Abstract
In this paper, we report the surface assembly of a functional minicellulosome by using a synthetic yeast consortium. The basic design of the consortium consisted of four different engineered yeast strains capable of either displaying a trifunctional scaffoldin, Scaf-ctf (SC), carrying three divergent cohesin domains from Clostridium thermocellum (t), Clostridium cellulolyticum (c), and Ruminococcus flavefaciens (f), or secreting one of the three corresponding dockerin-tagged cellulases (endoglucanase [AT], exoglucanase [EC/CB], or β-glucosidase [BF]). The secreted cellulases were docked onto the displayed Scaf-ctf in a highly organized manner based on the specific interaction of the three cohesin-dockerin pairs employed, resulting in the assembly of a functional minicellulosome on the yeast surface. By exploiting the modular nature of each population to provide a unique building block for the minicellulosome structure, the overall cellulosome assembly, cellulose hydrolysis, and ethanol production were easily fine-tuned by adjusting the ratio of different populations in the consortium. The optimized consortium consisted of a SC:AT:CB:BF ratio of 7:2:4:2 and produced almost twice the level of ethanol (1.87 g/liter) as a consortium with an equal ratio of the different populations. The final ethanol yield of 0.475 g of ethanol/g of cellulose consumed also corresponded to 93% of the theoretical value. This result confirms the use of a synthetic biology approach for the synergistic saccharification and fermentation of cellulose to ethanol by using a yeast consortium displaying a functional minicellulosome.
According to the new Energy Policy Act, several billion gallons of renewable fuel must be produced by 2012, with most of that volume produced as biofuels from renewable biomass. Cellulosic biomass is the most abundant and sustainable material for biofuel production because of its high sugar content. It has been estimated that 1.4 billion tons of cellulosic biomass can be produced annually in the United States without affecting the food supply, animal feed, and fiber use (18). Ethanol is useful as an alternative transportation fuel and could lessen the nation's dependence on foreign oil (13).
Unfortunately, cost-effective production of ethanol from cellulosic biomass remains a major challenge, primarily due to its highly recalcitrant nature (10). Typically, the synergistic actions of endoglucanase, exoglucanase, and β-glucosidase are required for the complete hydrolysis of cellulose to glucose. The high cost associated with the use of a large quantity of enzymes required for efficient biomass conversion to fermentable sugars is a primary impeding factor. While the cost of ethanol production has become more competitive by combining cellulose saccharification and fermentation (SSCF), a new method known as consolidated bioprocessing (CBP), which further combines enzyme production with SSCF into a single process, has gained increasing recognition as a potential solution for the low-cost production of ethanol (12). However, a natural microorganism that possesses the capability for efficient enzyme production, cellulose saccharification, and ethanol fermentation remains elusive (23). In recent years, efforts have been made in engineering microorganisms toward the goal of consolidated bioprocessing (4). In particular, Saccharomyces cerevisiae is an attractive engineering candidate due to its high ethanol productivity and high inherent ethanol tolerance (16). However, many past attempts based on either secretion of cellulases or surface display of cellulases have resulted in relative low ethanol productivity (2, 3, 7, 8).
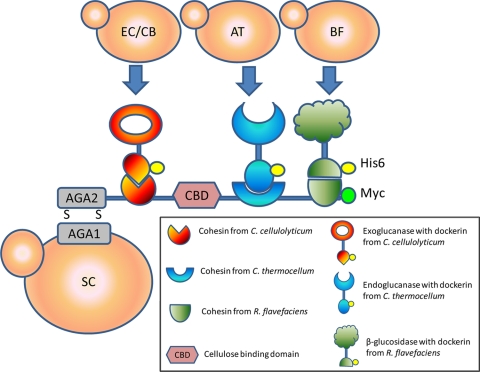
In nature, anaerobic microorganisms have developed an elaborate enzyme complex known as cellulosome for efficient hydrolysis of cellulose. This highly ordered structure allows the assembly of multiple enzymes in close proximity to the substrate, resulting in a high level of enzyme-substrate-microbe synergy (6). Our group reported recently the functional assembly of minicellulosomes on the yeast surface and demonstrated an up-to-3-fold increase in ethanol production from phosphoric acid-swollen cellulose (PASC) compared with free enzymes (20). A similar enhancement in ethanol production has also been reported by the Zhao group, who used an engineered yeast strain coexpressing a displayed miniscaffoldin and three different cellulases (22). However, coexpression of all four components in a single strain resulted in relatively low levels of exoglucanase and β-glucosidase, probably due to the heavy metabolic burden and potential jamming of the secretion machinery. To address these issues, we report here the use of a synthetic yeast consortium composed of one strain displaying the miniscaffoldin and three strains secreting dockerin-tagged cellulases for the functional presentation of minicellulosomes on the yeast surface (Fig. 1). By exploiting the specific interaction of the three separate cohesin-dockerin pairs employed, optimal performance can be obtained by fine-tuning the required ratio of cellulase-secreting cells and miniscaffoldin-displaying cells. The resulting consortium is capable of producing ethanol directly from PASC.
