Abstract
Schwanniomyces occidentalis β-fructofuranosidase (Ffase) releases β-fructose from the nonreducing ends of β-fructans and synthesizes 6-kestose and 1-kestose, both considered prebiotic fructooligosaccharides. Analyzing the amino acid sequence of this protein revealed that it includes a serine instead of a leucine at position 196, caused by a nonuniversal decoding of the unique mRNA leucine codon CUG. Substitution of leucine for Ser196 dramatically lowers the apparent catalytic efficiency (kcat/Km) of the enzyme (approximately 1,000-fold), but surprisingly, its transferase activity is enhanced by almost 3-fold, as is the enzymes' specificity for 6-kestose synthesis. The influence of 6 Ffase residues on enzyme activity was analyzed on both the Leu196/Ser196 backgrounds (Trp47, Asn49, Asn52, Ser111, Lys181, and Pro232). Only N52S and P232V mutations improved the transferase activity of the wild-type enzyme (about 1.6-fold). Modeling the transfructosylation products into the active site, in combination with an analysis of the kinetics and transfructosylation reactions, defined a new region responsible for the transferase specificity of the enzyme.
β-Fructofuranosidases (EC 3.2.1.26) are enzymes of biotechnological interest that catalyze the release of β-fructose from the nonreducing termini of various β-d-fructofuranoside substrates. In general, they exhibit a high degree of sequence homology, and based on their amino acid sequences, they fall into family 32 of the glycosyl-hydrolases (GH), along with invertases, inulinases, and fructosyltransferases (http://www.cazy.org). The GH32 family has been studied intensely, and some three-dimensional structures are now available, such as that of inulinase from Aspergillus awamorii (26), fructan-exohydrolase from Cichorium intybus (CiFEH) (34, 36), or invertase from Thermotoga maritima (2, 3) and Arabidopsis thaliana (35). These proteins contain a five-blade β-propeller N-terminal catalytic module and a C-terminal β-sandwich domain (19). Multiple-sequence alignment of GH32 proteins, which are included in the GH-J clan together with the GH68 proteins of the inulosucrase family, reveals the presence of three conserved motifs, each containing a key acidic residue (in boldface) implicated in substrate binding and hydrolysis: Asn-Asp-Pro-Asn-Gly (NDPNG), Arg-Asp-Pro (RDP), and Glu-Cys (EC) (28). These conserved residues are implicated in a double-displacement reaction in which a covalent glycosyl-enzyme intermediate is formed. Thus, the catalytic mechanism proposed for the Saccharomyces cerevisiae invertase implies that Asp23 (NDPNG) acts as a nucleophile and Glu204 (EC) acts as the acid/base catalyst (29), whereas Asp309 (RDP) of Acetobacter diazotropicus levansucrase influences the efficiency of sucrose hydrolysis (7) and Arg188 and Asp189 of the latter motif define the substrate binding and specificity of exoinulinase from A. awamorii toward fructopyranosyl residues (26).
As well as hydrolyzing sucrose, β-fructofuranosidases may also catalyze the synthesis of short-chain fructooligosaccharides (FOS), in which one to three fructosyl moieties are linked to the sucrose skeleton by different glycosidic bonds, depending on the source of the enzyme (12, 21, 31). FOS act as prebiotics, and they exert a beneficial effect on human health, participating in the prevention of cardiovascular diseases, colon cancer, and osteoporosis (16). Currently, FOS are mainly produced by Aspergillus fructosyltransferase in industry (10, 31), providing a mixture of FOS with an inulin-type structure that contains β-(2→1)-linked fructose oligomers (1F-FOS: 1-kestose or nystose). Curiously, when the link between two fructose units (6F-FOS: 6-kestose) or between fructose and the glucosyl moiety (6G-FOS: neokestose) involves a β-(2→6) link, the prebiotic properties of the FOS may be enhanced beyond that of commercial FOS (23).
The yeast Schwanniomyces occidentalis (also called Debaryomyces occidentalis) produces a number of extracellular enzymes that make it of interest in biotechnology. Several of its amylolytic enzymes have been characterized, including amylases and glucoamylase (1, 9), as well as an invertase (17). In addition, we also characterized an extracellular β-fructofuranosidase (Ffase) from this yeast that hydrolyzes sucrose, 1-kestose, and nystose (5). This enzyme exhibited a transfructosylating activity that efficiently produces the trisaccharides 6-kestose and 1-kestose in the ratio 3:1, generating the highest 6-kestose yield yet reported, as far as we know. The Ffase three-dimensional structure has recently been solved (6) and represented as a homodimer, each modular subunit arranged like other GH32 enzymes. The Asp50 (NDPNG) and Glu230 (EC) located at the center of the propeller are the catalytic residues implicated in substrate binding and hydrolysis, whereas Arg178 and Asp179 form the RDP motif (6).
The genetic codes of some yeasts incorporate certain variations. For example, while CUG was believed to be a universal codon for leucine, in the cytoplasm of certain species of the genus Candida (15) it encodes a serine, as in Pichia farinosa (33). The reassignment of this codon is mediated by a novel serine-tRNA that acquired a leucine 5′-CAG-3′ anticodon (25).
Here, we show that deviation from the standard use of the CUG leucine codon to encode serine was correlated with the transferase capacity and specificity of the Ffase enzyme. Indeed, the S196L substitution enhanced the transferase activity of the enzyme 3-fold. Several site-directed mutants were generated and characterized to study their transferase capacities. These results are considered on the basis of the enzymes' three-dimensional structure, which enables a novel putative binding site of sucrose that serves as a water substitute donor in the hydrolytic reaction yielding the tranglycosylation product 6-kestose to be identified.
MATERIALS AND METHODS
Materials, organisms, transformations, and growth conditions.
1-Kestose [α-d-glucopyranosyl-(1→2)-β-d-fructofuranosyl-(1→2)-β-d-fructofuranose] and nystose [α-d-glucopyranosyl-(1→2)-β-d-fructofuranosyl-(1→2)-β-d-fructofuranosyl-(1→2)-β-d-fructofuranose] were obtained from TCI Europe (Zwijndrecht, Belgium). The S. occidentalis strains employed were ATCC 26077, ATCC 20499, and ATCC 26076. S. cerevisiae EUROSCARF Y02321 [BY4741; mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YIL162w(SUC2)::kanMX4] (accession code Y02321) was used as the expression host, and it was transformed by the standard lithium acetate method. Yeasts were grown at 29°C on YEPD (1% yeast extract, 2% peptone, 2% glucose) or inulin-based medium (2% yeast extract, 1.5% inulin). SC(U)D or SC(U)S medium (0.67% yeast nitrogen base [YNB], 0.1% leucine, 0.05% histidine and methionine, 2% glucose [D] or 2% sucrose [S]) was used to select transformants, and YPGal (1% yeast extract, 1% peptone, 2% galactose) was used to induce protein expression in S. cerevisiae. Growth was monitored spectrophotometrically at a wavelength of 600 nm (A600). Escherichia coli DH5α [λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1] was used for DNA manipulation and amplification by standard techniques.
