scenarios
| Pathogen exposure and risk scenario | Range of infection risk/10,000 exposed people with rainwater tanks from single event | % of SEQ population exposed to pathogensa | Range of infection risk/10,000 people in SEQ from single event | No. of events/yrb | Range of infection risk/yr (no./10,000 people in SEQ) |
|---|---|---|---|---|---|
| Salmonella spp. | |||||
| Liquid ingestion via drinking | 6.9 × 101-3.8 × 102 | 0.77 | 5.4 × 10−1-2.9 × 10° | 18.3 | 9.8 × 10°-5.3 × 101 |
| Liquid ingestion via hosing | 7.0 × 10−2-4.1 × 10−1 | 3.66 | 3.0 × 10−3-1.5 × 10−2 | 5.2 | 1.0 × 10−2-8.0 × 10−2 |
| Aerosol ingestion via showering | 1.3 × 10−1-7.7 × 10−1 | 0.77 | 1.0 × 10−3-5.0 × 10−3 | 18.3 | 1.9 × 10−2-1.0 × 10−1 |
| Aerosol ingestion via hosing | 1.4 × 10−4-7.9 × 10−4 | 3.66 | 5.0 × 10−6-2.9 × 10−5 | 5.2 | 2.6 × 10−5-1.5 × 10−4 |
| G. lamblia | |||||
| Liquid ingestion via drinking | 1.1 × 102-6.8 × 102 | 1.01 | 1.1 × 10°-6.9 × 10° | 18.3 | 2.0 × 101-1.3 × 102 |
| Liquid ingestion via hosing | 1.1 × 10−1-7.1 × 10−1 | 4.76 | 5.0 × 10−3-3.4 × 10−2 | 5.2 | 3.0 × 10−2-1.8 × 10−1 |
| Aerosol ingestion via showering | 2.1 × 10−1-1.3 × 10° | 1.01 | 2.1 × 10−3-1.3 × 10−2 | 18.3 | 3.9 × 10−2-2.4 × 10−1 |
| Aerosol ingestion via hosing | 2.2 × 10−4-1.3 × 10−3 | 4.76 | 1.0 × 10−5-6.5 × 10−5 | 5.2 | 5.3 × 10−5-3.3 × 10−4 |
| L. pneumophila | |||||
| Aerosol inhalation via showering | 3.0 × 10−2-8.6 × 10−2 | 0.46 | 1.4 × 10−4-4.0 × 10−4 | 18.3 | 2.6 × 10−3-7.3 × 10−3 |
| Aerosol inhalation via hosing | 1.8 × 10−2-5.1 × 10−2 | 2.20 | 4.0 × 10−4-1.1 × 10−3 | 5.2 | 2.1 × 10−3-5.8 × 10−3 |
Estimated for each risk scenario as % of rainwater tanks in which the pathogen was detected by binary PCR × % of urban SEQ households that use the rainwater as specified in the risk scenario.
The number of days or events per year when exposure occurs is estimated assuming that pathogens are present in the tank water only 5% of the time. Drinking 1 liter and showering are assumed to be daily events, while hosing is assumed to be a biweekly event.
Infection risk for SEQ population.
