Abstract
Here, we report cloning of cyanobacterial genes encoding pteridine glycosyltransferases that catalyze glucosyl or xylosyl transfer from UDP-sugars to tetrahydrobiopterin. The genes were cloned by PCR amplification from genomic DNA which was isolated from culture and environmental samples and overexpressed in Escherichia coli for an in vitro activity assay.
Tetrahydrobiopterin (BH4) is well known among pteridine compounds as a cofactor for aromatic amino acid hydroxylases and nitric oxide synthases in animals (19). Pteridine glycosides such as biopterin and 6-hydroxymethylpterin glycosides have been found in cyanobacteria and anaerobic photosynthetic bacteria (2, 4, 5, 8, 11, 13, 15, 17, 21). Although the function of these glycosides remains unknown, they are abundant and ubiquitous in cyanobacteria, implying some essential role (3, 6, 16-18, 21). There is a group of enzymes, named pteridine glycosyltransferases (PGTs), known to catalyze a variety of glycosyl transfers to pteridines. The first PGT isolated from the cyanobacterium Synechococcus sp. PCC 7942 was shown to catalyze a glucosyl transfer from UDP-glucose to BH4 and was therefore named UDP-glucose:BH4 glucosyltransferase (BGluT) (7). After cloning of the gene encoding BGluT (6), a PGT that catalyzes the transfer of glucuronic acid for cyanopterin synthesis was identified (12). In addition, there are many putative PGT homologs encoded in bacterial genomes, although their exact catalytic functions have not been determined. We recently found that BGluT is useful for the simultaneous detection of oxidized and reduced forms of BH4 in animal samples (14). Glycosyltransferases are also being studied intensively for applications in the design of novel pharmaceutical derivatives (1, 10). We were thus encouraged to find PGTs with new substrate specificities or enzymatic properties not only for study of protein structure and function but also for application in BH4 research. In this study, we succeeded in cloning four cyanobacterial genes encoding PGTs with either glucosyl- or xylosyltransferase activity, and here we report the results.
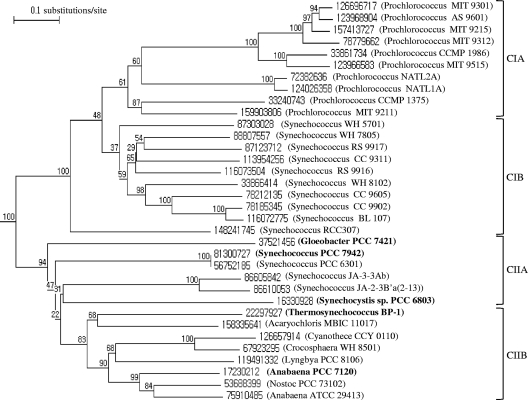
PGT genes were cloned from Arthrospira platensis CY-007 (obtained through Hawaii Oceanic Institute sampling) and Arthrospira maxima CY-049 (UTEX 2342), which were cultured in the Korea Marine Microalgae Culture Center, and from environmental DNA sequences (designated UCNR-001 and UCNR-002) isolated from wild algal mats in the Nakdong River, South Korea. In order to amplify conserved internal sequences of the unknown PGT genes, degenerate PCR primers were designed from the nucleotide sequences of cyanobacterial PGT homologs using GeneFisher2 (9). A protein homology search with BGluT against the bacterial genome database in NCBI revealed more than a hundred PGT homologs. When a phylogenetic tree was constructed from the putative sequences, there was a separate group comprising cyanobacterial PGTs. Figure 1 shows the cyanobacterial cluster, in which members shared sequence identities of more than 34%. Because the degenerate primers