Plasmids, cloning, and mutagenesis.
Genomic DNA was isolated from yeast using standard techniques. The S. occidentalis INV (X17604) gene was obtained by PCR using genomic DNA and the primers SOINVatg (+1 to +22), 5′-CGGGATCCATGGTACAAGTTTTAAGTGTAT-3′, and SOINVter (+1586 to +1608), 5′-CCTCGAGCTACTTATTTAGTTCTCTAATGA-3′, which include BamHI and XhoI recognition sequences (in boldface), respectively. The 1.6-kb PCR products were treated with BamHI-XhoI, inserted into the pST-Blue1 vector (Perfectly Blunt Cloning Kit; Novagen), and sequenced (SIDI, Universidad Autónoma de Madrid, Madrid, Spain). Plasmids for sequence analysis were purified with the Wizard Plus SV Minipreps kit (Promega) according to the manufacturer's protocol. Some sequence changes were found in the INV gene, and the new sequence was denominated as Ffase. For heterologous expression, the Ffase gene from S. occidentalis ATCC 26077 was amplified using the primers FfpYES-B (+1 + 25), 5′-TAGGATCCAACATGGTACAAGTTTTAAGTGTATTAG-3′, and FfpYES-X (+1591 + 1614), 5′-CATCTCTAGACTAGCCCTACTTATTTAGTTCTCT-3′. Restriction sites for BamHI and XbaI (shown in boldface) were included in these primers to clone the PCR product into the pYES2.0 shuttle vector (Invitrogen) under the control of the GAL1 promoter, thereby generating the Ffase-pYES construct. This plasmid was used as a template for L196S, L196E, W47Y, N49S, N52S, S111T, K181F, and P232V site-directed mutagenesis using specific primers (Table 1) and a method described previously (6). The PCR product was incubated with DpnI for 2 h to digest the parental DNA, and 5 μl of this reaction mixture was used directly to transform E. coli. DNA sequencing was used to verify that only the desired mutation was present in the amplified products.
TABLE 1.
Oligonucleotides employed for mutagenesis
| Name | Sequence (5′→3′)a |
|---|---|
| 196S F | GTTGTTTCAAAATCGCAAGAGTACAAAATTCAAATTTTTGG |
| 196S R | CGATTTTGAAACAACCATGATCCATTGATTTGAATCTTCATGC |
| 196E F | GTTGTTGAAAAATCGCAAGAGTACAAAATTCAAATTTTTGG |
| 196E R | CGATTTTTCAACAACCATGATCCATTGATTTGAATCTTCATGC |
| W47Y F | GAAAAAGGATATATGAATGATCGAATGGTCTATTTCTACGATAA |
| W47Y R | GATCATTCATATATCCTTTTTCCGGAGTAAAATGAATTAATGGCCGGTTATAC |
| N49S F | GGATGGATGTCAGATCCGAATGGTCTATTCTACGAT |
| N49S R | ATTCGGATCTGACATCCATCCTTTTTCCGGAGTAAA |
| N52S F | GAATGATCCGAGTGGTCTATTCTACGATAAAACTGCTAAGCTTTG |
| N52S R | GAATAGACCACTCGGATCATTCATCCATCCTTTTCCGGAGTAAATAC |
| S111T F | GGTATCTTTACTGGTAGTATTGTCGTTGACCATAATAATACCTCT |
| S111T R | AATACTACCAGTAAAGATACCTTCATTATCGTGTTCAGGTCCAAT |
| K181F F | CGTGATCCATTCGTTTTCTGGCATGAAGATTCAAAT |
| K181F R | CCAGAAAACGAATGGATCACGGAATTGGTTTGAGGA |
| P232V F | TATGAATGTGTCGGTTTAATTGAAGTTCCTATTGAGAATTCAGAC |
| P232V R | AATTAAACCGACACATTCATACTGATTTCCGTAATAACCAGAAGA |
The underlined nucleotides indicate the position of the altered codon.
Protein purification, detection, and quantification.
β-Fructofuranosidase from S. occidentalis ATCC 26077 (Ffase) was purified as described elsewhere (27). Basically, yeast was grown in inulin-based medium (1 liter), and the culture filtrate (1.4 × 103 U ml−1; 8.9 mg ml−1) was concentrated, fractioned, and dialyzed (against buffer A: 20 mM Tris-HCl, pH 7.0) using a VivaFlow 50 system (Vivascience). The resulting fraction (60.7 × 103 U ml−1; 54.7 mg ml−1) was applied to a DEAE-Sephacel column equilibrated with buffer A, and the proteins were eluted with a 0 to 0.5 M NaCl gradient in buffer A. The active fractions eluting in 0.15 M NaCl were pooled, dialyzed in buffer A, and concentrated (105.8 × 103 U ml−1; 1 mg ml−1). For proteins expressed in S. cerevisiae, the optimum conditions for expression were defined in a time course, and the heterologous proteins were analyzed by measuring the fructofuranosidase activity (sucrose hydrolysis) and/or in Western blots. Yeasts were pregrown in SC(U)D medium, and they were then grown in YPGal medium (1 liter) to the beginning of the stationary phase (A600 = 6 to 7). The culture filtrates (0.6 to 12 U ml−1; about 15 mg ml−1) were then concentrated (20 to 360 U ml−1; about 6 mg ml−1) and applied to a DEAE-Sephacel chromatography column (see above). The active fractions eluted in 0.1 M NaCl were pooled, dialyzed, concentrated (0.05 to 0.8 U ml−1; about 0.7 mg ml−1), and stored at −70°C as described elsewhere (6). Coomassie-stained SDS-PAGE (8%) of the samples confirmed the purity of the protein fraction.