Of the 82 tanks tested, 10 (12.2%), 13 (15.9%), and 6 (7.3%) were positive for Salmonella spp., G. lamblia, and L. pneumophila, respectively, on at least one sampling occasion. By multiplying the percentage of positive tanks for each pathogen by the proportion of the urban population that have a tank and use the water for drinking and/or hosing, the fraction of the urban SEQ population potentially exposed to each pathogen was calculated to range from 0.46% to 4.76% across the different scenarios (Table 6). By multiplying the infection risk per 10,000 exposed persons per event by the fraction of the population that was exposed to each pathogen, the risks of infection from Salmonella spp., G. lamblia, and L. pneumophila per 10,000 persons in urban SEQ per event were calculated. Although the percentage of persons in urban SEQ drinking or showering with tank water was 4-fold less than that of those using tank water for hosing, the infection risk from liquid ingestion via drinking was still several orders of magnitude greater than the risks indicated for the other scenarios (Table 6). Finally, by multiplying the infection risk per 10,000 persons in urban SEQ per event by number of such exposure events per year, the infection risk per 10,000 persons in urban SEQ per year was calculated. Both Salmonella spp. and G. lamblia showed infection risks with similar orders of magnitude for each scenario (Table 6), with values up to 1.2 × 102 for liquid ingestion via drinking, 2.4 × 10−1 for aerosol ingestion via showering, 1.8 ×10−1 for liquid ingestion via hosing, and 3.4 × 10−4 for aerosol ingestion via hosing. L. pneumophila showed much lower infection risks for aerosol inhalation, with a maximum value of 7.3 × 10−3. Using a threshold value of one extra infection per 10,000 persons per year, it is apparent that of all the scenarios considered, only those involving liquid ingestion via drinking present an unacceptable level of risk from infection. The calculations for infection based on the “worst-case” assumptions indicated that if undisinfected rainwater is ingested by drinking, then the infection incidence is expected to range from 9.8 × 10° to 5.4 × 101 (Salmonella spp.) and from 2.0 × 101 to 1.3 × 102 (G. lamblia) cases per 10,000 persons in urban SEQ per year.
Monte Carlo analysis.
Monte Carlo analysis of the risks of Salmonella sp. and G. lamblia infection from drinking roof-harvested rainwater indicated 18% and 27% probabilities, respectively, of obtaining one or more extra infections per 10,000 persons in urban SEQ. The mean numbers of infections per 10,000 persons in urban SEQ across all 500,000 simulations were 1.2 × 101 (for Salmonella spp.) and 3.1 × 101 (G. lamblia). Using the assumptions defined in the methodology for relating infection to illness incidence, these values would equate to 9.8 × 10° to 5.4 × 101 cases (mean from Monte Carlo analysis of 1.2 × 101) of salmonellosis and 1.0 × 101 to 6.5 × 101 cases (mean from Monte Carlo analysis of 1.6 × 101) of giardiasis per 10,000 persons in urban SEQ per year.
DISCUSSION
Study approach.
In this study, qPCR methods were used to quantify bacterial and protozoal pathogens in a large number of water samples (n = 214) from roof-harvested rainwater tanks. One advantage of PCR methods is that they could be used to detect and quantify specific pathogens with greater specificity than with traditional culture-based methods for the detection of pathogens in water (35, 51). In addition, qPCR detection of pathogens is rapid compared to traditional culture-based methods. The PCR methods used in this study were rigorously evaluated prior to being used to detect and quantify these pathogens in tank rainwater samples. The specificities of primers were determined against known microbial genomes and sequences with the Basic Local Alignment Search Tool (BLAST) program to ensure that no homology with known gene sequences of other microorganisms commonly found in water was detected. The cross-reactivity of each primer set was also evaluated by testing DNAs isolated from other nontarget species of microorganisms commonly found in water (2). The primers used in this study did not amplify any PCR products other than those products that were expected. An experiment was conducted to determine the potential presence of PCR-inhibitory substances in rainwater samples collected from three different tanks. The results indicated that the tested rainwater samples were free of PCR-inhibitory substances (2).
Sources of contamination.