FIG. 1.
Surface assembly of a functional minicellulosome through intracellular complementation, using a synthetic yeast consortium. The basic design consisted of four different engineered yeast strains capable of either displaying a trifunctional scaffoldin Scaf-ctf (SC) or secreting one of the three corresponding dockerin-tagged enzymes (endoglucanase [AT], exoglucanase [EC/CB], or β-glucosidase [BF]).
MATERIALS AND METHODS
Strains, plasmids, and media.
Escherichia coli strain JM109 [recA1 endA1 supE44 hsdR17 gyrA96 thi relA1 λ− Δ(lac-proAB) (F′ traD36 proAB lacIq lacZΔM15)] was used as a host for recombinant DNA manipulation. S. cerevisiae strain EBY100 (MATa AGA1::GAL1-AGA1::URA3 ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL) was used for displaying the scaffoldin and the secretion of β-glucosidase. S. cerevisiae strain BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) was used for secretion of the remaining enzymes. E. coli strains were grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl) with 100 μg/liter of ampicillin when required. Except for the consortium experiments, yeast strains were either grown in YPD medium (2% dextrose, 1% yeast extract, 2% peptone) or SDC medium (2% glucose, 0.67% yeast nitrogen base, 0.5% Casamino Acids). The filamentous fungus Trichoderma reesei used for mRNA extraction was cultured in potato dextrose agar medium (Difco Laboratories) containing 0.4% potato starch, 2% glucose, and 2% agar at 25°C.
Plasmid construction and transformation.
Primers used for plasmid construction are provided in Table S1 of the supplemental material. Construction of the surface display vector pSctf was previously described (20). To construct the secretion plasmid pAt, a 1,338-bp fragment of the Clostridium thermocellum endoglucanase CelA gene was amplified by PCR using FAt and RAt as primers and the vector pETAt as the template. The amplified fragment was cloned into the ClaI and XhoI sites of plasmid pCEL15 (15) under the control of a constitutive PGK promoter. Plasmid pEc, encoding a His6-tagged exoglucanase (CelE) from Clostridium cellulolyticum, was generated by PCR from pETEc (9) using primers FEc and REc. The amplified fragment was cloned into ClaI-XhoI-linearized plasmid pCEL15 to obtain pEc. The secretion vector pCBH2c was constructed using a two-step procedure. First, the dockerin domain of C. cellulolyticum was amplified by PCR using primers FDc and RDc and ligated into the BglII and XhoI sites of pCEL15 to form pDc. The gene coding for the cellobiohydrolase CBHII was amplified from the total RNA extracted from T. reesei by reverse transcription-PCR using primers FCBH2 and RCBH2. The resulting product was digested and ligated into the ClaI and BamHI sites of pDc to form pCBH2c. To generate plasmid pBGLf, the Ruminococcus flavefaciens dockerin domain amplified from pETGf (6) using primers FDf and RDf was ligated into pBGL encoding a β-glucosidase gene from Thermoascus aurantiacus (11). All yeast transformations were performed according to the standard lithium acetate procedure as described elsewhere (1). All the recombinant strains used in this research are summarized in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain | Host/plasmid | Promoter | Marker | Tag | Description |
|---|---|---|---|---|---|
| CE | BY4742/pCEL15 | PGK | URA | His6 | Secretes a small peptide (negative control) |
| AT | BY4742/pAt | PGK | URA | His6 | Secretes endoglucanase At (CelA from C. thermocellum with its native dockerin) |
| EC | BY4742/pEc | PGK | URA | His6 | Secretes exoglucanase Ec (CelE from C. cellulolyticum with its native dockerin) |
| CB | BY4742/pCBH2c | PGK | URA | His6 | Secretes cellobiohydrolase CBHc (CBHII from T. reesei fused with a dockerin from C. cellulolyticum) |
| BF | EBY100/pBGLf | GAP | TRP | His6 | Secretes β-glucosidase Bglf (Bg1I from T. aurantiacus fused with a dockerin from R. flavefaciens) |
| SC | EBY100/pScaf3 | GAL | TRP | C-myc | Displays the miniscaffoldin Sacf-ctf |
Development of synthetic consortia and fermentation.