The polyclonal invertase antiserum used for Western blotting was generated by injecting rabbits with commercially available S. cerevisiae invertase (Novozymes), using 1 mg of protein in Freund's complete adjuvant for the initial injection. Decreasing amounts of protein were subsequently injected in Freund's incomplete adjuvant at the following intervals: 2 weeks, 500 μg; 1 month, 200 μg; 2 months, 100 μg. The immune serum was clarified using an extracellular protein extract from S. cerevisiae Y02321, including the pYES2.0 vector. The anti-INV antibody obtained was used at a dilution of 1:2,000. For immunoblots, extracellular proteins were prepared from cultures growing at an A600 of 6 to 7 and were then concentrated and partly fractionated by filtration through Microcon YM-10 membranes. The proteins were resolved electrophoretically and transferred to Immobilon-P membranes (Millipore) that were probed with the invertase antibody. Binding of the antibody was detected using a secondary goat anti-rabbit IgG coupled to horseradish peroxidase (GE Healthcare, Amersham) used as indicated by the manufacturer. The Ffase associated with the cellular fraction was assayed after the addition of glass beads and after five cycles of agitation in a vortex mixer for 1 min, as indicated (21). Protein concentrations were measured photometrically at 280 nm (NanoDrop spectrophotometer ND-1000).
MALDI-TOF analysis.
Proteins were excised from SDS-PAGE gels, digested with trypsin, and identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) (Autoflex; Bruker, Bremen, Germany) at the Proteomic Service of the Centro de Biología Molecular Severo Ochoa (Madrid, Spain). The tryptic peptide map obtained was assigned by comparing their masses with those calculated from theoretical tryptic digestion. The assignment was verified by analyzing the peptide by reverse-phase LC coupled to MS (RP-LC/MS) using a Deca XP mass spectrometer and a ThermoHypersil (0.18- by 150-mm) C18 column. The mass spectrometer was operated in the selected tandem MS (MS/MS) ion-monitoring mode, and the spectra from the peptide were analyzed by assigning the fragments to the candidate sequence after calculating the series of theoretical fragmentations.
Enzyme and kinetic analyses.
Ffase (hydrolytic activity) was assayed by the dinitrosalicylic acid (DNS) method adapted to a 96-well microplate, and the optimal parameters were determined as described elsewhere (5). Standard Ffase activity was measured using sucrose as a substrate, and 1 U of activity was defined as the amount capable of catalyzing the formation of 1 μmol of reducing sugar per minute. For all kinetic analyses, the velocity was measured in triplicate with 5 × 10−2 to 5 × 10−4 mg ml−1 of enzyme and 0 to 500 mM substrate. Reactions were performed over 20 min in 100 mM sodium acetate at the optimum pH and the temperature defined for each variant. The plotting and analysis of the curves were carried out using SigmaPlot software (version 7.101), and the kinetic parameters were calculated by fitting the initial rate values to the Michaelis-Menten equation. Ffase activity was also detected by zymogram analysis using nondenaturing gradient gels (4 to 15%; Bio-Rad) stained with 1% (wt/vol) 2,3,5-triphenyltetrazolium chloride (5). FOS production was analyzed by high-performance liquid chromatography after incubating 0.3 U of enzyme and 1 M (342 g liter−1) sucrose, as indicated previously (5).
Computer analysis and molecular modeling.
The nucleotide and amino acid sequences were analyzed using programs in the GCG Sequence Analysis Software Package (available from the University of Wisconsin) in conjunction with sequence data from the Swiss-Prot, GenBank, and EMBL databases. The structural analysis was carried out using the O program (14), and the figures were obtained with PYMOL (http://pymol.sourceforge.net/). The transfructosylating product 6-kestose was modeled from the experimental 6-kestose crystal structure coordinates extracted from the Cambridge Structural Database (CSD Refcode CELGIJ), superimposing its terminal fructose moiety onto the fructose found in the Ffase crystal. The 1-kestose substrate was modeled into the Ffase active site as inferred from structural superimposition of the Cichorium intybus fructan exohydrolase (CiFEH)-1-kestose coordinates (Protein Data Bank [PDB] code 2AEZ) onto the Ffase-fructose complex (PDB code 3KF3).
Nucleotide sequence accession number.
The sequence reported here was submitted to the EMBL database (GenBank accession no. CQ890277).
RESULTS
The sequence of the β-fructofuranosidase from S. occidentalis contains some unexpected changes.
Initial studies of an invertase from S. occidentalis showed that the active form of this enzyme was a homodimer consisting of two subunits, each containing 533 amino acids, whose sequence was deduced after analysis of the INV gene (18). We used PCR to isolate this gene and found that the sequence amplified showed marked differences from that previously reported, including four changes in the amino acid sequence encoded (Fig. 1A). Identical results were found in two other S. occidentalis strains (see Materials and Methods). The new INV gene isolated encoded a putative protein of 535 amino acids (instead of 533), and it included the three conserved acidic residues in the motifs shared by all proteins of the GH32 family (19, 28), NDPNG, RDP, and EC (Asp50, Asp179, and Glu230, respectively). To verify that the β-fructofuranosidase from S. occidentalis (Ffase) with transfructosylating activity that we had characterized previously (5) was encoded by the sequence analyzed, the enzyme was purified and analyzed by MALDI-TOF and fingerprinting (data not shown). Most of the masses retrieved (42% coverage) coincided with the translation of the new DNA sequence. However, when an unexpected peptide of 2,005 m/z was analyzed using microspray-ion trap MS (data not shown), the Ffase sequence included a serine instead of a leucine residue at position 196 (Fig. 1B). This change was caused by nonuniversal decoding of the unique mRNA CUG leucine codon, corresponding to 586CTG588 in the DNA sequence analyzed.
FIG. 1.
(A) Alignment of the regions showing differences between the β-fructofuranosidase from S. occidentalis described here (Ffase) and that previously reported (P24133). The differences are indicated in boldface. The Glu230 involved in sucrose hydrolysis, which is not present in P24133, is marked with an asterisk, and the Ser196 modified to Leu196 in S. cerevisiae is underlined. The superscript numbers indicate the amino acid positions. (B) Ffase was purified from S. occidentalis and digested with trypsin. The sequence of the peptide of 2,005 m/z identified by MALDI-TOF and generated by microspray-ion trap MS analysis is shown.
The nonuniversal decoding of the leucine CUG codon affects Ffase activity.