Rainwater tanks may become contaminated due to material being washed into the tank from the roof and gutters following rain events. The primary sources of pathogens are likely to be from fecal materials from birds, lizards, and possums which have access to the roof. Indeed, an anecdote from the study was that tank water contamination was evident from the observation of obvious accumulation of bird feces under a television antenna located on the roof area connected to the tank. One rainwater sample in this study was positive for C. jejuni mapA genes. C. jejuni is recognized as one of the etiologic agents of acute diarrheal disease (rather than general Campylobacter spp.) and could potentially be from bird feces (32, 56). However, other potential sources, such as feces of possums or lizards, cannot be ruled out. Campylobacter spp. could not be isolated from possum feces in New Zealand (11), while the presence of Campylobacter spp. in roof-harvested rainwater samples in New Zealand has been reported (45). The Salmonella spp. detected in rainwater samples could potentially be from bird feces (21). Both Legionella spp. and Salmonella spp. have previously been detected in roof-harvested rainwater cisterns and/or in tanks, using culture-based methods, in the United States, in New Zealand, and in the tropics (7, 47). The Giardia lamblia β-giardin gene, which was detected in 21 out of 214 samples (9.8%) tested in phases one and two in this study, has also been reported to have a high prevalence (45%) in rainwater cisterns in the U.S. Virgin Islands (9). However, no Cryptosporidium parvum was detected in this study, unlike in the study by Crabtree et al. (9), which found that 23% of rainwater cisterns in the U.S. Virgin Islands were contaminated with this pathogen. Possums are known to be carriers of Giardia cysts (36), and thus it could be that these possums are a major source of Giardia spp. in roof-harvested rainwater in Queensland while wild birds or other animals are the source of the Cryptosporidium spp. in the other studies. Another potential source which deserves investigation is the fruit bat, since there are several fruit bat colonies in the Brisbane area and these bats fly over and drop feces on house roofs.
Occurrence of pathogens.
It is expected that pathogens present in feces on a roof would succumb to desiccation, high temperatures, and UV radiation. However, pathogens that become associated with organic matter in the gutters could be protected and thus have a longer survival capacity. These protected pathogens would have a greater potential to be washed in a viable and infective state into the tank during a rainfall event than pathogens deposited directly on the roof. Hence, pathogen contamination of rainwater tanks may be episodic and would be dependent on the time between deposition on roofs and rainfall events. A critical piece of information required for QMRA was the proportion of time that the pathogens were present in rainwater tanks. The occurrence of pathogens in roof-harvested rainwater is not well documented in research literature. Most studies collected one-off samples from rainwater tanks and screened for a number of pathogens using traditional culture- and PCR-based methods (2, 47). However, the results of the 3-month sampling study suggest that it is unlikely that pathogens will be present in tanks all the time, with pathogens occurring only up to 5% of the time. Fewtrell and Kay (16), who undertook a recent risk assessment analysis with respect to Campylobacter spp. in toilets flushed with roof-harvested rainwater, assumed that C. jejuni was present in tank water from 0 to 10% of the time.
It must be noted that the presence of pathogens in the tank water is likely to be strongly related to rainfall events. This is because pathogens in feces deposited on the roof are generally washed off to the tanks after rainfall events. Rainwater samples in phase one were collected immediately after rainfall events (data not shown), while only the first samples in phase two (3-month survey) were collected following rainfall. Dry conditions accompanied sample collection throughout the remainder of the survey period. The highest levels of contamination were indeed shown by samples that were taken immediately following rainfall events (data not shown). This suggests that for areas with frequent light rains throughout the year, pathogens may be present for a larger proportion of the time than what we have assumed for SEQ in our calculations (i.e., 5%). Therefore, it is possible that we are underestimating the risk of infection in our calculations. This warrants a more rigorous survey of rainwater tanks (possibly 1 year of sampling) to obtain more accurate information regarding the occurrence of pathogens. Understanding pathogen inactivation dynamics inside rainwater tanks may provide important information on the occurrence of pathogens. We are currently investigating the inactivation of Salmonella spp. in rainwater tanks in SEQ.
Viability and infectivity of pathogens.
One major limitation of the PCR method is that it does not provide information regarding the viability and infectivity of detected pathogens, which is critical for QMRA analysis. In the absence of such data, it was assumed that 100% of the PCR-detected cells and cysts are viable and infective. This assumption is likely to overestimate the risk of infection, as 100% of the PCR-detected cells and cysts may not be viable. However, the overestimation of risk could be preferable to an underestimation obtained via culture-based methods (52). This is a critical issue and warrants more rigorous investigation with environmental water samples where pathogens might inactivate rapidly due to environmental factors such as temperature, predation, and sunlight. To overcome this problem, qPCR could be integrated with cell culture (12) or dyes (i.e., ethidium monoazide and propidium monoazide, which penetrates only dead cells) (41, 44) in order to obtain information regarding the viability of bacterial and protozoal pathogens.
Health risks associated with roof-harvested rainwater.
