Abstract
The majority of environmental microorganisms cannot be grown by traditional techniques. Here we employed, and contrasted with conventional plating, an alternative approach based on cultivation of microorganisms inside diffusion chambers incubated within natural samples, followed by subculturing in petri dishes. Using this approach, we isolated microorganisms from subsurface sediments from the Field Research Center (FRC) in Oak Ridge, TN. The sediments were acidic and highly contaminated with uranium, heavy metals, nitrate, and organic pollutants. Phylogenetic analysis of 16S rRNA gene sequences revealed clear differences between diversity of isolates obtained by the diffusion chamber approach and those obtained by conventional plating. The latter approach led to isolation of members of the Alpha- and Gammaproteobacteria, Actinobacteria, and Verrucomicrobia. Isolates obtained via the diffusion chamber approach represented the Alpha-, Beta-, and Gammaproteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes. Notably, one-third of the isolates obtained by the new method were closely related to species known from previous molecular surveys conducted in the FRC area. Since the initial growth of microorganisms inside diffusion chambers occurred in the presence of the environmental stress factors, we expected the isolates we obtained to be tolerant of these factors. We investigated the physiologies of selected isolates and discovered that the majority were indeed capable of growth under low pH and/or high concentrations of heavy metals and nitrate. This indicated that in contrast to conventional isolation, the diffusion chamber-based approach leads to isolation of species that are novel, exhibit tolerance to extant environmental conditions, and match some of the species previously discovered by molecular methods.
Most microorganisms found in environmental samples cannot be cultivated using conventional laboratory methods, an observation made repeatedly over the past hundred years (see references 3, 8, 9, 16, 20, 24, 36, 47, 48, and 49, among many other works). Recently, significant efforts have been made to improve microbial recovery by developing novel cultivation approaches involving single-cell encapsulation, utilizing low substrate concentrations, manipulating medium composition, including addition of signaling compounds, increase in incubation time, and in situ cultivation (4, 6, 7, 10, 11, 15, 21, 22, 38, 39, 41, 44). In spite of significant progress in bringing new species into culture, more than half of the bacterial phyla still do not have cultured representatives (16, 24, 36), and the majority of environmental species remain unexplored. Gaining access to these microorganisms is of substantial basic and applied importance.
This is particularly true in relation to bioremediation and contaminated sites, for which our basic understanding would be improved by a greater knowledge of the microorganisms active at the site. For example, many microbial species from contaminated sites are known exclusively from molecular surveys. They are only somewhat similar (e.g., ≤95% rRNA gene homology) to cultured isolates and are often assigned to an unclassified group (1, 2, 5, 13, 30, 34, 37, 42). Thus, the roles of these species in key processes of bioremediation, such as biosorption, bioaccumulation, biotransformation, and biomineralization, remain to be elucidated. This lack of knowledge calls for new approaches, such as metagenomics, proteogenomics, other functional approaches (e.g., see references 23, 45, and 46), and targeted efforts aiming to bring such species into culture to explore their functional roles, abilities, and adaptations to survive under stress.
We have developed a new approach for microbial cultivation by growing formerly uncultivated species in situ, followed by growth ex situ (4, 22). Here we apply this method to isolate microorganisms from the U.S. Department of Energy's Field Research Center (FRC) in Oak Ridge, TN. The subsurface sediment in this area is highly contaminated with radionuclides, heavy metals, nitrate, and organic compounds (Table 1) (http://www.esd.ornl.gov/orifrc/). Previous isolation experiments focused on anaerobic bacteria and resulted in the isolation of strains belonging to frequently cultured phyla (13, 27, 40, 42). However, molecular characterization of the microbial communities in the subsurface sediment and groundwater revealed the presence of diverse communities comprising a significant number of as yet uncultivated bacteria (e.g., see references (1 and 13). Here we show that our new cultivation methodology helps overcome these limitations by bringing into culture novel species that (i) belong to rarely cultured phyla, (ii) match species previously detected by molecular surveys, and (iii) are environmentally relevant because they exhibit tolerance to the conditions prevalent in their original environment, such as high concentrations of nitrate and heavy metals and low pH.
TABLE 1.