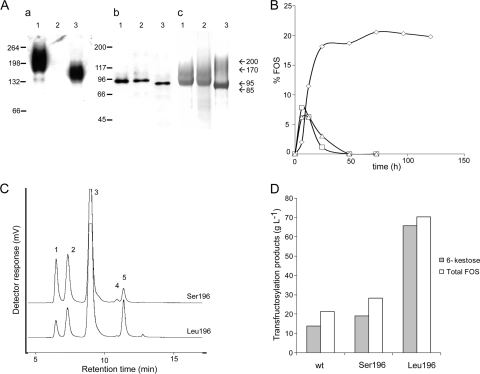
The functionality of the gene isolated here (Ffase) was verified through its heterologous expression using an S. cerevisiae strain that was unable to use sucrose as a carbon source. Although the Ser196 residue (CUG = Ser) is included in a nonconserved region of the GH32 family, we mutated the Ffase triplet 586CTG588 to TCA, which encodes serine in the standard code, in order to obtain a heterologously expressed protein with the wild-type amino acid sequence (Ffase-Ser196). Genes encoding Ffase-Leu196 and Ffase-Ser196 were included in S. cerevisiae, and as expected, both complemented the lack-of-growth phenotype in sucrose of the host strain. Maximum levels of hydrolytic activity (approximately 12 U ml−1) were detected in the Ffase-Ser196 culture filtrates at the beginning of the stationary phase (12 to 16 h of growth; A600 = 6), whereas activity fell to near 30% in cultures older than 30 h. Slightly lower activity (≤8 U) was found in the cell-associated fraction during these phases. However, replacing the Ser196 residue by leucine promoted a severe decrease in the levels of hydrolytic activity in both fractions analyzed (≤1 U ml−1), and it was not detected in gels under nondenaturing conditions (Fig. 2A, a). A priori, these results suggest that Ffase-Leu196 has weaker activity, secretion, or stability or a combination of these characteristics. Analysis of the extracellular Ffase using an anti-INV antibody that recognized this protein in its native and heterologously produced forms (Fig. 2A, b) showed that they were secreted in similar amounts. As expected, glycosylation is not the same in the two yeast species (6), and the proteins expressed in S. cerevisiae had a molecular mass of 95 kDa, at least 10 kDa higher than the protein expressed in S. occidentalis. Moreover, there was a marked difference in their estimated molecular masses on gels under nondenaturing conditions (Fig. 2A, a).
FIG. 2.
Analysis of the enzymes expressed in S. occidentalis (wild type) or in S. cerevisiae (Ffase-Ser196 and Ffase-Leu196). (A) Zymogram (a), Western blot (b), and purification of proteins (c) from S. cerevisiae expressing Ffase-Ser196 (lanes 1) or Ffase-Leu196 (lanes 2) and S. occidentalis (lanes 3). The hydrolase activity of 10 μg of total extracellular proteins was revealed in situ using sucrose as the substrate (a), and 1 μg of each protein was immunoblotted and probed with anti-INV antibodies (b). The purified proteins (10 μg) were subjected to SDS-PAGE and Coomassie stained (c). The numbers on the left (a and b) indicate the positions of molecular mass standards in kDa, and the numbers on the right (c) indicate the molecular masses assigned to the Ffase under the different conditions assayed. (B) Time course of FOS production catalyzed by the Ffase enzymes expressed in S. occidentalis (triangles) and in S. cerevisiae containing Ser (squares) or Leu (rhombus) at position 196. The data are represented as the percentage of FOS (wt/wt) in the total sugar composition of the reaction mixture. The total reaction volume was 2 ml, and 0.3 U of purified enzyme in 0.2 M sodium acetate buffer, pH 5.6, was used. The reaction temperature was 50°C for the wild-type and Ffase-Ser196 enzymes and 45°C for the Ffase-Leu196 variant. (C) HPLC chromatogram corresponding to the reactions of maximum FOS production obtained with the Ffase-Ser196 (6 h) and Ffase-Leu196 (72 h) expressed in S. cerevisiae. The detector response scales for both chromatograms were the same: 1, fructose; 2, glucose; 3, sucrose; 4, 1-kestose; and 5, 6-kestose. (D) Maximum FOS and 6-kestose concentrations (g liter−1) produced by the wild-type (wt), Ffase-Ser196 (Ser196), and Ffase-Leu196 (Leu196) enzymes.
The S. occidentalis enzyme and the two proteins expressed in S. cerevisiae were highly purified (Fig. 2A, c), and some of their biochemical characteristics were studied. In general, and as expected, the enzymes displayed classical Michaelis-Menten kinetics (data not shown), and no significant changes were found in the kinetic behavior of the wild-type and the heterologously expressed Ffase-Ser196 (Table 2). However, Ffase-Leu196 had a drastically lower apparent catalytic efficiency than the wild-type enzyme (kcat/Km, approximately 1/1,000), and the difference was greater as the size of the substrate analyzed increased (sucrose to nystose). In addition, replacing Ser196 with leucine affected other biochemical characteristics of the enzyme's hydrolytic activity, such as pH and temperature dependence. Thus, while no changes were found in the optimal pH (5.5) and temperature (50 to 55°C) of the Ffase-Ser196 expressed in the two yeast species, the optimal temperature of the Ffase-Leu196 fell by 10°C (to 40 to 45°C) and the pH by 0.5 (to 5.0) (data not shown).
TABLE 2.
Kinetic parameters of the wild-type and the heterologously expressed Ffase variants, including the residue Ser or Leu196a
| Substrate | Variant | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|
| Sucrose | Wt | 7.5 ± 1 | 185 ± 9 | 24.6 |
| Ser196 | 8.2 ± 1.4 | 173.3 ± 16 | 21.1 | |
| Leu196 | 32.9 ± 5.6 | 0.93 ± 0.06 | 0.028 | |
| 1-Kestose | Wt | 2.3 ± 0.6 | 55.3 ± 5 | 24 |
| Ser196 | 2.6 ± 0.7 | 51 ± 5 | 19.6 | |
| Leu196 | 22.6 ± 3.6 | 0.2 ± 0.006 | 0.009 | |
| Nystose | Wt | 2.5 ± 0.6 | 102 ± 8 | 41.1 |
| Ser196 | 2.2 ± 0.4 | 86 ± 4 | 39.1 | |
| Leu196 | 21.5 ± 2.3 | 0.22 ± 0.03 | 0.01 |
kcat values were calculated assuming a molecular mass of 170 kDa for the Ffase expressed in S. occidentalis (wild type) and 200 kDa for the enzyme expressed in S. cerevisiae containing Ser or Leu196. The values are given ± standard errors based on the curve fitting using SigmaPlot.
Substitution at Ser196 modifies the transferase activity of Ffase.