The results of QMRA based on the assumptions discussed above indicate that the only likely risk encountered from the roof-harvested rainwater samples was from drinking water contaminated with Salmonella spp. and G. lamblia. L. pneumophila, at the levels detected in the roof-harvested rainwater samples, did not present a threat for uses of tank water as potable water. Uses of the tank water as nonpotable water also presented no threat to human health at the pathogen numbers detected. This indicates that there is little risk from using roof-harvested rainwater as nonpotable water, which is important given that 260,000 subsidies have been granted to SEQ households for installing rainwater tanks.
The numbers of illnesses per 10,000 persons in urban SEQ per year ranged from 9.8 × 10° to 5.4 × 101 (mean from Monte Carlo analysis of 1.2 × 101) for salmonellosis and from 1.0 × 101 to 6.5 × 101 cases (mean from Monte Carlo analysis of 1.6 × 101) for giardiasis. The incidences of these diseases reported in the Notifiable Diseases Surveillance System Database (http://www9.health.gov.au/cda/Source/CDA-index.cfm) are 5.7 cases of salmonellosis per 10,000 people in Queensland and up to 5 cases of giardiasis per 10,000 people in other states (giardiasis is not a notifiable disease in SEQ) over the past 10 years. Hence, the QMRA suggests that the additional use of roof-harvested rainwater in urban SEQ may in fact substantially increase the incidences of salmonellosis and giardiasis. No such rise in reported cases of salmonellosis in SEQ over recent years is apparent.
A number of explanations for this discrepancy are possible. There is a naturally high incidence of gastroenteritis in the community (e.g., 8,000 cases per 10,000 people per year), which may mask the actual disease (25). Before the disease can be reported in the Notifiable Diseases Surveillance System Database, it must first be identified, and not every individual will seek medical attention if the illness is mild and lasts only for a few days. Consequently, the incidence of disease indicated in the Notifiable Diseases Surveillance System Database is at best a minimum value and may be substantially underestimating actual disease incidence. Hall et al. (24) estimated that between 8 and 11% of Campylobacter- and Salmonella-related illnesses are reported.
Since 10% of the population in Australia currently use roof-harvested rainwater as a major source of their drinking water and an additional ∼5% use roof-harvested rainwater for showering, toilet flushing, and clothes laundering (1), we would expect that if rainwater tanks were a public health hazard, then ample evidence for this would exist today. A literature search indicated that to date, several disease outbreaks and clinical cases associated with rainwater consumption have been reported worldwide (5, 8, 38, 47). Australian epidemiological studies have failed to show a significant rise in cases of gastroenteritis due to drinking of roof-harvested rainwater. An epidemiological study in South Australia among 4- to 6-year-old children suggested that roof-harvested rainwater posed no increased risk of gastroenteritis compared with mains water (26). Recently, a double-blinded, randomized controlled trial among nonimmunocompromised rainwater drinkers in Adelaide using sham or active water treatment units also showed no significant increase in the incidence of highly credible gastroenteritis from the consumption of untreated rainwater (42). However, such results should be interpreted with care due to the lack of sensitivity of the epidemiological tool to detect gastroenteritis (31). In addition, considering the high costs and time required, epidemiological studies with sources such as roof-harvested rainwater may not be practical for the sensitive detection of gastroenteritis in the community. Heyworth et al. (26) also pointed out that their data could also have reflected a level of acquired immunity among regular users of roof-harvested rainwater and therefore may not reflect the actual risk to new users.
Another limitation is that cases of gastroenteritis due to drinking untreated rainwater could actually be masked by the background levels of gastroenteritis from other sources, such as consumption of food and community-based infections. Hellard et al. (25) suggested that the incidence of gastroenteritis in the community could be as high as 8,000 cases per 10,000 people per year.