Yeast strains EBY100 harboring either pSctf or pBGLf (carrying a Trp1 marker) and BY4742 harboring either pCEL15, pAt, pCBH2c, or pEc (carrying a URA3 marker) were first precultured in SDC medium at 30°C for 18 h. For coculturing of the synthetic consortia, the initial strains were mixed to the desired ratio into 200 ml SGC medium (20.0 g/liter galactose, 6.7 g/liter yeast nitrogen base without amino acids, 5.0 g/liter Casamino Acids) supplemented with 10 mM CaCl2 to an optical density at 600 nm (OD600) of 1 and grown for 48 h at 20°C.
Resting cell assays.
For the resting cell assays, PASC was prepared from Avicel PH101 (Sigma) according to the method of Walseth (21) and used as the substrate. Cells from the different consortia were first washed once with buffer containing 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 10 mM CaCl2 and resuspended in 20 mM Tris-HCl buffer (pH 6.0) supplemented with 10 mM CaCl2 and 10 g/liter PASC to a final OD of 50. Samples were collected periodically and immediately mixed with 3 ml of DNS reagent (10 g/liter dinitrosalicylic acid, 10 g/liter sodium hydroxide, 2 g/liter phenol, 0.5 g/liter sodium sulfite) to determine the level of reducing sugar. After incubating at 95°C for 10 min, 1 ml of 40% Rochelle salts was added to fix the color before measuring the absorbance of the supernatants at 575 nm. The glucose concentration was determined using a glucose HK assay kit from Sigma.
Fermentation.
Cells from the different consortia were washed and resuspended in 10 ml SDC medium containing 6.7 g/liter yeast nitrogen base without amino acids, 20 g/liter Casamino Acids, and 10 g/liter PASC as the sole carbon source to a final OD of 50 or 15 mg (cell dry weight)/ml deducing sugars, and glucose concentrations were measured by the methods described above. The amount of residual cellulose was measured by the phenol-sulfuric acid method as described by Dubois et al. (5). The ethanol concentration was measured by gas chromatography (model 6890; Hewlett Packard) using a flame ionization detector and an HP-FFTP column.
Immunofluorescence microscopy.
Immunofluorescence microscopy was performed as described previously (20). Briefly, cells were washed with phosphate-buffered saline (PBS) and resuspended in PBS containing 1 mg/ml of bovine serum albumin (BSA). Either anti-His6 or anti-Myc antibody was added and incubated for 1 h with occasional mixing. Alexa Fluor 488-conjugated anti-mouse IgG was added after washing and resuspending in PBS with 1 mg/ml of BSA. Images were acquired by using a fluorescence microscope (Olympus BX51) after washing with PBS three times. Whole-cell fluorescence was measured using a fluorescence microplate reader (Synergy4; BioTek, VT) with an excitation wavelength at 485 nm and an emission wavelength at 535 nm.
Enzyme activity assay.
To assay the functional secretion of β-glucosidase, 100 μl of a 10 mM concentration of the fluorescent substrate p-4-methylumbellifery-β-d-glucopyranoside was added to 100 μl of culture medium and incubated at 37°C for 1 h. The activity was confirmed by detecting the fluorescence under UV light. To test the successful secretion of functional AT, EC, and CBHC in the medium, a modified Congo red staining method was employed (19). Briefly, carboxymethyl cellulose (CMC) was added to 100 μl of culture medium to a concentration of 1% and incubated at 37°C for 2 h. Thereafter, 20 μl of Congo red solution (0.1% Congo red in 50 mM potassium phosphate buffer [pH 6.5]) was used for staining. The culture medium from BY4742/pCEL15 was used as a negative control in all cases.
RESULTS
Secretion of dockerin-tagged endoglucanase, exoglucanase, and β-glucosidase.