The Ffase from S. occidentalis presents a transfructosylating activity that produces mainly 6-kestose (6F-FOS) and then 1-kestose (1F-FOS) in a 3:1 ratio (5). Therefore, we decided to examine the transferase capacities of the two heterologously expressed enzymes (Fig. 2B). After a 6- to 12-h reaction, a maximum FOS concentration of about 25 g liter−1 was obtained using the enzyme containing Ser196 expressed in both yeast species (S. cerevisiae and S. occidentalis), corresponding to approximately 7% (wt/wt) of the total sugar composition in the mixture. Surprisingly, Ffase-Leu196 showed enhanced transferase capacity (almost 3-fold), and 39 g liter−1 FOS was produced after a 12-h reaction, which corresponded to 11.5% (wt/wt) of the total sugar composition. The maximum concentration of FOS was reached in 72 h, and it was 70.3 g liter−1, which corresponded to almost 21% (wt/wt) of the sugar in the mixture. In addition, the replacement of Ser196 by leucine increased the 6-kestose/1-kestose ratio about 7-fold [β-(2→6)-linked FOS:β-(2→1)-linked FOS ratio], which meant that there was virtually no significant production of 1-kestose (4.6 g liter−1) (Fig. 2C and D).
Levansucrases from Bacillus subtilis (24) and Gluconoazetobacter diazotrophicus (22) synthesize levans directly from sucrose, and these polymers are basically formed by β-(2→6)-linked fructose units. A position equivalent to that of Ser196 in Ffase is occupied by glutamate in these two levansucrases, and thus, we decided to evaluate the effect of the S196E substitution on Ffase activity. Although extracellular Ffase-Glu196 protein was evident in immunoblots probed with the anti-INV antibodies (data not shown), no Ffase activity was detected in any of the cellular fractions analyzed.
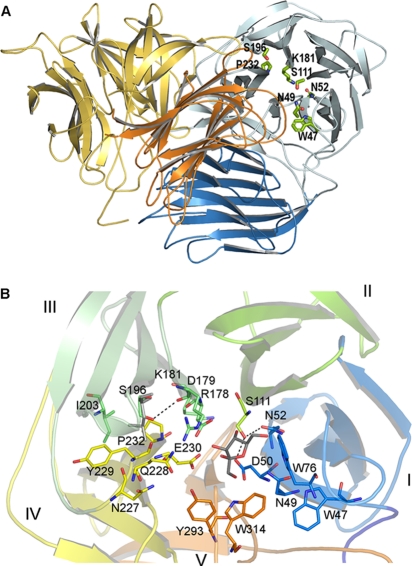
As mentioned above, the catalytic domain of each subunit folds into a β-propeller that is assembled from five blades (I to V), each composed of four antiparallel β-strands. Given the position of Ser196 in the Ffase dimer (Fig. 3A), the Ser196 residue is located in blade III, forming a hydrogen bond with Arg178 from the RDP motif (Fig. 3B).
FIG. 3.
(A) Cartoon of the three-dimensional structure of the S. occidentalis Ffase. Two monomers (blue and orange) associate to form a tight dimer through mainly polar and hydrophobic interactions. The two domains within each monomer are shaded distinctly. The residues investigated in this work are represented as sticks. (B) The catalytic domain folds into a propeller made up of five blades (I to V), each represented in a different color. The residues mentioned in the text are shown as sticks with the same color code. Fructose in the Ffase active site is represented in grey. Asp50 (NDPNG), Asp179 (RDP), and Glu230 (EC) are the catalytic residues. The putative position of a modeled Leu196 is represented in grey.
Improving transferase activity by site-directed mutagenesis.
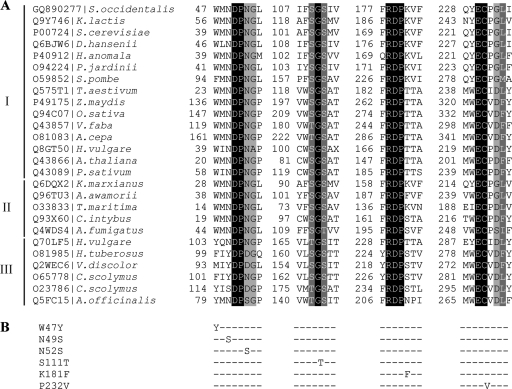
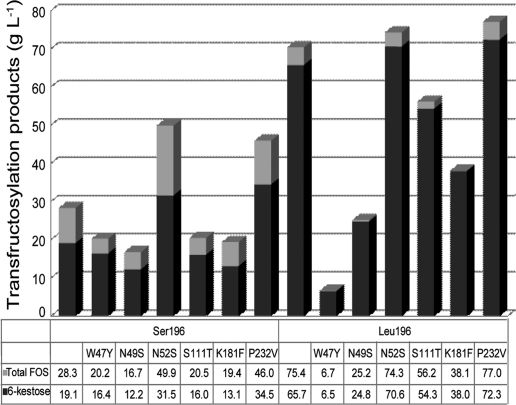
A multiple alignment of invertases and fructosyltransferases from different organisms available in the EMBL/SWISS-PROT databases clearly revealed the presence of some well-conserved regions, enabling the positions of residues that could be related to their activity to be established (Fig. 4). Indeed, variations in the amino acid sequence adjacent to the active nucleophile site ([WM]NDPNG) have been associated with the transglycosylation capacity of some invertases from plants, such as onion (30) and wheat (32). Accordingly, the residues Trp47, Asn49, and Asn52 from the S. occidentalis enzyme were mutated, and the activity of the new enzymes generated was analyzed. Mutant enzymes with a W47Y, N49S, or N52S substitution had significantly higher Km values for sucrose on both a Leu196 and a Ser196 protein background (Table 3). In addition, the transfructosylating activities of W47Y and N49S mutants were greatly reduced (Fig. 5), possibly as a consequence of the strong fall in catalytic efficiency (approximately 300- to 5,000-fold for Ffase-Leu196 and approximately 11- to 78-fold for Ffase-Ser196) (Table 3). In contrast, the N52S mutation increased the transfructosylating activity of the Ffase-Ser196 variant by approximately 1.8-fold.
FIG. 4.
(A) Multiple alignments of Ffase and several GH32 proteins in the regions surrounding the sites mutated. The accession code and organism source are indicated in the first column. Identical residues are shaded in black, whereas conserved and semiconserved residues are shaded in dark and light grey, respectively. I, β-fructofuranosidase, EC 3.2.1.26; II, β-fructosidase, EC 3.2.1.80; III fructosyltransferases 2.4.1 (99/100/243); K. lactis, Kluyveromyces lactis; D. hansenii, Debaryomyces hansenii; P. jardinii, Pichia jardinii; S. pombe, Schizosaccharomyces pombe; T. aestivum, Triticum aestivum; Z. maydis, Zea maydis; O. sativa, Oryza sativa; V. faba, Vicia faba; A. cepa, Allium cepa; H. vulgare, Hordeum vulgare; P. sativum, Pisum sativum; K. marxianus, Kluyveromyces marxianus; A. fumigatus, Aspergillus fumigatus; H. tuberosus, Helianthus tuberosus; V. discolor, Viguiera discolor; C. scolymus, Cichorium scolymus; A. officinalis, Asparagus officinalis. (B) Amino acid changes included in the Leu or Ser196 background.