Similarly, the methodology used to estimate health risk could have inflated the risk calculated due to the assumption of 100% of pathogen cells or cysts being viable and infective. The QMRA also did not take into account households that used effective disinfection treatment of rainwater before using it as potable water. Treatments such as UV disinfection or boiling of the water before its use as potable water would eliminate/reduce exposure of individuals to pathogens and hence infection. Another factor is the possibility of individuals acquiring immunity to certain pathogens due to frequent exposure. However, to counterbalance this, no attempt was made to include the greater infection risk to the elderly or immunocompromised for a given dose, since the dose-response relationships described by Haas et al. (23) that were used in the QMRA were based on healthy adults and these relationships were applied uniformly across the population. There are uncertainties about the dose-infection response relationship and its relationship to illness response, particularly in view of the epidemiology-based relationship for Salmonella spp. (15). There were also uncertainties in the measurement of the proportion of time that pathogens occurred in the tank due to the bimonthly sampling regime. A more rigorous estimation of this value is warranted, since any errors will have a direct effect on the risk calculation (e.g., risk would increase approximately 20-fold if pathogens are present 100% of the time). Nevertheless, until more data become available to reduce some of these uncertainties, the results indicate that it would be prudent to disinfect roof-harvested rainwater, such as by the installation of a UV disinfection unit, boiling, or other forms of disinfection, before using it as potable water, especially for drinking.
Conclusions.
Recent water restrictions in several capital cities in Australia and drought conditions have resulted in the installation of rainwater tanks at rates not seen before. The increasing role being played by rainwater tanks in water security in SEQ, including the mandating of rainwater tanks for all new developments in SEQ, means that tank and roof hygiene will assume greater importance in the future. Therefore, the development of a robust methodology for the assessment of the possible health risk from roof-harvested rainwater is essential. We believe that the methodology developed so far provides a step towards achieving this objective, but further refinements will be needed to provide a better estimate of health risk. It is evident that further information relating to the occurrence of pathogens throughout the year and the viability of pathogens in roof-harvested rainwater tanks is needed. Currently, a study is being designed in which a number of rainwater tanks will be surveyed for a year for the presence of pathogens in order to obtain information regarding their seasonal persistency and variability. Culture-based methods and qPCR methods incorporating dyes such as propidium iodide and propidium monoazide will be incorporated into the methodology to provide information on the viability of the cells detected in water samples.
Current estimates of health risk suggests that it would be prudent to disinfect roof-harvested rainwater, such as by the installation of a UV disinfection unit, boiling, or other forms of disinfection, before using it as potable water, especially for drinking. This would be especially prudent for the elderly and immunocompromised. Maintenance of good roof and gutter hygiene and elimination of overhanging tree branches and other structures where possible to prevent the congregation of animals are also recommended. Inclusion of giardiasis in the notifiable disease list in Queensland should be considered, given that G. lamblia was found in rainwater tank samples.
Supplementary Material
Acknowledgments
This study, a collaborative research project between DERM and the Queensland University of Technology (QUT), was funded by the Queensland Department of Environment and Resource Management (DERM).
We thank Robert Huston, University of Queensland (UQ), for arranging access to some of the tanks and those residents of Queensland who provided tank water samples.
Footnotes
Published ahead of print on 17 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.ABS. 2007. Environmental issues: people's views and practices (no. 4602.0). Australian Bureau of Statistics, Canberra, Australia.