To enable the complete hydrolysis of cellulose to glucose, three different yeast strains were engineered to secrete either an endoglucanase CelA from C. thermocellum (A), an exoglucanase CelE from C. cellulolyticum (E), or β-glucosidase BglI from Thermoascus aurantiacus (Bgl). For specific docking of secreted enzymes onto the surface display miniscaffoldin (Scaf-ctf), three different dockerins from C. thermocellum (t), C. cellulolyticum (c), and R. flavefaciens (f) were used to generate dockerin-tagged enzymes, resulting in At, Ec, and Bglf, respectively. All three dockerin-tagged enzymes were secreted using an α-factor secretion peptide and flanked by a His6 tag.
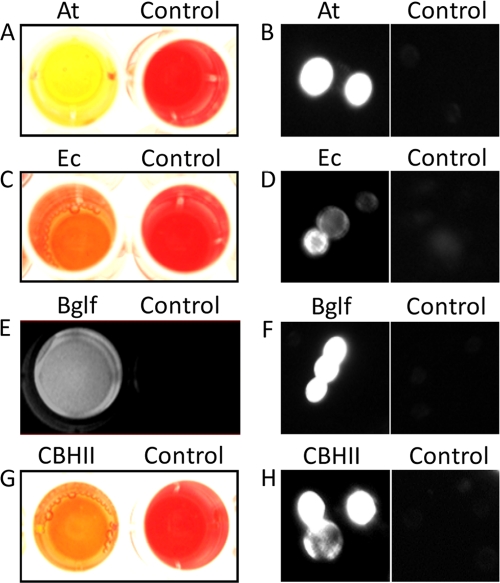
To examine the functional secretion of At, the culture medium for BY4742/pAt was introduced into a 1% CMC solution for 2 h and stained with Congo red. A clear color change from red to yellow confirmed At activity (Fig. 2A), while no color change was observed with the culture medium of cells displaying Scaf-ctf. In addition, functionality of the dockerin domain in At was confirmed by incubating the culture medium with cells displaying Scaf-ctf, followed by immunofluorescence microscopy using the anti-His6 antibody (Fig. 2B). Similarly, the activity of secreted Ec was confirmed by observing a color change from red to orange with the Congo red assay (Fig. 2C). Secretion of Bglf was confirmed by detecting a strong fluorescence signal upon addition of the fluorescent substrate p-4-methylumbellifery-β-d-glucopyranoside to the culture medium of EBY100/pBGLf (Fig. 2E). Moreover, detectable fluorescence was observed on the Scaf-ctf-displaying cells only after incubation with the culture medium of either BY4742/pEc or EBY100/pBGLf, again confirming the functionality of the dockerin domains of the two secreted enzymes (Fig. 2D and F). Collectively, these results confirmed the successful secretion of functional dockerin-tagged enzymes from the three engineered yeast strains.
FIG. 2.
Secretion of dockerin-tagged enzymes. (A to D) Enzyme activities of different secretion strains in the growth medium were examined by using either CMC (A to C) or p-4-methylumbellifery-β-d-glucopyranoside (D) as the substrate. For the CMC assay, Congo red was added as an indicator. (E to G) Binding of secreted enzymes onto surface-displayed Scaf-ctf was confirmed by immunofluorescence microscopy. Cells were probed with anti-C-His6 serum and fluorescently stained with a goat anti-mouse IgG conjugated with Alexa Fluor 488. The growth medium for EBY100/pScat-ctf was used as a control in all cases.
Functional assembly of each enzyme onto the surface-displayed scaffoldin.
With the successful secretion of all dockerin-tagged enzymes, the feasibility of recruiting these engineered yeast strains into a synthetic consortium system was examined. For the initial experiments, four different consortia with different cell populations—a strain displaying Scaf-ctf (SC), a strain carrying pCEL15 (CE) as a nonsecretion control, an At-secreting strain (AT), an Ec-secreting strain (EC), and a Bglf-secreting strain (BF)—were created to test the ability of the consortia to hydrolyze PASC and to produce ethanol. By exploiting the specific interactions of the different dockerin-cohesin pairs, we expected the spontaneous self-assembly of a functional minicellulosome onto the displayed Scaf-ctf (see Fig. S1 in the supplemental material). All consortia developed are listed in Table 2.
TABLE 2.