TABLE 3.
Kinetic parameters of mutant enzymesa
| Ffase | Mutation | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|
| Ser196 | Control | 8.2 ± 1.4 | 173.3 ± 16 | 21.1 |
| W47Y | 71.4 ± 10.7 | 135.4 ± 13 | 1.89 | |
| N49S | 551.5 ± 99 | 149.8 ± 14 | 0.27 | |
| N52S | 28.04 ± 5.7 | 45.3 ± 7.7 | 1.62 | |
| S111T | 26.0 ± 3.9 | 517 ± 103 | 19.9 | |
| K181F | 57.1 ± 9.1 | 42.8 ± 7.3 | 0.74 | |
| P232V | 19.8 ± 3.9 | 5.8 ± 1.1 | 0.29 | |
| Leu196 | Control | 32.9 ± 5.6 | 0.93 ± 0.06 | 0.028 |
| W47Y | 5000 ± 200 | 0.027 ± 0.004 | 5.4 × 10−6 | |
| N49S | 368 ± 30 | 0.032 ± 0.004 | 8.7 × 10−5 | |
| N52S | 5600 ± 700 | 32.3 ± 4.2 | 0.006 | |
| S111T | 19.8 ± 6.6 | 0.39 ± 0.1 | 0.020 | |
| K181F | 51.4 ± 6.8 | 0.22 ± 0.06 | 0.004 | |
| P232V | 23.7 ± 5.5 | 0.47 ± 0.05 | 0.019 |
Ffase Ser196 or Leu196 (control) was used to generate the mutants. Sucrose was used as a substrate. kcat values were calculated assuming a molecular mass of 200 kDa for the enzyme. The values are expressed ± standard errors based on the curve fitting using SigmaPlot.
FIG. 5.
FOS produced by Ffase-Ser196 and Ffase-Leu196 mutants. The reactions were carried out on sucrose. The concentrations of the total FOS (light grey) and 6-kestose (dark grey) obtained are indicated.
Different plant invertases are characterized by single-amino-acid substitutions in the conserved EC(P/V) sequence, which includes the acid/base catalyst (11). A proline is present in extracellular invertases and exoinulinases, whereas a valine occupies this position in vacuolar invertases (4) and fructosyltransferases (Fig. 4). The Pro238Val substitution in the extracellular invertase from Chenopodium rubrum produced a decrease (28%) in the rate of raffinose cleavage (4). Similarly, the P232V substitution also produced a notable reduction in the catalytic efficiency of Ffase (approximately 32% on a Leu196 and 98% on a Ser196 background), although, interestingly, it increased the transfructosylase activity of the Ser196 variant by 1.6-fold (62%). Finally, positions Ser111 and Lys181 were mutated in Ffase, according to the homologous residues in Aspergillus sp. enzymes (Fig. 4), currently the main industrial FOS producers (10, 31). However, substitutions S111T and K181F produced mutant enzymes with reduced (K181F) or unaffected (S111T) catalytic efficiencies (Table 3) without improving the transferase activity (Fig. 5).
DISCUSSION
One aim of this study was to identify amino acid residues involved in the transferase capacity of Ffase, as well as to generate improved enzymes with potential applications in FOS production. Accordingly, we show that a single-amino-acid substitution involving the nonuniversal decoding of the leucine CUG codon as serine (S196L) switches the enzymatic activity of the β-fructofuranosydase from S. occidentalis, reducing the hydrolase activity and increasing the transferase capacity approximately 3-fold. We also show that the N52S and P232V substitutions improve the transferase activity of the wild-type enzyme by about 1.6-fold.
A remarkable feature of yeasts is the wide variation in their genomic G+C contents. It has been proposed that the codon usage bias is generally correlated with the overall genome G+C content (8) and that the codons favored are due to the tDNA gene copy number (20). CUG is a rare leucine codon in S. cerevisiae (G+C, 40%), and it is also a rare codon for serine in S. occidentalis (G+C, 36%) (13). The decoding of the leucine CUG codon as serine has been reported previously in the cytoplasm of some Candida species (15) and in P. farinosa (33). However, our study of the Ffase sequence provides the first direct evidence for CUG decoding as serine in S. occidentalis. In this context, it has not been possible to express the β-glucuronidase (gusA) or β-lactamase (bla) gene or the phleomycin resistance-conferring gene (Tn5ble) from E. coli in S. occidentalis (13). Thus, an appropriate DNA composition and codon usage bias might be important parameters for efficient expression of heterologous genes in this host. However, these three genes contain various CUG codons (19 in gusA, 7 in bla, and 11 in Tn5ble), and their alternative decoding as serine could be relevant in the functioning of these proteins when expressed in S. occidentalis.
In terms of Ffase activity, the S196L substitution might be expected to position the longer leucine side chain (β-propeller, blade III) in a hydrophobic pocket surrounded by Ile203 (also in blade III) and Tyr229 and Pro232 (from blade IV) (Fig. 3B). This modification may produce a local rearrangement that would affect not only the position of Arg178 from the RDP motif, but also the conformation of the first strand of blade IV (Tyr229-Pro238), where the catalytic Glu230 is located. Consequently, such modifications may be deleterious for hydrolytic activity. However, the improvement in the Ffase transferase/hydrolase ratio observed upon Ser/Leu replacement indicates that this region might influence the retention time of sucrose in the enzyme active site and the subsequently higher probability of finding a sucrose acceptor instead of a water molecule, leading to transglycosylation.
When we consider all the residues and positions investigated in this study (Fig. 3 and 4), it is noteworthy that the mutations affecting the biosynthetic activity of Ffase are situated in two regions. The N52S mutant is located in the (WM)NDPNG motif, a segment that includes the nucleophile that is involved in developing transfructosylating activity in plant enzymes. The mutations involving the Ser196 residue lie in a second region situated at the opposite side of the active-site cleft, in the environment of the acid/base catalyst within the EC(P/V) motif (Fig. 3B). Independently, the S196L and P232V replacements introduce some structural changes in this area, leading to a general increase in FOS production. Moreover, the specificity for 6-kestose is enhanced by these substitutions, and consequently, this region may be envisaged as a potential novel acceptor site for binding sugar instead of water, promoting subsequent transferase activity.