Chemical characteristics of groundwater in borehole FW111, maximum values registered in area 3, and ranges used in experiments in this study
| Contaminant or parameter | Concn (mg/liter) or pH range in FW111 groundwatera | Maximum concn (mg/liter) or pH value in area 3a | Exptl concn (mg/liter) or pH range |
|---|---|---|---|
| Aluminum | 90.2-123 | ||
| Cadmium | 0.08-0.09 | 4.45 | 0.56-5.6 |
| Chromium | 0.66-0.69 | ||
| Cobalt | 0.49-0.59 | 2.32 | 0.29-2.9 |
| Nickel | 7.02-7.1 | 18.9 | 2.9-29 |
| Uranium | 15.97-17.03 | 68.51 | 4.76-47.6 |
| Carbon (total) | 22.68 | ||
| Nitrate | 1,541-1,575 | 54,540 | 3,100-31,000 |
| pH | 2.99-3.52 | 2.8 | 3.5-7 |
Data from the FRC website; note that some of the values were reported after this study had been completed and were not available to determine the range of concentrations used in the present experiments.
MATERIALS AND METHODS
Sampling.
The FRC area includes places with contaminated subsurface sediment and groundwater, as well as uncontaminated background sites. A detailed description of physical and chemical characteristic of the environment can be found on the FRC website (http://www.esd.ornl.gov/orifrc/). In September 2004, we obtained a subsurface sediment core from borehole FW111 (area 3) that covered the depth from 4.25 m to 9.75 m. The core was homogenized in segments in an anaerobic glove box at Oak Ridge National Laboratory using methods essentially the same as those described previously (34) and transported chilled and under argon to Northeastern University in Boston, MA. Upon arrival, the material was wetted with deionized water to replace moisture lost from evaporation and stored in plastic containers; the same containers were used to deploy diffusion chambers during growth experiments.
Media.
In microbial growth experiments, we used the following media (all components were from Difco [Becton & Dickinson, Company, Sparks, MD]; concentrations are (vol/wt): A, no supplements; CY, 0.01% Bacto Casamino Acids and 0.01% Bacto Yeast Extract; CYE, 0.01% Bacto Casamino Acids, 0.01% Bacto Yeast Extract, and 0.01% humic acids. The media were supplemented with 10% cold water extract from the sediment to better mimic the natural environment. This extract was prepared by mixing sediment and deionized water at a 1:10 (vol/wt) ratio, vortexing for 1 min, spinning down (10 min, 11,000 rpm), and filter sterilization using 0.22-μm-pore-size filters. All media were solidified with 1.5% Bacto agar. Colonies grown on these media were subcultured for purification in 1.5% Bacto agar plates supplemented with lysogeny, also known as Luria-Bertani, broth (LB) at 10% of the manufacturer's suggested concentration (2.5 g/liter; Sigma Aldrich, St. Louis, MO).
Cultivation and isolation.
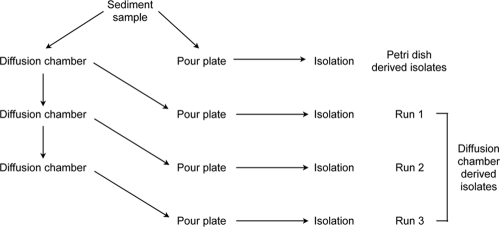
The overall experimental scheme of cultivation experiments is presented in Fig. 1. A 1-g subsample from the upper part of the crushed core was diluted with cold-water extract and mixed with the medium A, CY, or CYE to obtain a final dilution of 10−4 to 10−6. The mixtures were used to inoculate pour plates and diffusion chambers. The diffusion chambers were prepared as described previously (4, 22). In short, a 0.03-μm-pore-size polycarbonate membrane (GE Osmonics Labstore, Minnetonka, MN) was glued to the bottom side of a stainless steel O-ring using silicone glue. This formed a small chamber that was filled with 3 ml of one of the above mixes. After the agar mixture solidified, a second membrane was glued to the upper surface of the O-ring to seal the chamber. The chambers were incubated for 4 weeks on top of the wetted sediment in the lab and flipped on a regular basis to prevent the buildup of anoxic conditions. After incubation, the chambers were removed and opened, and the agar with the grown material was taken out of the chamber. This material was homogenized by passaging it through a syringe with a 25-gauge needle, diluted with cold-water extract, mixed with agar to obtain the same dilution as in the original sample (10−4 to 10−6), and used to inoculate a new diffusion chamber and a parallel pour plate. After the second round of cultivation, this procedure was repeated once more, resulting in three rounds of in situ incubation and three runs of cultivation on pour plates (Fig. 1). Colonies growing in the subcultured pour plates were further subcultured by streaking on 0.1× LB plates for subsequent isolation and identification. For each cultivation round, we set up 6 to 12 diffusion chambers and an equal number of pour plates.