- 2.Ahmed, W., F. Huygens, A. Goonetilleke, and T. Gardner. 2008. Real-time PCR detection of pathogenic microorganisms in roof-harvested rainwater in Southeast Queensland, Australia. Appl. Environ. Microbiol. 74:5490-5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed, W., A. Goonetilleke, and T. Gardner. 2010. Implications of faecal indicator bacteria for the microbiological assessment of roof-harvested rainwater quality in Southeast Queensland, Australia. Can. J. Microbiol. 56:471-479. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong, T. W., and C. N. Haas. 2008. Legionnaires’ disease: evaluation of a quantitative microbial risk assessment model. J. Water Health 6:149-166. [DOI] [PubMed] [Google Scholar]
- 5.Ashbolt, R., and M. D. Kirk. 2006. Salmonella Mississippi infections in Tasmania: the role of native Australian animals and untreated drinking water. Epidemiol. Infect. 134:1257-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernander, R., J. E. D. Palm, and S. G. Svärd. 2001. Genome ploidy in different stages of the Giardia lamblia life cycle. Cell. Microbiol. 3:55-62. [DOI] [PubMed] [Google Scholar]
- 7.Broadhead, A. N., A. Negron-Alvira, L. A. Baez, T. C. Hazen, and M. J. Canoy. 1998. Occurrence of Legionella species in tropical rain water cisterns. Caribb. J. Sci. 24:71-73. [Google Scholar]
- 8.Brodribb, R., P. Webster, and D. Farrell. 1995. Recurrent Campylobacter fetus subspecies fetus bacteraemia in a febrile neutropaenic patient linked to tank water. Commun. Dis. Intell. 19:312-313. [Google Scholar]
- 9.Crabtree, K. D., R. H. Ruskin, S. B. Shaw, and J. B. Rose. 1995. The detection of Cryptosporidium oocysts and Giardia cysts in cistern water in the U.S. Virgin Islands. Water Res. 30:208-216. [Google Scholar]
- 10.Delgado-Viscogliosi, P., T. Simonart, V. Parent, G. Marchand, P. E. Pierlot, et al. 2005. Rapid method for enumeration of viable Legionella pneumophila and other Legionella spp. in water. Appl. Environ. Microbiol. 71:4086-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devane, M. L., C. Nicol, A. Ball, J. D. Klena, P. Scholes, J. A. Hudson, M. G. Baker, B. J. Gilpin, N. Garrett, and M. G. Savill. 2005. The occurrence of Campylobacter subtypes in environmental reservoirs and potential transmission routes. J. Appl. Microbiol. 98:980-990. [DOI] [PubMed] [Google Scholar]
- 12.Di Giovanni, G. D., F. H. Hashemi, N. J. Shaw, F. A. Abrams, M. W. LeChevallier, and M. Abbaszadegan. 1999. Detection of infectious Cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture PCR. Appl. Environ. Microbiol. 65:3427-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillaha, T. A., and W. J. Zolan. 1985. Rainwater catchment water quality in Micronesia. Water Res. 19:741-746. [Google Scholar]
- 14.Engleberg, N. C., C. Carter, D. R. Weber, N. P. Cianciotto, and B. I. Eisenstein. 1989. DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect. Immun. 57:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.FAO/WHO. 2002. Risk assessments of Salmonella in eggs and broiler chickens. Microbiological risk assessment series 2. World Health Organization, Food and Agriculture Organization of the United Nations, Geneva, Switzerland.
- 16.Fewtrell, L., and D. Kay. 2007. Quantitative microbial risk assessment with respect to Campylobacter spp. in toilets flushed with harvested rainwater. 21:275-280. [Google Scholar]
- 17.Fey, A., S. Eichler, S. Flavier, R. Christen, M. G. Hofle, and C. A. Guzman. 2004. Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water using salmonella as a model organism. Appl. Environ. Microbiol. 70:3618-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Gardiner, A. 2009. Domestic rainwater tanks: usage and maintenance patterns in Southeast Queensland. AWA Water 36:151-156. [Google Scholar]
- 20.Gerba, C. P., J. B. Rose, and C. N. Haas 1996. Quantitative microbial risk assessment for reclaimed wastewater, p. 254-260. In Water Tech Sydney. AWWA, St. Leonard's, Australia.
- 21.Gupta, B. R., and J. C. Verma. 1989. Prevalence of Salmonella from avian sources. Ind. J. Comp. Microbiol. Immunol. Infect. Dis. 10:140-145. [Google Scholar]
- 22.Guy, R. A., P. Payment, U. J. Krull, and P. A. Hörgen. 2003. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 69:5178-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas, C. N., J. B. Rose, and C. P. Gerba. 1999. Quantitative microbial risk assessment. John Wiley & Sons, Inc., New York, NY.
- 24.Hall, G., J. Raupach, and K. Yohannes. 2006. An estimate of under-reporting of foodborne notifiable diseases: Salmonella, Campylobacter, Shiga toxin producing E. coli (STEC). National Centre for Epidemiology and Population Health Working Paper no. 52. http://nceph.anu.edu.au/Publications/Working_Papers/WP52.pdf. (Accessed July 2009.)