designed from all of the cyanobacterial PGTs were too highly degenerate, the cluster was divided into four subgroups, as shown in Fig. 1: this division allowed primers to be designed for each of the four subgroups. The PGTs in subgroup I were clearly distinguishable from the others, because they all originated from marine picocyanobacteria, which are abundant in the pelagic realm. Subgroup I could be divided further into two groups comprising PGTs from either Prochlorococcus species (CIA) or marine Synechococcus species (CIB). Subgroup II was also divided into two groups, CIIA, consisting mostly of PGTs from Synechococcus species, and CIIB, containing the other PGTs. Among the primers designed for each subgroup, those for the CIIA and CIIB subgroups successfully amplified DNA sequences of the expected sizes. The primer sequences were 5′-GTTCAGGAWTAGGAGGTGGAGT-3′ (CIIA-forward)/5′-CGCYTCAATWGCTACATTTCCA-3′ (CIIA-reverse) and 5′-ACGACTGGCTMYCGYTTTAYCTGA-3′ (CIIB-forward)/5′-GCYTCCACCCAYTTRGGGGTCA-3′ (CIIB-reverse). Based on the determined partial gene sequences, additional sets of primer pairs were designed for the inverse PCR method (20). The sequences were 5′-GATGAACTACAACAGGGTCTGCGTC-3′ (CY-007 forward)/5′-CGGCTTTTTAAGGCTTTTGCCATATTC-3′ (CY-007 reverse), 5′-GTCTGCGTGAATGTCGAGG-3′ (CY-047 forward)/5′-ATGACCTCGGCTGTGTAAG-3′ (CY-047 reverse), and 5′-CCTACAAAAAGAGCTAGGCGACTGTTTTG-3′ (UCNR forward)/5′-CCAAAGAAACGGAAGCCATGCTG-3′ (UCNR reverse). Total genomic DNA samples were partially digested with RsaI and then self-ligated to be used as templates for PCR amplification with the primer pairs. The amplified DNA sequences revealed the missing 5′- and 3′-end sequences of the genes.
FIG. 1.
Neighbor-joining phylogenetic tree of cyanobacterial PGT protein sequences, identified by NCBI accession numbers. Bootstrap values are presented at the nodes. The names of strains whose PGTs are characterized are in bold.
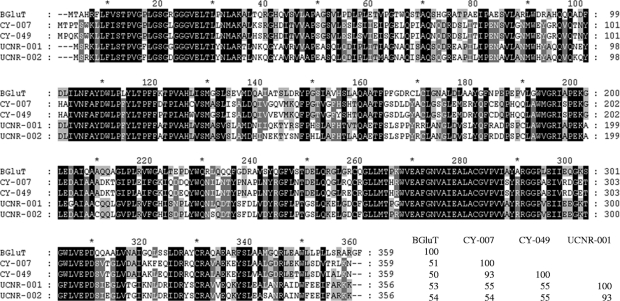
The deduced protein sequences were multiply aligned with BGluT (Fig. 2). Amino acid identities for all sequences in pairwise comparisons are given as percentages in Fig. 2. Recently, draft assemblies of the genome sequences of Arthrospira platensis strain Paraca and Arthrospira maxima CS-328 (UTEX 2342) were announced. The annotated PGT (GenBank accession no. EDZ91868) of Arthrospira maxima CS-328 was identical to the PGT of CY-049 at both the amino acid and nucleotide levels, proving that the two organisms originated from the same UTEX stock (UTEX 2342). On the other hand, the PGTs of Arthrospira platensis strains Paraca and CY-007 were different at nine individual nucleotides, resulting in seven amino acid differences. A phylogenetic analysis showed that CY-007 and CY-049 PGTs belonged to the CIIB subgroup and that UCNR-001 and UCNR-002 PGTs clustered in the CIIA subgroup (data not shown).
FIG. 2.
Alignment of multiple PGT sequences. Conserved sequences are shaded at four levels using GeneDoc software. At the end of the alignment, amino acid identities in percentages are given for all sequences in pairwise comparisons.