FIG. 1.
Flow chart of steps in microbial isolation.
Species identification.
Microbial identification was performed by sequencing a part of the 16S rRNA gene (25). The colony material was used directly as a template for PCR. On several occasions, this approach failed to amplify the gene. In these cases, colony DNA was isolated using a Qiagen blood and tissue kit (Qiagen, Valencia, CA) according to the manufacturer's recommendations. The 16S rRNA gene was amplified with a Taq polymerase (Promega, Madison, WI) using the eubacterial primers 27F (AGA GTT TGA TCC TGG CTC AG) and 1492R (GGT TAC CTT GTT ACG ACT T) (25). The PCR products were purified and sequenced commercially (Seqwright, Houston, TX) by fluorescent dye terminator sequencing. The PCR products from all samples were sequenced using the internal primer 357F (CCT ACG GGA GGC AGC AG) (31), resulting in sequences with a length of 500 to 900 bp. Additional sequencing with the primers 518R (ATT ACC GCG GCT GCT GG) (31) and 1492R (25) was carried out for selected isolates to obtain nearly complete 16S rRNA sequences from those isolates (see File S1 in the supplemental material). The sequences were edited using the 4Peaks software program (by A. Griekspoor and T. Groothuis [http://mekentosj.com/science/4peaks/]), aligned to the ARB database (26), and added to the phylogenetic tree using the parsimony addition tool. The sequences were grouped into strains if different by no more than one nucleotide and into species if different by less than 3% of nucleotides (43). For comparative purposes, the sequences obtained from the same sampling site by rRNA gene surveys (1, 2, 5, 13, 30, 34, 37, 42) were added to the phylogenetic tree as well. The strains that were chosen for the physiological characterization have been used to construct a phylogenetic tree in ARB (23).
Physiological characterization.
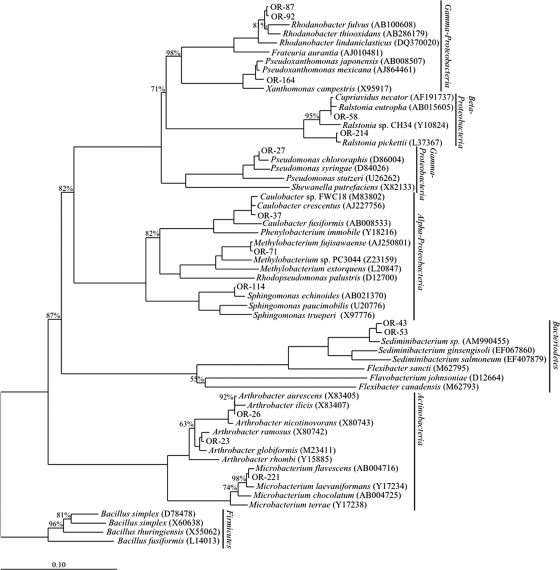
Fourteen strains were chosen for physiological characterization (Fig. 2). The experiments were conducted in triplicate or quadruplicate in 96-well plates containing 150 μl of 0.1× LB at pH 7. The medium was supplemented with nitrate (50, 100, 250, or 500 mM), uranium (20, 50, 100, or 200 μM), nickel (50, 100, 250, or 500 μM), cobalt (5, 10, 25, or 50 μM), or cadmium (5, 10, 25, and 50 μM). In selected experiments, the pH of the medium was adjusted to 3.5, 4, 4.5, 5, or 7, using HEPES (Fluka; via Sigma-Aldrich, St. Louis, MO) (pH 7) or Homopipes (Fluka; via Sigma-Aldrich, St. Louis, MO) (pH 3.5 to 5) as buffering agents. The plates were inoculated with 15 μl of cell suspension from overnight liquid cultures grown in 0.1× LB at 27°C with shaking at 200 rpm. Incubations were performed at 27°C in a plate reader with the incubation module (Biotek Synergy HT), with the optical density at 600 nm (OD600) measured every 10 min for 24 h.