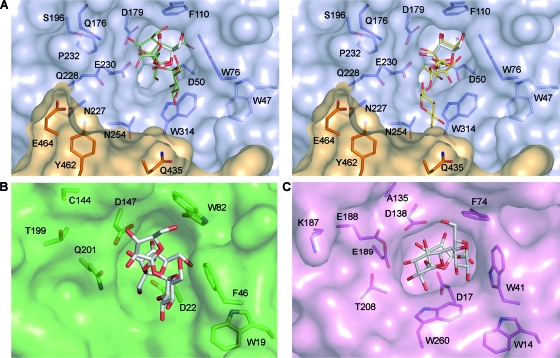
In an attempt to understand the possible molecular basis for this particular behavior, active sites from Ffase and other structurally known GH32 enzymes were analyzed, and the transfructosylating products 1-kestose and 6-kestose were modeled into the active enzyme site (Fig. 6A, left and right). On inspection, one wall of the active-site cleft (at the right) is hydrophobic, and it is contoured by aromatic residues that are conserved among structurally known GH32 enzymes. The roles of these residues in substrate binding can be illustrated by the fact that the terminal sugar unit of raffinose and 1-kestose stack against the Phe46 and Trp41 in the plant and bacterial enzymes (Fig. 6B and C). In contrast, there is more variation in the opposite wall containing the acid/base catalyst Glu230, which in the case of Ffase is also shaped by the contiguous subunit within the dimer. The residues from the environment of the EC motif, such as Asn227 and Gln228, are in close proximity to the sucrose moiety of the putative 6-kestose molecule. Therefore, they appear to be properly positioned to influence sucrose binding, and consequently, the enhanced specificity for 6-kestose upon changes in this region can be understood.
FIG. 6.
(A) Close-up view of the active site in S. occidentalis Ffase in a complex with fructose (PDB code 3KF3). Putative positions for 1-kestose (green; left) and 6-kestose (yellow; right) transfructosylation products were modeled into the active site of Ffase. (B and C) The fructan 1-exohydrolase IIa from C. intybus (CiFEH) complexed with 1-kestose (PDB code 2AEZ) (B) and the invertase from T. maritima complexed with raffinose (PDB code 1W2T) (C). The ligands found in the crystals are represented as white sticks. As can be seen in panel A, the active site of Ffase (blue) is also shaped by the adjacent subunit (orange), and it defines subsites in addition to those found in the bacterial and plant enzymes.
In summary, our results suggest a novel substrate donor-binding site located near the EC motif that would yield the β-(2→6)-linked transfructosylation product. This novel site may contribute to the fact that the enzyme from S. occidentalis is the highest producer of 6-kestose known.
Acknowledgments
This work was supported by grants from the Spanish Ministry of Education and Science (BIO2007-67708-C04-03 and -04) and by an institutional grant from the Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa. M.A.B. and M.D.A. were supported by the Spanish Comunidad de Madrid program and by an FPU fellowship from the Spanish Ministry of Education and Science, respectively.
We thank Ivana Janatova for help at the start of this research and Francisco J. Plou for his assistance with the HPLC analysis.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Abarca, D., M. Fernández-Lobato, L. del Pozo, and A. Jiménez. 1991. Isolation of a new gene (SWA2) encoding an alpha amylase from Schwanniomyces occidentalis and its expression in Saccharomyces cerevisiae. FEBS Lett. 279:41-44. [DOI] [PubMed] [Google Scholar]
- 2.Alberto, F., C. Bignon, G. Sulzenbacher, B. Henrissat, and M. Czjzek. 2004. The three-dimenisonal structure of invertase (β-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases. J. Biol. Chem. 279:18903-18910. [DOI] [PubMed] [Google Scholar]
- 3.Alberto, F., E. Jordi, B. Henrissat, and M. Czjzek. 2006. Crystal structure of inactivated Thermotoga maritima invertase in complex with the trisaccharide substrate raffinose. Biochem. J. 395:457-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altenbach, D., E. Rudiño-Pineira, C. Olvera, T. Boller, A. Wiemken, and T. Ritsema. 2009. An acceptor-substrate binding site determining glycosyl transfer emerges from mutant analysis of a plant vacuolar invertase and a fructosyltransferase. Plant Mol. Biol. 69:47-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Álvaro-Benito, M., M. de Abreu, L. Fernández-Arrojo, F. J. Plou, J. Jiménez-Barbero, A. Ballesteros, J. Polaina, and M. Fernández-Lobato. 2007. Characterization of a β-fructofuranosidase from Schwanniomyces occidentalis with transfructosylating activity yielding the prebiotic 6-kestose. J. Biotechnol. 132:75-81. [DOI] [PubMed] [Google Scholar]
- 6.Álvaro-Benito, M., A. Polo, B. González, M. Fernández-Lobato, and J. Sanz-Aparicio. 2010. Structural and kinetic analysis of Schwanniomyces occidentalis invertase reveals a new oligomerization pattern and the role of its supplementary domain in substrate binding. J. Biol. Chem. 285:13930-13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batista, F. R., L. Hernández, J. R. Fernández, J. Arrieta, C. Menéndez, R. Gómez, Y. Támbara, and T. Pons. 1999. Substitution of Asp-309 by Asn in the Arg-Asp-Pro (RDP) motif of Acetobacter diazotropicus levansucrase affects sucrose hydrolysis, but not enzyme specificity. Biochem. J. 337:503-506. [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, S. L., W. Lee, A. K. Hottes, L. Shapiro, and H. H. McAdams. 2004. Codon usage between genomes is constrained by genome-wide mutational processes. Proc. Natl. Acad. Sci. U. S. A. 101:3480-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowhanick, T. M., I. Russell, S. W. Scherer, G. G. Stewart, and V. L. Seligy. 1990. Expression and regulation of glucoamylase from the yeast Schwanniomyces castelli. J. Bacteriol. 172:2360-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghazi, I., L. Fernández-Arrojo, H. Garcia-Arellano, M. Ferrer, A. Ballesteros, and F. J. Plou. 2007. Purification and kinetic characterizaton of a fructosyltransferase from Aspergillus aculeatus. J. Biotechnol. 128:204-211. [DOI] [PubMed] [Google Scholar]
- 11.Goetz, M., and T. Roitsch. 1999. The different pH optima and substrate specificities of extracellular and vacuolar invertases from plants are determined by a single amino-acid substitution. Plant J. 20:707-711. [DOI] [PubMed] [Google Scholar]