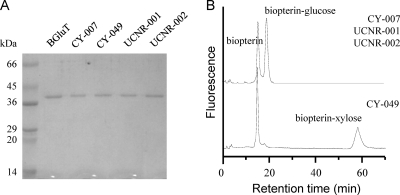
In order to identify the catalytic function of the putative PGTs, the recombinant proteins were produced in Escherichia coli. The complete open reading frame (ORF) sequences were amplified by PCR from the genomic DNA samples, cloned into the pGEM-T vector, and subsequently cloned as NdeI/BamHI restriction fragments into pET-28b (for CY-007 and CY-049 sequences) or pET-15b (for UCNR-001 and UCNR-002 sequences). E. coli BL21(DE3)/pLysS transformants were induced with 0.05 to 0.2 mM isopropyl-β-d-thiogalactopyranoside and were cultured for 8 h at 22°C. The recombinant proteins were purified by chromatography on Ni-nitrilotriacetic acid gel according to the instructions of the manufacturer (Qiagen). The proteins were eluted with 250 mM imidazole, dialyzed against a mixture of 20 mM Tris-HCl (pH 7.5) and 30% (vol/vol) glycerol, and stored in aliquots at −70°C until use. Purification of the proteins was confirmed by electrophoresis on an SDS-polyacrylamide gel (Fig. 3A). BGluT from a previous purification was used (6). Aliquots of PGT were assayed at 37°C for 10 min in a reaction mixture of 100 μl containing 50 mM sodium phosphate, pH 7.5, 10 mM MnCl2, 0.2% ascorbic acid, 1 μM BH4 (Schircks Lab, Switzerland), and 100 μM UDP-glucose or UDP-xylose. The reaction mixture was combined with an equal volume of acidic iodine solution (2% KI and 1% I2 in 1 N HCl) for 1 h in the dark. After centrifugation, the supernatant was mixed in a 10:1 volume ratio with 5% ascorbic acid and subjected to high-performance liquid chromatography (HPLC). HPLC was performed with a Gilson 321 pump equipped with an Inertsil ODS-3 column (150 by 2.3 mm; particle size, 5 μm [GL Science, Japan]) and a fluorescence detector (Shimadzu RF-10AXL). Pteridines were eluted with 10 mM potassium phosphate buffer (pH 6.0) at a flow rate of 1.2 ml/min and were monitored at excitation and emission wavelengths of 350 and 450 nm, respectively.
FIG. 3.
Analysis of purified recombinant PGTs on an SDS-12.5% polyacrylamide gel (A) and HPLC analysis of the enzymatic products (B).
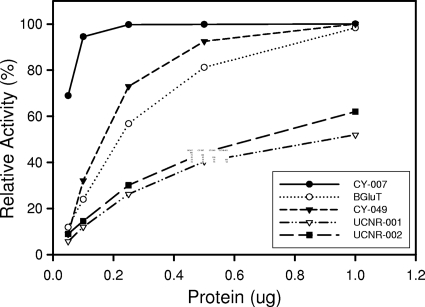
The enzymatic products of PGTs (Fig. 3) appeared only when enzymes were incubated with BH4 as a sugar acceptor and either UDP-glucose (for CY-007, UCNR-001, and UNCR-002 PGTs) or UDP-xylose (for CY-049 PGT) as a sugar donor. HPLC analysis of cultured CY-007 and CY-049 cells confirmed the presence of the corresponding biopterin glycosides (data not shown), supporting the conclusion that the PGTs exhibited genuine in vivo activities. This is the first report of a gene encoding a PGT that catalyzes xylosyl transfer to BH4. Although the data are not shown here, we found additional xylosyl transfer PGTs in Anabaena sp. PCC 7120, Gloeobacter violaceus PCC 7421, and Thermosynechococcus elongatus BP-1, whose genomic sequences were determined. The putative PGT genes (represented in Fig. 1) were amplified by PCR from the genomic DNA, which was a kind gift from the Kazusa DNA Research Institute (http://genome.kazusa.or.jp/cyanobase/). The recombinant proteins for the in vitro activity assay were prepared by cloning the genes into pET-28b and overexpressing the proteins in E. coli according to the same procedures performed for the other PGTs. Interestingly, CY-007 and CY-049 PGTs exhibited different substrate specificities, although they share 93% protein sequence identity, and they also had higher specific activities than the other PGTs (Fig. 4). The three-dimensional structures of the proteins are currently being investigated to further understanding of the structural properties involved. Considering the cyanobacterial PGTs hitherto identified, there seems to be little correlation between their substrate preferences and phylogenetic classification. However, the CI group PGTs, which diverged early from the CII group PGTs, might have some distinctive features. Finally, the successful cloning of PGT genes from environmental DNA allows for potentially new PGTs to be isolated from cyanobacteria, which are abundant in nature.