FIG. 2.
Neighbor-joining phylogenetic tree showing the position of isolates chosen for physiological investigation.
Evaluation of physiological data.
Microbial growth rates were determined by calculating the slope of the natural log transformation of the OD600 values plotted against time. The growth rates were used to determine the influence of the stress factors on the short-term growth of the isolates by comparing the growth rate in the presence of a given stress factor to that at pH 7 with no stress (control). The statistical significance of the differences was assessed using Student's t test.
RESULTS
In total, we isolated 71 microbial strains, which grouped into 58 species. Of these, 61 strains and 50 species were obtained via the diffusion chamber-based approach (Table 2; see also File S1 in the supplemental material). The diffusion chamber-grown isolates represented six microbial phyla; the standard cultivation approach produced representatives of four phyla. Strains from the Alpha- and Gammaproteobacteria and the Actinobacteria were isolated by both approaches, whereas representatives of the Betaproteobacteria, Bacteroidetes, and Firmicutes were unique to the diffusion chamber-based technique.
TABLE 2.
Phylogenetic affiliations of the strains obtained by conventional and diffusion chamber-based techniques
| Classification | No. of strains isolated by: |
Total no. of strains isolated | |||||
|---|---|---|---|---|---|---|---|
| Conventional petri dish method | Diffusion chamber-based approach |
||||||
| Chamber run |
All chamber runs | Both methods | |||||
| 1 | 2 | 3 | |||||
| Alphaproteobacteria | 3 | 18 | 2 | 9 | 29 | 1 | 32 |
| Betaproteobacteria | 3 | 1 | 7 | 11 | 11 | ||
| Gammaproteobacteria | 1 | 4 | 2 | 6 | 7 | ||
| Bacteroidetes | 4 | 4 | 4 | ||||
| Verrucomicrobia | 1 | 1 | |||||
| Actinobacteria | 6 | 7 | 3 | 10 | 1 | 16 | |
| Firmicutes | 1 | 1 | 1 | ||||
| Total | 11 | 36 | 3 | 22 | 61 | 72 | |
We chose 14 isolates from five microbial phyla for physiological characterization. The selected strains exhibited robust growth and were closely related to species previously detected in the FRC area by molecular methods (Tables 3 and 4). Ten of these strains were obtained exclusively via the diffusion chamber-based approach, one was unique to standard cultivation, and three were isolated by both approaches.
TABLE 3.
Phylogenetic relationships between isolates obtained in this study and organisms detected by 16S rRNA gene surveys of the FRC area
| Strain no. | Closest cultured relative | Sim (%)a | Cult-metb | Obtained fromc: |
|||
|---|---|---|---|---|---|---|---|
| Con area | Area 1 | Area 2 | Area 3 | ||||
| OR212 | Blastobacter denitrificans | 100 | C | + | |||
| OR160 | Rhodopseudomonas palustris | 99.9 | C | + | |||
| OR74 | Bradyrhizobium elkanii | 99.4 | C | + | + | ||
| OR71 | Methylobacterium radiotolerans | 99.6 | P/C | + | + | ||
| OR37 | Caulobacter sp. FWC41 | 99.9 | C | + | |||
| OR205 | Bacterium Ellin 5130 | 99.1 | C | + | |||
| OR214 | Ralstonia pickettii | 100 | C | + | + | + | + |
| OR58 | Cupriavidus necator | 99.9 | C | + | |||
| OR189 | Betaproteobacterium B7 | 98.9 | C | + | |||
| OR202 | Imtechium assamiensis | 99.2 | C | + | + | ||
| OR175 | MBTE degrading bacterium | 95.5 | C | + | |||
| OR27 | Pseudomonas chloroaphis | 100 | P | + | |||
| OR164 | Pseudoxanthomonas japonensis | 99.3 | C | + | |||
| OR87 | Rhodanobacter thiooxydans | 98.4 | C | + | + | ||
| OR92 | Rhodanobacter thiooxydans | 99.2 | C | + | + | ||
| OR113 | Rhodanobacter sp. | 96.3 | C | + | |||
| OR43 | Sediminibacterium sp. | 98.7 | C | + | |||
| OR53 | Sediminibacterium sp. | 97.9 | C | + | |||
| OR221 | Microbacterium laevaniformans | 99.8 | C | + | |||
| OR30 | Arthrobacter sp. SMCC ZAT262 | 98.7 | P | + | + | ||
| OR23 | Arthrobacter ramosus | 100 | P | + | + | ||
| OR28 | Arthrobacter sp. | 99.0 | P | + | |||
| OR26 | Arthrobacter aurescens | 99.7 | P | + | |||
| OR12 | Arthrobacter aurescens | 98.9 | C | + | |||
| OR33 | Arthrobacter nicotinovorans | 100 | C | + | |||
| OR82 | Rhodococcus erythropolis | 99.9 | C | + | |||
Sim, similarity of the 16S rRNA gene sequences.