- 25.Hellard, M. E., M. I. Sinclair, A. B. Forbes, and C. K. Fairley. 2001. A randomised blinded, controlled trial investigating the gastrointestinal health effects of drinking water quality. Environ. Health Pers. 109:773-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyworth, J. S., G. Glonek, E. J. Maynard, P. A. Baghurst, and J. Finlay Jones. 2006. Consumption of untreated tank rainwater and gastroenteritis among young children in South Australia. Int. J. Epidemiol. 35:1051-1058. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann, W., T. B. Martonen, and R. C. Graham. 1989. Predicted deposition of nonhygroscopic aerosols in the human lung as a function of subject age. J. Aerosol Med. 2:49-68. [Google Scholar]
- 28.Holberton, D. V., and J. Marshall. 1995. Analysis of consensus sequence patterns in Giardia cytoskeleton gene promoters. Nucleic Acids Res. 23:2945-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holländer, R., M. Bullermann, C. Grob, H. Hartung, K. König, F.-K. Lücke, and E. Nolde. 1996. Microbiological and hygienic aspects of the use of rainwater as process water for toilet flushing, garden irrigation and laundering. Gesundheitswesen 58:288-293. (In German.) [PubMed] [Google Scholar]
- 30.Hörman, A., R. Rimhannen-Finne, L. Maunula, C.-H. von Bonsdorff, N. Torvela, A. Heikinheimo, and M.-L. Hänninen. 2004. Campylobacter spp., Giardia spp., Cryptosporidum spp., noroviruses, and indicator organisms in surface water in southwestern Finland, 2000-2001. Appl. Environ. Microbiol. 70:87-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hrudey, S. E., and E. J. Hrudey. 2004. Safe drinking water: lessons from recent outbreaks in affluent countries. IWA Publishing, London, United Kingdom.
- 32.Kapperud, G., and O. Rosef. 1983. Avian wildlife reservoir of Campylobacter fetus subsp. jejuni, Yersinia spp., and Salmonella spp. in Norway. Appl. Environ. Microbiol. 45:375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keating, G. A., and T. E. McKone. 1993. Measurements and evaluation of the water-to-air transfer and air concentration of trichloroethylene in a shower chamber. Modeling of indoor air quality and exposure. American Society for Testing Materials, Philadelphia, PA.
- 34.Lye, D. J. 2002. Health risks associated with consumption of untreated water from household roof catchment system. J. Am. Water Res. Assoc. 38:1301-1306. [Google Scholar]
- 35.Malorny, B., C. Löfström, M. Wagner, N. Krämer, and J. Hoorfar. 2008. Enumeration of Salmonella bacteria in food and feed samples by real-time PCR for quantitative microbial risk assessment. Appl. Environ. Microbiol. 74:1299-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marino, M. R., T. J. Brown, D. C. Waddington, R. E. Brockie, and P. J. Kelly. 1992. Giardia intestinalis in North Island possums, house mice and ship rats. N. Z. Vet. J. 40:24-27. [DOI] [PubMed] [Google Scholar]
- 37.Meera, V., and M. Ahammed. 2006. Water quality of rooftop harvesting systems: a review. J. Water Supply Res. Technol. 55:257-268. [Google Scholar]
- 38.Merritt, A., R. Miles, and J. Bates. 1999. An outbreak of Campylobacter enteritis on an island resort, north Queensland. Commun. Dis. Intell. 23:215-219. [DOI] [PubMed] [Google Scholar]
- 39.Oliver, J. D. 2000. The public health significance of viable but nonculturable bacteria, p. 277-300. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, DC.
- 40.O'Toole, J., K. Leder, and M. Sinclair. 2008. A series of exposure experiments—recycled water and alternative water sources. A. Aerosolsizing and endotoxin experiments. Research report 45. CRC for Water Quality and Treatment, Adelaide, Australia.