- 12.Gutiérrez-Alonso, P., L. Fernández-Arrojo, F. J. Plou, and M. Fernández-Lobato. 2009. Biochemical characterization of a β-fructofuranosidase from Rhodotorula dairenensis with transfructosylating activity. FEMS Yeast Res. 9:768-773. [DOI] [PubMed] [Google Scholar]
- 13.Janatova, I., P. Costaglioli, J. Wesche, J. M. Masson, and E. Meilhoc. 2003. Development of a reporter system for the yeast Schwanniomyces occidentalis: influence of DNA composition and codon usage. Yeast 20:687-701. [DOI] [PubMed] [Google Scholar]
- 14.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 15.Jukes, T. H., and S. Osawa. 1996. CUG codons in Candida spp. J. Mol. Evol. 42:321-322. [DOI] [PubMed] [Google Scholar]
- 16.Kaur, N., and A. K. Gupta. 2002. Applications of inulin and oligofructose in health and nutrition. J. Biosci. 27:703-714. [DOI] [PubMed] [Google Scholar]
- 17.Klein, R. D., M. R. Deibel, Jr., J. L. Sarcich, H. A. Zurcher-Neely, I. M. Reardon, and R. L. Heinrikson. 1989. Purification and characterization of invertase from a novel industrial yeast, Schwanniomyces occidentalis. Prep. Biochem. 19:293-319. [DOI] [PubMed] [Google Scholar]
- 18.Klein, R. D., R. A. Poorman, M. A. Favreau, M. H. Shea, N. T. Hatzenbuhler, and S. C. Nulf. 1989. Cloning and sequence analysis of the gene encoding invertase from the yeast Schwanniomyces occidentalis. Curr. Genet. 16:145-152. [DOI] [PubMed] [Google Scholar]
- 19.Lammens, W., K. Le Roy, L. Schroeven, A. Van Laere, A. Rabijns, and W. Van den Ende. 2009. Structural insights into glycoside hydrolase family 32 and 68 enzymes: functional implications. J. Exp. Bot. 60:727-740. [DOI] [PubMed] [Google Scholar]
- 20.Lin, S. Y., J. K. Byrnes, J. K. Hwang, and W. H. Li. 2006. Codon-usage bias versus gene conversion in the evolution of yeast duplicate genes. Proc. Natl. Acad. Sci. U. S. A. 103:14412-14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linde, D., I. Macias, L. Fernández-Arrojo, F. J. Plou, A. Jiménez, and M. Fernández-Lobato. 2009. Molecular and biochemical characterization of a β-fructofuranosidase from Xanthophyllomyces dendrorhous. Appl. Environ. Microbiol. 75:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martínez-Fleites, C., M. Ortíz-Lombardía, T. Pons, N. Tarbouriech, E. J. Taylor, J. G. Arrieta, L. Hernández, and G. J. Davies. 2005. Crystal structure of levansucrase from the Gram-negative bacterium Gluconoazetobacter diazotrophicus. Biochem. J. 390:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marx, S. P., S. Winkler, and W. Hartmeier. 2000. Metabolization of β-(2,6)-linked fructose-oligosaccharides by different bifidobateria. FEMS Microbiol. Lett. 182:163-169. [DOI] [PubMed] [Google Scholar]
- 24.Meng, G., and K. Fütterer. 2003. Structural framework of fructosyltransfer in Bacillus subtilis levansucrase. Nat. Struct. Biol. 10:935-941. [DOI] [PubMed] [Google Scholar]
- 25.Miranda, I., R. Silva, and M. A. S. Santos. 2006. Evolution of the genetic code in yeasts. Yeast 23:203-213. [DOI] [PubMed] [Google Scholar]
- 26.Nagem, R. A. P., A. L. Rojas, A. M. Golubev, O. S. Korneeva, E. V. Eneyskaya, A. A. Kulminskaya, K. N. Neustroev, and I. Polikarpov. 2004. Crystal structure of exo-inulinase from Aspergillus awamori: the enzyme fold and structural determinants of substrate recognition. J. Mol. Biol. 344:471-480. [DOI] [PubMed] [Google Scholar]
- 27.Polo, A., M. Álvaro-Benito, M. Fernández-Lobato, and J. Sanz-Aparicio. 2009. Crystallization and preliminary X-ray diffraction analysis of the fructofuranosidase from Schwanniomyces occidenalis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65:1162-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pons, T., D. G. Naumoff, C. Martínez-Fleites, and L. Hernández. 2004. Three acidic residues are at the active site of a β-propeller architecture in glycoside hydrolase families 32, 43, 62 and 68. Proteins 54:424-432. [DOI] [PubMed] [Google Scholar]
- 29.Reddy, A., and F. Maley. 1996. Studies on identifying the catalytic role of Glu-204 in the active site of yeast invertase. J. Biol. Chem. 271:13953-13958. [DOI] [PubMed] [Google Scholar]
- 30.Ritsema, T., L. Hernández, A. Verhaar, D. Altenbach, T. Boller, A. Wiemken, and S. Smeekens. 2006. Developing fructan-synthesizing capability in a plant invertase via mutations in the sucrose-binding box. Plant J. 48:228-237. [DOI] [PubMed] [Google Scholar]
- 31.Sangeetha, P. T., M. N. Ramesh, and S. G. Prapulla. 2005. Fructooligosaccharide production using fructosyl transferase obtained from recycling culture of Aspergillus oryzae CFR 202. Process Biochem. 40:1085-1088. [Google Scholar]
- 32.Schroeven, L., W. Lammens, A. Van Laere, and W. Van den Ende. 2008. Transforming wheat vacuolar invertase into a high affinity sucrose:sucrose 1-fructosyltransferase. New Phytol. 180:822-831. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, C., T. Kashiwagi, and K. Hirayama. 2002. Alternative CUG codon usage (Ser for Leu) in Pichia farinosa and the effect of a mutated killer gene in Saccharomyces cerevisiae. Protein Eng. 15:251-255. [DOI] [PubMed] [Google Scholar]
- 34.Verhaest, M., W. Van den Ende, K. Le Roy, C. J. De Ranter, A. Van Laere, and A. Rabijns. 2005. X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus. Plant J. 41:400-411. [DOI] [PubMed] [Google Scholar]
- 35.Verhaest, M., W. Lammens, K. Le Roy, B. De Conick, C. J. De Ranter, A. Van Laere, W. Van den Ende, and A. Rabijns. 2006. X-ray diffraction structure of a cell-wall invertase from Arabidopsis thaliana. Acta Crystallogr. D Biol. Crystallogr. 62:1555-1563. [DOI] [PubMed] [Google Scholar]
- 36.Verhaest, M., W. Lammens, K. Le Roy, C. J. De Ranter, A. Van Laere, A. Rabijns, and W. Van den Ende. 2007. Insights into the fine architecture of the active site of chycory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor. New Phytol. 174:90-100. [DOI] [PubMed] [Google Scholar]