Cult-met, cultivation method used. P, pour plate; C, diffusion chamber.
TABLE 4.
Levels of stress factors leading to growth arrest of selected strains isolated in this study
| Strain | pH of medium |
Concn of: |
|||||
|---|---|---|---|---|---|---|---|
| Unbuffered | Buffered | NO3− (mM) | Uranium (μM) | Nickel (μM) | Cobalt (μM) | Cadmium (μM) | |
| Alphaproteobacteria | |||||||
| OR37 | 3.5 | 4 | 250 | >200 | >500 | >50 | >50 |
| OR71 | 3.5 | 4 | 250 | >200 | >500 | >50 | 5 |
| OR114 | 3.5 | 3.5 | 250 | >200 | 500 | >50 | >50 |
| Betaproteobacteria | |||||||
| OR58 | 3.5 | 4 | 500 | 200 | 500 | >50 | >50 |
| OR214 | <3.5 | <3.5 | 500 | 200 | 500 | >50 | >50 |
| Gammaproteobacteria | |||||||
| OR27 | <3.5 | 3.5 | 500 | >200 | 500 | >50 | >50 |
| OR164 | 4.5 | 5 | 250 | 100 | 250 | 50 | 25 |
| OR87 | 3.5 | 4 | 250 | 200 | 250 | >50 | 25 |
| OR92 | 3.5 | 3.5 | 250 | 200 | 100 | >50 | 25 |
| Bacteroidetes | |||||||
| OR43 | 4 | 4 | 100 | >200 | >500 | >50 | 5 |
| OR53 | 4 | 4 | 100 | >200 | 500 | >50 | 5 |
| Actinobacteria | |||||||
| OR221 | <3.5 | <3.5 | >500 | >200 | 500 | >50 | >50 |
| OR23 | 4.5 | 4.5 | 250 | 100 | 250 | >50 | 5 |
| OR26 | 4 | 4.5 | 500 | 200 | 500 | >50 | >50 |
The tested strains fell into three groups based on their growth response to the different stress factors. The first group was tolerant to most stress factors at the environmentally relevant concentrations. This group was represented by a single, diffusion chamber-grown isolate, OR221, closely related to Microbacterium laevaniformans (Actinobacteria). Its growth was not inhibited even at the concentrations of the stress factors beyond the range detected at the site of its isolation (e.g., Ni, Co, and Cd) (Table 4). Isolates from the second group were tolerant to many stress factors but inhibited by some at concentrations close to their environmental maxima. The strains from the second group, three members of the Alphaproteobacteria, OR37, OR71, and OR114, and two of the Betaproteobacteria, OR58 and OR214, were inhibited only by the lowest pH tested, high concentrations of nitrate, and selected heavy metals. All strains in this group were isolated using the diffusion chamber-based cultivation approach; one of them was also isolated in conventional petri dishes. The third group of isolates exhibited a notable lack of tolerance to environmentally relevant concentrations of several stress factors, typically nitrate, uranium, and pH. This group was represented by a mix of strains obtained by both cultivation strategies used here.