- 41.Rawsthorne, H., C. N. Dock, and L. A. Jaykus. 2009. PCR-based method using propidium monoazide to distinguish viable from nonviable Bacillus subtilis spores. Appl. Environ. Microbiol. 75:2936-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigo, S., M. Sinclair, A. Forbes, D. Cunliffe, and K. Leder. 19 August 2010, posting date. Drinking rainwater: a double-blinded, randomised controlled study of water treatment filters and gastroenteritis incidence. Am. J. Public Health doi: 10.2105/AJPH.2009.185389. [DOI] [PMC free article] [PubMed]
- 43.Rose, J. B., C. N. Haas, and S. Regli. 1991. Risk assessment and control of waterborne giardiasis. Am. J. Public Health 81:709-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rudi, K., B. Moen, S. M. Dromtrop, and A. L. Holck. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex cells. Appl. Environ. Microbiol. 71:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savill, M. G., J. A. Hudson, A. Ball, J. D. Klena, P. Scholes, R. J. White, R. E. McCormack, and D. Jankovic. 2001. Enumeration of Campylobacter in New Zealand recreational and drinking waters. J. Appl. Microbiol. 91:38-46. [DOI] [PubMed] [Google Scholar]
- 46.Schlesinger, R. B. 1985. Comparative deposition of inhaled aerosols in experimental animals and humans. A review. J. Toxicol. Environ. Health 15:197-214. [DOI] [PubMed] [Google Scholar]
- 47.Simmons, G., V. Hope, G. Lewis, J. Whitmore, and G. Wanzhen. 2001. Contamination of potable roof-collected rainwater in Auckland, New Zealand. Water Res. 35:1518-1524. [DOI] [PubMed] [Google Scholar]
- 48.Simmons, G., S. Jury, C. Thornley, D. Harte, J. Mohiuddin, and M. Taylor. 2008. A legionnaires disease outbreak: a water blaster and roof-collected rainwater systems. Water Res. 42:1449-1458. [DOI] [PubMed] [Google Scholar]
- 49.Spiller, D. 2008. Urban water pricing and supply. Water for today, water for tomorrow: establishment and operation of the SEQ Water Grid. Aust. Econ Rev. 41:420-427. [Google Scholar]
- 50.Tanaka, H., T. Asano, E. D. Schroeder, and G. Tchobanoglous. 1998. Estimating the safety of wastewater reclamation and reuse using enteric virus monitoring data. Water Environ. Res. 28:39-51. [Google Scholar]
- 51.Toze, S. 1999. PCR and the detection of microbial pathogens in water and wastewater. Water Res. 33:3040-3045. [Google Scholar]
- 52.Toze, S., E. Bekkle, D. Page, J. Sidhu, and M. Shackleton. 2010. Use of static quantitative microbial risk assessment to determine pathogen risks in an unconfined carbonate aquifer used for managed aquifer recharge. Water Res. 44:1038-1049. [DOI] [PubMed] [Google Scholar]
- 53.U.S. EPA. 1992. Guidelines for reuse (manual). EPA/625/R-92/004. U.S. EPA, Washington, DC.
- 54.U.S. EPA. 2004. Estimated per capita water ingestion and body weight in the United State—an update. Based on data collected by the United States Department of Agriculture's 1994-1996 and 1998 continuing survey of food intakes by individuals. EPA-822-R-00-001. U.S. EPA, Office of Water, Office of Science and Technology, Washington, DC.
- 55.Veazie, L., I. Brownlee, and H. J. Sears. 1979. An outbreak of gastroenteritis associated with Giardia lamblia, p. 174-191. In W. Jakubowski and J. C. Hoff (ed.), Waterborne transmission of giardiasis. U.S. EPA, Washington, DC.
- 56.Waldenström, J., T. Broman, I. Carlsson, D. Hasselquist, R. P. Achterberg, J. A. Wagenarra, and B. Olsen. 2002. Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl. Environ. Microbiol. 68:5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, V. L. 2000. Legionella pneumophila, p. 2424-2435. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed., vol. 2. Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 58.Zhou, Y., J. M. Benson, C. Irvin, H. Irshad, and Y.-S. Cheng. 2007. Particle size distribution and inhalation dose of shower water under selected operating conditions. Inhalation Toxicol. 19:333-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.