Consortia generated in this study
| Consortium | Populations |
|---|---|
| C1 | SC, CE, CE, CE |
| C2 | SC, AT, CE, CE |
| C3 | SC, AT, EC, CE |
| C4 | SC, AT, EC, BF |
| C5 | SC, AT, CB, CE |
| C6 | SC, AT, CB, BF |
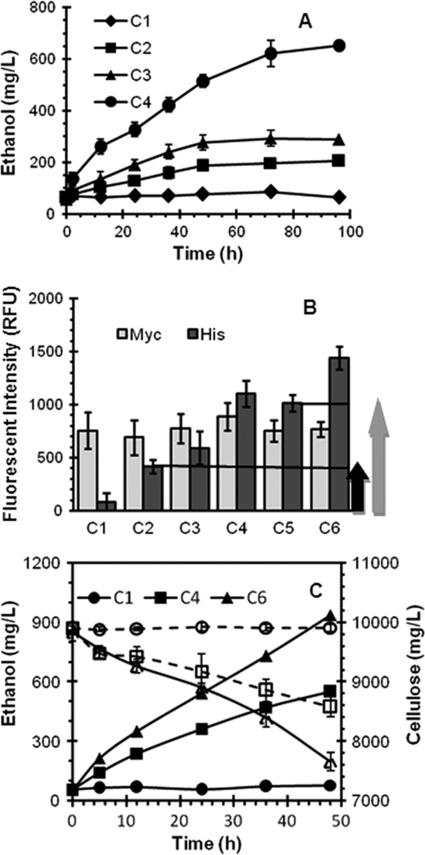
Different populations of cells were cocultured, washed, and resuspended in SDC medium to a final OD of 50. As shown in Fig. 3A, the consortium composed of SC, CE, and AT (C2) showed a noticeable increase in ethanol production compared to the control consortium (SC and CE) secreting no enzyme (C1). This result clearly suggests the functional assembly of secreted At onto the displayed Scaf-ctf, as demonstrated by the ability to hydrolyze PACS and subsequent ethanol production. The surface assembly of At was further verified by immunofluorescence microscopy using both anti-C-myc and anti-His antibodies (Fig. 3B). While the fluorescent intensity detected for the C-myc tag was similar between the two consortia (C1 and C2), only the consortium containing AT (C2) showed an appreciable level of His tag fluorescence, confirming the docking of secreted At onto the surface-displayed Scaf-ctf.
FIG. 3.
Cellulose hydrolysis and ethanol production by different synthetic consortia (see Table 2). Different consortia were established using a strain displaying Scaf-ctf (SC), a strain carrying pCEL15 (CE) as a control, an At-secreting strain (AT), an Ec-secreting strain (EC), a CHBc-secreting strain (CB), and/or a Bglf-secreting strain (BF). (A) Ethanol production from PASC by different synthetic consortia. (B) Docking of At, Ec, and/or BglF onto Sacf-ctf-displaying cells in different consortia. Cells were probed with either anti-C-myc or anti-C-His6 serum and fluorescently stained with a goat anti-mouse IgG conjugated with Alexa Fluor 488. Whole-cell fluorescence was determined using a fluorescence microplate reader. The dark arrow and gray arrow indicate the increase in fluorescence intensity after At and CBHII docking. (C) Cellulose hydrolysis (dashed lines) and ethanol production (solid lines) from PASC by a consortium composed of SC, AT, CB, and BF.
Consistent with the expected enhancement in glucose liberation by the addition of exoglucanase, ethanol production (Fig. 3A) was slightly increased for the consortium containing both AT and EC (C3). However, a further 3-fold increase in ethanol production was observed for the consortium (C4) containing all the functional populations (SC, AT, EC, and BF) required to assemble the trifunctional minicellulosome (Fig. 3A). This increase in ethanol production was expected, as the presence of β-glucosidase is essential for complete hydrolysis of cellulose into glucose. By probing the His tag from each secreted cellulase, the level of enzyme incorporation onto the surface for different consortia could be directly assessed using whole-cell fluorescence measurements. Although the incorporation of secreted Ec (C3) and Bglf (C4) was confirmed by an increase in the His tag fluorescence intensity, the level of Ec incorporation was substantially lower than that of At or Bglf (Fig. 3B). This low level of Ec incorporation is consistent with the modest increase in ethanol production, suggesting that the efficiency of the consortium could be further improved by increasing the exoglucanase activity.