DISCUSSION
Microbial cultivation from sites with significant contamination, such as FRC sites, has been difficult and sometimes even unsuccessful (12). Cultivation attempts undertaken at several FRC sites have produced limited numbers of microbial isolates, although isolation of several nitrate and uranium reducers, such as Geobacter and Rhodanobacter and other species, have been reported (35). Additionally, standard cultivation is known to be biased since it produces isolates that are not necessarily representative of all active, dominant community members. Here we applied a novel cultivation strategy in an attempt to isolate microbial species from FRC sites contaminated with a range of pollutants, including heavy metals (Table 1). This strategy is based on in situ incubation in diffusion chambers (22), which in other applications produced culture collections significantly different from those obtained by conventional techniques (4). This approach includes a step of growth in situ and thus by definition leads to isolation of species that do have a capacity to grow in their contaminated habitat. It follows that, in contrast to conventional cultivation, often criticized for isolating rare and environmentally irrelevant species, at least some isolates obtained using this method must represent the active component of the target community. If so, our isolates should exhibit an adequate level of tolerance to the stress factors characteristic of the natural habitat. We therefore explored physiological response of selected strains to stress factors and confirmed that many of them are indeed tolerant to the high concentrations of nitrate, heavy metals, and low pH characteristic of their environment.
In this study, the number of strains obtained by the diffusion chamber method was considerably larger than the number of strains obtained by direct plating of samples. Perhaps a more interesting aspect of our collection is its species composition. Apart from covering more microbial phyla, many of the diffusion chamber-reared isolates closely matched species previously detected via molecular signatures in the same general area and its vicinity (1, 2, 5, 13, 30, 34, 37, 42). The comparison reveals that 26 of the 71 isolated strains belong to the same species identified by their 16S rRNA signatures in previous surveys of these sites (Table 3). The overwhelming majority of these strains (20 out of 26) were obtained using the diffusion chamber approach (Table 3).
Of particular interest are three 16S rRNA surveys that have been conducted in the area of our cultivation study (FRC's area 3) (1, 13, 37), including a study that sampled the same depth (1). In the latter study, the authors constructed a clone library consisting of 29 bacterial operational taxonomic units (OTUs) based on >98% rRNA gene sequence identity. We were able to cultivate six of them, from three different phyla (Betaproteobacteria, Bacteroidetes, and Actinobacteria). Five out of these six strains were obtained via an in situ growth method. Fields et al. (13) identified 46 OTUs, and we were able to isolate three of them, belonging to the Alpha-, Beta-, and Gammaproteobacteria; two of the three were obtained by the diffusion chamber-based approach. The latter two included Ralstonia isolates with 16S rRNA gene sequences closely related to sequences retrieved multiple times in both the above studies.
The recovery of multiple species previously known from molecular (but not cultivation) surveys of the area is encouraging and indicates progress toward a better recovery of the microbial diversity. This is in agreement with our earlier observations, which indicated that the diffusion chamber-based method allows access to a larger diversity of environmental microorganisms than more traditional methods currently in use (4). Therefore, the use of the diffusion chamber likely leads to unique culture collections that are different from those produced by conventional cultivation.
Since our method includes a step in which the microorganisms are grown in situ, at least some of the obtained isolates must be able to multiply under environmental conditions. This is important because direct plating may produce isolates from cells present in the environment transiently or in the form of resistant spores only and the incidence of which was an accident of dispersal. Such isolates are likely to be irrelevant to the function of the target community. Unlike culture collections produced by more standard approaches, ours must be less impacted by this shortcoming, as evidenced by the fact that many diffusion chamber-reared strains are tolerant of the stress factors characteristic of the FRC. One isolate, OR221, closely related to M. laevaniformans, was unique among our isolates since it exhibited growth in the presence of all the different stress factors tested and at all concentrations, including those in excess of environmentally relevant ones (Table 4). OR221 is also related to three Microbacterium oxydans isolates (16S rRNA gene sequence similarity from 98.8 to 99.2%) obtained from the groundwater of the radioactive waste depository Tomsk-7 in Siberia, Russia (32). These (Siberian) isolates tolerate 4 to 20 times higher concentrations of uranium, nickel, and cadmium than those used in our experiments. The same isolates were able to precipitate uranium at pH values close to those in the area of our study (29, 32), pointing to a possibility that OR221 may also be capable of uranium immobilization in situ.