FIG. 4.
Comparative analysis of PGT activities. The maximal activity (100%) corresponds to complete glycosylation of 1 μM BH4 in the reaction mixture, which contained 0.5 mM UDP-xylose for CY-049 PGT or 0.5 mM UDP-glucose for the other PGTs. The mixtures were incubated for 10 min with the indicated amounts of proteins.
Nucleotide sequence accession numbers.
The complete ORF sequences were deposited in GenBank under accession numbers GU812289 (for strain CY-007), GU812291 (for strain CY-049), GU812292 (for the UCNR-001 sequence), and GU812293 (for the UCNR-002 sequence).
Acknowledgments
This work was supported by a 2006 grant from Inje University.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Blanchard, S., and J. S. Thorson. 2006. Enzymatic tools for engineering natural product glycosylation. Curr. Opin. Chem. Biol. 10:263-271. [DOI] [PubMed] [Google Scholar]
- 2.Cha, K. W., W. Pfleiderer, and J. J. Yim. 1995. Pteridines, part CVI. Isolation and characterization of limipterin (1-O-(L-erythro-biopterin-2′-yl)-β-N-acetylglucosamine) and its 5,6,7,8-tetrahydro derivative from green sulfur bacterium Chlorobium limicola f. thiosulfatophilum NCIB 8327. Helv. Chim. Acta 78:600-614. [Google Scholar]
- 3.Cha, E.-Y., J. S. Park, S. Jeon, J. S. Kong, Y. K. Choi, J.-Y. Ryu, Y.-I. Park, and Y. S. Park. 2005. Functional characterization of the gene encoding UDP-glucose:tetrahydrobiopterin α-glucosyltransferase in Synechococcus sp. PCC 7942. J. Microbiol. 43:191-195. [PubMed] [Google Scholar]
- 4.Cho, S. H., J. U. Na, H. Youn, C. S. Hwang, C. H. Lee, and S. O. Kang. 1998. Tepidopterin, 1-O-(L-threo-biopterin-2′-yl)-N-acetylglucosamine from Chlorobium tepidum. Biochim. Biophys. Acta 1379:53-60. [DOI] [PubMed] [Google Scholar]
- 5.Choi, Y. K., Y. K. Hwang, Y. H. Kang, and Y. S. Park. 2001. Chemical structure of 1-O-(L-erythro-biopterin-2′-yl)-α-glucose isolated from a cyanobacterium Synechococcus sp. PCC 7942. Pteridines 12:121-125. [Google Scholar]
- 6.Choi, Y. K., Y. K. Hwang, and Y. S. Park. 2001. Molecular cloning and disruption of a novel gene encoding UDP-glucose:tetrahydrobiopterin α-glucosyltransferase in the cyanobacterium Synechococcus sp. PCC 7942. FEBS Lett. 502:73-78. [DOI] [PubMed] [Google Scholar]
- 7.Chung, H. J., Y.-A. Kim, Y. J. Kim, Y. K. Choi, Y. K. Hwang, and Y. S. Park. 2000. Purification and characterization of UDP-glucose:tetrahydrobiopterin glucosyltransferase from Synechococcus sp. PCC 7942. Biochim. Biophys. Acta 1524:183-188. [DOI] [PubMed] [Google Scholar]
- 8.Forrest, H. S., and C. Van Baalen. 1970. Microbiology of unconjugated pteridines. Annu. Rev. Microbiol. 24:91-108. [DOI] [PubMed] [Google Scholar]
- 9.Giegerich, R., F. Meyer, and C. Schleiermacher. 1996. GeneFisher—software support for the detection of postulated genes. Proc. Int. Conf. Intell. Syst. Mol. Biol. 4:68-77. [PubMed] [Google Scholar]