One major benefit of the consortium system is our ability to modulate any given population without affecting the other cell populations. To test this hypothesis, a cellobiohydrolase/exoglucanase CBHII from T. reesei (CBH), which has been shown to have high-level secretion in S. cerevisiae (17), was tagged with the C. cellulolyticum dockerin domain (CBHc) and functionally secreted from strain BY4742 (CB) (Fig. 2G and H). As shown in Fig. 3B, compared to the consortium with EC (C4), the new consortia (C5 and C6) using CB showed a higher level of His tag fluorescence intensity, indicating the improved docking level of CBHc. This enhanced level of CBHc docking was accompanied by a corresponding increase in PASC hydrolysis and ethanol production (Fig. 3C). An ethanol level of 930 mg/liter was achieved after 50 h, which corresponds to 83% of the theoretical value (0.43 g of ethanol/g of cellulose). These results confirm the cooperative action between each population in the consortium and the capabilities of the surface-assembled minicellulosomes for cellulosic ethanol production.
Systematic improvement of the synthetic consortia for producing cellulosic ethanol.
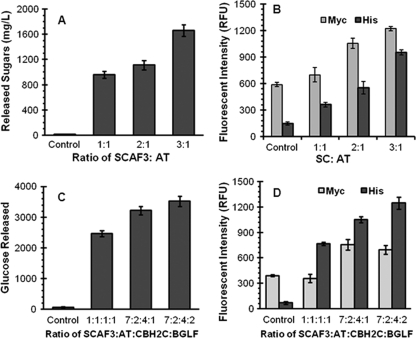
The results presented here demonstrate the feasibility of using the yeast consortium for ethanol production. However, the ethanol yield is lower than our previously reported yield when using in vitro enzyme loading (20). This can be attributed to a lower percentage of cells displaying the miniscaffoldin (18% to 60%) and the lower efficiency of enzyme docking (less than a 1:3 ratio between the C-myc tag and His tag signal). To address these problems, a systematic improvement of the synthetic consortium was investigated. By exploiting the modular nature of the consortium design, we can easily fine-tune the ratio of different populations by controlling the initial inoculation ratio. To investigate this possibility, we first optimized the level of reducing sugar production with this strategy. Initially, cultures with different inoculation ratios of SC and AT were harvested after 48 h of coculturing and resuspended in buffer containing PASC. As shown in Fig. 4A, the reducing sugar concentration increased with increasing SC:AT ratio, reaching the highest level at a ratio of 3:1, above which no further improvement was observed (data not shown). This increase in reducing sugar production was accompanied by a corresponding increase in the C-myc signal (Fig. 4B), consistent with the expected higher percentage of cells displaying Scaf-ctc. Moreover, the higher level of His tag signal suggests improved binding of the secreted At onto the displayed Scaf-ctf due to the higher cohesin/dockerin ratio. This result clearly illustrates our ability to improve the performance of the consortium simply by adjusting the inoculation ratio.
FIG. 4.
Systematic optimization of different cell populations in the synthetic consortium. (A and B) Effects of the SC:AT ratio on reducing sugars production (A) and At binding (B). (C and D) Effects of the SC:AT:CB:BF ratio on glucose production (C) and enzyme binding (D). Enzyme binding was determined by immunofluorescence microscopy. Cells were probed with either anti-C-myc or anti-C-His6 serum and fluorescently stained with a goat anti-mouse IgG conjugated with Alexa Fluor 488. Whole-cell fluorescence was determined using a fluorescence microplate reader.
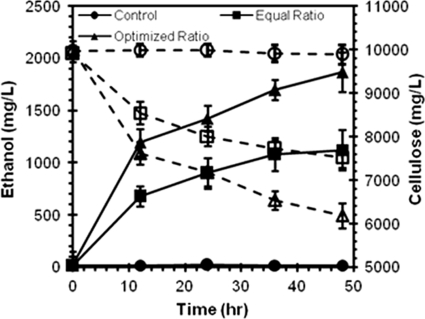
Using a similar strategy, we attempted to fine-tune the ratio of CB and BF in the consortium while keeping the ratio of SC to AT close to 3:1. As shown in Fig. 4C glucose production could be further improved by increasing both the CB and BF ratios, and the optimized consortium which released the highest level of glucose was found to consist of a SC:AT:CB:BF ratio of 7:2:4:2. This enhancement was the result of both a larger fraction of cells displaying Scaf-ctf (35%) and an increased amount of enzymes docked onto the minicellulosome (Fig. 4D). To investigate whether the improved glucose release in the resting cell assay could be directly translated into ethanol production, anaerobic fermentations using PASC were conducted. As shown in Fig. 5, the optimized consortium showed an almost-2-fold increase in both cellulose hydrolysis and ethanol production. The ethanol yield of 0.475 g of ethanol/g of sugar consumed also corresponds to 93% of the theoretical value and is comparable to the value obtained with the in vitro enzyme loading experiments (20).