Another isolate showing a high tolerance to heavy metals was OR37, a close relative of Caulobacter crescentus (Table 4). This corroborates earlier findings that C. cresentus tolerates high concentrations of uranium (18, 19), possibly due to four metal ion efflux systems coded in its genome (33). Two additional isolates, OR58 and OR214, also showed a high tolerance to the majority of the heavy metals (Table 4). These isolates are closely related to Ralstonia metallidurans, known to possess two plasmids carrying metal resistance genes (28).
Interestingly, among the isolates obtained by the diffusion chamber approach, there are several that ex situ did not exhibit significant tolerance to several of the extant stress factors. For example, isolates OR87 and OR92, likely from the genus Rhodanobacter, while tolerant of acidic conditions, grew only moderately in the presence of any of the heavy metals tested. Strains of Rhodanobacteria have also been recovered by the Kostka group (Florida State University) from the FRC site (14) and detected in a recent metagenomic study (17). They are also capable of growth on nitrate and at pH 4. Other strains, such as OR43 and OR53, appear to have opposite adaptations: they exhibited a tolerance of high concentrations of various heavy metals but appeared to be very sensitive to increased concentrations of nitrate and pH below 4.5.
It is unclear how a species can grow during in situ incubations if this species is only tolerant of a subset of the in situ challenges. It is possible that microorganisms might be able to modify their microenvironments, conditioning them for other species. For example, an organism capable of metal reduction could locally reduce the metal concentration, opening a way for nontolerant species to grow in the vicinity. Such synergies remain largely hypothetical, and their potential importance in bioremediation processes makes them an exciting subject for future research.
Supplementary Material
Acknowledgments
We thank David Watson and Lisa Fagan of ORNL for sediment sampling and collaboration, John M. Price from the Office of Environmental Health and Safety for his support with the handling of the sediment samples at Northeastern University, and Veronica Godoy from Northeastern University for technical support and advice.
This research was funded by the Environmental Remediation Sciences Program (ERSP), Office of Biological and Environmental Research (OBER), U.S. Department of Energy, grants DE-FG02-04ER63782 to K.L. and DE-FG02-04ER63782 to S.S.E. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725.
This is contribution no. 272 of the Marine Science Center, Northeastern University, Nahant, MA.
Footnotes
Published ahead of print on 24 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abulencia, C. B., D. L. Wyborski, J. A. Garcia, M. Podar, W. Chen, S. H. Chang, H. W. Chang, D. Watson, E. L. Brodie, T. C. Hazen, and M. Keller. 2006. Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl. Environ. Microbiol. 72:3291-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akob, D. M., H. J. Mills, and J. E. Kostka. 2007. Metabolically active microbial communities in uranium-contaminated subsurface sediments. FEMS Microbiol. Ecol. 59:95-107. [DOI] [PubMed] [Google Scholar]
- 3.Amann, J. 1911. Die direkte Zählung der Wasserbakterien mittels des Ultramikroskops. Centralbl. Bakteriol. 29:381-384. [Google Scholar]
- 4.Bollmann, A., K. Lewis, and S. S. Epstein. 2007. Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl. Environ. Microbiol. 73:6386-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodie, E. L., T. Z. Desantis, D. C. Joyner, S. M. Baek, J. T. Larsen, G. L. Andersen, T. C. Hazen, P. M. Richardson, D. J. Herman, T. K. Tokunaga, J. M. Wan, and M. K. Firestone. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruns, A., H. Cypionka, and J. Overmann. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 68:3978-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bussmann, I., B. Philipp, and B. Schink. 2001. Factors influencing the cultivability of lake water bacteria. J. Microbiol. Methods 47:41-50. [DOI] [PubMed] [Google Scholar]