- 10.Griffith, B. R., J. M. Langenhan, and J. S. Thorson. 2005. ‘Sweetening’ natural products via glycorandomization. Curr. Opin. Biotechnol. 16:622-630. [DOI] [PubMed] [Google Scholar]
- 11.Hatfield, D. L., C. Van Baalen, and H. S. Forrest. 1961. Pteridines in blue green algae. Plant Physiol. 36:240-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang, Y. K., J. Y. Kang, H. J. Woo, Y. K. Choi, and Y. S. Park. 2002. Functional investigation of a gene encoding pteridine glycosyltransferase for cyanopterin synthesis in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1570:141-144. [DOI] [PubMed] [Google Scholar]
- 13.Ikawa, M., J. J. Sasner, J. F. Haney, and T. L. Foxall. 1995. Pterins of the cyanobacterium Aphanizomenon flos-aquae. Phytochemistry 38:1229-1232. [Google Scholar]
- 14.Kim, H.-L., D. H. Kim, Y. K. Lee, S. O. Park, Y.-W. Lee, O.-S. Kwon, and Y. S. Park. 2010. An enzymatic method to distinguish tetrahydrobiopterin from oxidized biopterins using UDP-glucose:tetrahydrobiopterin glucosyltransferase. Anal. Biochem. 397:79-83. [DOI] [PubMed] [Google Scholar]
- 15.Lee, H. W., C. H. Oh, A. Geyer, W. Pfleiderer, and Y. S. Park. 1999. Characterization of a novel unconjugated pteridine glycoside, cyanopterin, in Synechocystis sp. PCC 6803. Biophys. Biochim. Acta 1410:61-70. [DOI] [PubMed] [Google Scholar]
- 16.Maclean, F. I., Y. Fujita, H. S. Forrest, and J. Myers. 1965. Photosynthetic phosphorylation: stimulation by pteridines and a comparison with phosphodoxin. Science 149:636-639. [DOI] [PubMed] [Google Scholar]
- 17.Noguchi, Y., A. Ishii, A. Matsushima, D. Haishi, K. Yasumuro, T. Moriguchi, T. Wada, Y. Kodera, M. Hiroto, H. Nishimura, M. Sekine, and Y. Inada. 1999. Isolation of biopterin-α-glucoside from Spirulina (Arthrospira) platensis and its physiologic function. Mar. Biotechnol. 1:207-210. [DOI] [PubMed] [Google Scholar]
- 18.Saito, T., H. Ishikura, Y. Hada, K. Fukui, Y. Kodera, A. Matsushim, and Y. Inada. 2003. Photostabilization of phycocyanin and anthocyanin in the presence of biopterin-α-glucoside from Spirulina platensis under ultraviolet ray. Dyes Pigm. 56:203-207. [Google Scholar]
- 19.Thöny, B., G. Auerbach, and N. Blau. 2000. Tetrahydrobiopterin biosynthesis, regeneration, and functions. Biochem. J. 347:1-16. [PMC free article] [PubMed] [Google Scholar]
- 20.Triglia, T., M. G. Peterson, and D. J. Kemp. 1988. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 16:8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wachi, Y., J. G. Burgess, K. Iwamoto, N. Yamada, N. Nakamura, and T. Matsunaga. 1995. Effect of ultraviolet-A (UV-A) light on growth, photosynthetic activity and production of biopterin glucoside by the marine UV-A resistant cyanobacterium Oscillatoria sp. Biochim. Biophys. Acta 1244:165-168. [DOI] [PubMed] [Google Scholar]