FIG. 5.
Cellulose hydrolysis (dashed lines) and ethanol production (solid lines) from PASC by the optimized consortium.
DISCUSSION
Cellulose, a major component of the plant cell wall, is the most abundant renewable carbon source in nature that can be enzymatically degraded for ethanol production. However, the high cost of cellulases needed for complete hydrolysis is still one of the major obstacles in the quest for an economically feasible cellulose-based ethanol process (14). Cellulosome is a multicomponent enzyme complex that has been extensively investigated in recent years because of its intriguing potential in providing synergistic and highly efficient degradation of cellulose. Progress has been made in engineering yeast cells to display minicellulosome structures, toward the goal of CBP (20, 22). However, the two previous studies based on either in vitro loading of enzymes or simultaneous display of scaffoldin and secretion of cellulases in a single host cell presented problems. In the case of in vitro enzyme loading, the process cannot truly be CBP because separate E. coli cultivations are necessary. For the single host cell system, the inability to fine-tune levels of the three cellulases resulted in a highly uneven distribution toward endoglucanase. As a result, a substantially lower theoretical yield of 62% cellulose-to-ethanol conversion was obtained, compared to over 95% in the case of in vitro enzyme loading.
To address these problems, we engineered a synthetic yeast consortium capable of surface assembly of a functional minicellulosome via intracellular complementation. The basic design consisted of four different engineered yeast strains capable of either displaying the miniscaffoldin or secretion of one of the three required dockerin-tagged enzymes (endoglucanase, exoglucanase, or β-glucosidase). There are several unique features of our consortium system. First, the dockerin-cohesin pairs used in the present study are from three different species, which enable specific interactions between each dockerin-tagged enzyme and the displayed miniscaffoldin, resulting in highly controllable ordering of each enzyme in the minicellulosome structure. Second, by exploiting the modular nature of each population to provide a unique building block for the minicellulosome structure, the overall cellulosome assembly, cellulose hydrolysis, and ethanol production can be easily fine-tuned simply by adjusting the ratio of different populations in the consortium. As a result, the improved consortium consisted of a SC:AT:CB:BF ratio of 7:2:4:2 and produced almost twice the level of ethanol as a consortium with an equal proportion of the different populations. To our knowledge, this is the first successful report of the site-specific display of a multifunctional enzyme complex on the yeast surface through cooperative intracellular complementation using a synthetic consortium. Although our current study demonstrates the feasibility of assembling a functional minicellulosome consisting of only three enzymes on the yeast surface, there is no reason why other designer cellulosome structures cannot be similarly assembled by coordinating the required intracellular complementation within the consortium population.
Supplementary Material
Acknowledgments
We are grateful to Hisanori Tamaki for providing the plasmid pBGL and to I. S. Pretorius for providing the plasmid pCEL15.
This research was supported by grants from NSF (CBET 0903894) and DOE (EE0000988).
Footnotes
Published ahead of print on 1 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology, vol. 2. John Wiley & Sons, New York, NY.
- 2.Cho, K. M., Y. J. Yoo, and H. S. Kang. 1999. δ-Integration of endo/exo-glucanase and β-glucosidase genes into the yeast chromosomes for direct conversion of cellulose to ethanol. Enzyme Microb. Technol. 25:23-30. [Google Scholar]
- 3.Curry, C., N. Gilkes, G. Oneill, R. C. Miller, and N. Skipper. 1988. Expression and secretion of a Cellulomonas fimi exoglucanase in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 54:476-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Den Haan, R., S. H. Rose, L. R. Lynd, and W. H. van Zyl. 2007. Hydrolysis and fermentation of amorphous cellulose by recombinant Saccharomyces cerevisiae. Metab. Eng. 9:87-94. [DOI] [PubMed] [Google Scholar]
- 5.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 6.Fierobe, H. P., F. Mingardon, A. Mechaly, A. Belaich, M. T. Rincon, S. Pages, R. Lamed, C. Tardif, J. P. Belaich, and E. A. Bayer. 2005. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J. Biol. Chem. 280:16325-16334. [DOI] [PubMed] [Google Scholar]
- 7.Fujita, Y., J. Ito, M. Ueda, H. Fukuda, and A. Kondo. 2004. Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl. Environ. Microbiol. 70:1207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujita, Y., S. Takahashi, M. Ueda, A. Tanaka, H. Okada, Y. Morikawa, T. Kawaguchi, M. Arai, H. Fukuda, and A. Kondo. 2002. Direct and efficient production of ethanol from cellulosic material with a yeast strain displaying cellulolytic enzymes. Appl. Environ. Microbiol. 68:5136-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaudin, C., A. Belaich, S. Champ, and J. P. Belaich. 2000. CelE, a multidomain cellulase from Clostridium cellulolyticum: a key enzyme in the cellulosome. J. Bacteriol. 182:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Himmel, M. E., S. Y. Ding, D. K. Johnson, W. S. Adney, M. R. Nimlos, J. W. Brady, and T. D. Foust. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804-807. [DOI] [PubMed] [Google Scholar]
- 11.Hong, J., H. Tamaki, and H. Kumagai. 2007. Cloning and functional expression of thermostable beta-glucosidase gene from Thermoascus aurantiacus. Appl. Microbiol. Biotechnol. 73:1331-1339. [DOI] [PubMed] [Google Scholar]
- 12.Lynd, L. R., M. S. Laser, D. Brandsby, B. E. Dale, B. Davison, R. Hamilton, M. Himmel, M. Keller, J. D. McMillan, J. Sheehan, and C. E. Wyman. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169-172. [DOI] [PubMed] [Google Scholar]
- 13.Lynd, L. R., W. H. van Zyl, J. E. McBride, and M. Laser. 2005. Consolidated bioprocessing of cellulosic biomass: an update. Curr. Opin. Biotechnol. 16:577-583. [DOI] [PubMed] [Google Scholar]
- 14.McBride, J. E., J. J. Zietsman, W. H. Van Zyl, and L. R. Lynd. 2005. Utilization of cellobiose by recombinant β-glucosidase-expressing strains of Saccharomyces cerevisiae: characterization and evaluation of the sufficiency of expression. Enzyme Microb. Technol. 37:93-101. [Google Scholar]
- 15.Moses, G., R. R. Otero, and I. S. Pretorius. 2005. Domain engineering of Saccharomyces cerevisiae exoglucanases. Biotechnol. Lett. 27:355-362. [DOI] [PubMed] [Google Scholar]
- 16.Nevoigt, E. 2008. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 72:379-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penttilä, M. E., L. André, P. Lehtovaara, M. Bailey, T. T. Teeri, and J. K. C. Knowles. 1988. Efficient secretion of two fungal cellobiohydrolases by Saccharomyces cerevisiae. Gene 63:103-112. [DOI] [PubMed] [Google Scholar]
- 18.Perlack, R. D., L. L. Wright, A. F. Turhollow, R. L. Graham, B. J. Stokes, and D. C. Erbach. 2005. Biomass as feedstock for bioenergy and bioproducts industry: the technical feasibility of a billion-ton annual supply. U.S. Department of Energy, Oak Ridge, TN.
- 19.Thu, M., M. M. Oo, M. Myint, and S. S. Maw. 2008. Screening on cellulase enzyme activity of Aspergillus niger strains on cellulosic biomass for bioethanol production, p. 12-14. GMSARN International Conference on Sustainable Development: issues and prospects for the GMS. Greater Mekong Subregion Academic and Research Network, Pathumthani, Thailand.
- 20.Tsai, S. L., J. Oh, S. Singh, R. Z. Chen, and W. Chen. 2009. Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 75:6087-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walseth, C. S. 1952. Occurrence of cellulases in enzyme preparations from microorganisms. TAPPI J. 35:228-233. [Google Scholar]
- 22.Wen, F., J. Sun, and H. M. Zhao. 2010. Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl. Environ. Microbiol. 76:1251-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, Y. H. P., and L. R. Lynd. 2005. Cellulose utilization by Clostridium thermocellum: bioenergetics and hydrolysis product assimilation. Proc. Natl. Acad. Sci. U. S. A. 102:7321-7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.