- 8.Butkevich, V. S. 1932. Zür Methodik der bakterioloschen Meeresuntersuchungen und einige Angaben über die Verteilung der Bakterien im Wasser und in den Büden des Barents Meeres. Trans. Oceanogr. Inst. Moscow 2:5-39. (In Russian with German summary.) [Google Scholar]
- 9.Cholodny, N. 1929. Zur Methodik der quantitativen Erforschung des bakteriellen Planktons. Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. A 77:179-193. [Google Scholar]
- 10.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, K. E., S. J. Joseph, and P. H. Janssen. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71:826-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields, M. W., S. L. Carroll, W. Wu, M. Gentile, B. Gu, P. M. Jardine, C. S. Criddle, and J. Zhou. 2002. The use of denitrifying bacteria isolated from groundwater contaminated with nitrate and uranium for the denitrification of acidic groundwater, abstr. Q-441, p. 455. Abstr. 102nd Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 13.Fields, M. W., T. Yan, S. K. Rhee, S. L. Carroll, P. M. Jardine, D. B. Watson, C. S. Criddle, and J. Zhou. 2005. Impacts on microbial communities and cultivable isolates from groundwater contaminated with high levels of nitric acid-uranium waste. FEMS Microbiol. Ecol. 53:417-428. [DOI] [PubMed] [Google Scholar]
- 14.Green, S. J., O. Prakash, T. M. Gihring, D. M. Akob, P. Jasrotia, P. M. Jardine, D. B. Watson, S. D. Brown, A. V. Palumbo, and J. E. Kostka. 2010. Denitrifying bacteria isolated from terrestrial subsurface sediments exposed to mixed-waste contamination. Appl. Environ. Microbiol. 76:3244-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn, M. W., P. Stadler, Q. L. Wu, and M. Pockl. 2004. The filtration-acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J. Microbiol. Methods 57:379-390. [DOI] [PubMed] [Google Scholar]
- 16.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemme, C. L., Y. Deng, T. J. Gentry, M. W. Fields, L. Wu, S. Barua, K. Barry, S. G. Tringe, D. B. Watson, Z. He, T. C. Hazen, J. M. Tiedje, E. M. Rubin, and J. Zhou. 2010. Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J. 4:660-672. [DOI] [PubMed] [Google Scholar]
- 18.Hillson, N. J., P. Hu, G. L. Andersen, and L. Shapiro. 2007. Caulobacter crescentus as a whole-cell uranium biosensor. Appl. Environ. Microbiol. 73:7615-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, P., E. L. Brodie, Y. Suzuki, H. H. McAdams, and G. L. Andersen. 2005. Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J. Bacteriol. 187:8437-8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jannasch, H. W., and G. E. Jones. 1959. Bacterial populations in seawater as determined by different methods of enumeration. Limnol. Oceanogr. 4:128-139. [Google Scholar]
- 21.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 23.Kalyuzhnaya, M. G., A. Lapidus, N. Ivanova, A. C. Copeland, A. C. McHardy, E. Szeto, A. Salamov, I. V. Grigoriev, D. Suciu, S. R. Levine, V. M. Markowitz, I. Rigoutsos, S. G. Tringe, D. C. Bruce, P. M. Richardson, M. E. Lidstrom, and L. Chistoserdova. 2008. High-resolution metagenomics targets specific functional types in complex microbial communities. Nat. Biotechnol. 26:1029-1034. [DOI] [PubMed] [Google Scholar]
- 24.Keller, M., and K. Zengler. 2004. Tapping into microbial diversity. Nat. Rev. Microbiol. 2:141-150. [DOI] [PubMed] [Google Scholar]
- 25.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 26.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez, R. J., Y. Wang, M. A. Raimondo, J. M. Coombs, T. Barkay, and P. A. Sobecky. 2006. Horizontal gene transfer of PIB-type ATPases among bacteria isolated from radionuclide- and metal-contaminated subsurface soils. Appl. Environ. Microbiol. 72:3111-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mergeay, M., S. Monchy, T. Vallaeys, V. Auquier, A. Benotmane, P. Bertin, S. Taghavi, J. Dunn, D. van der Lelie, and R. Wattiez. 2003. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 27:385-410. [DOI] [PubMed] [Google Scholar]
- 29.Merroun, M., M. Nedelkova, A. Rossberg, C. Hennig, and S. Selenska-Pobell. 2006. Interaction mechanisms of bacterial strains isolated from extreme habitats with uranium. Radiochim. Acta 94:723-729. [Google Scholar]
- 30.Michalsen, M. M., A. D. Peacock, A. M. Spain, A. N. Smithgal, D. C. White, Y. Sanchez-Rosario, L. R. Krumholz, and J. D. Istok. 2007. Changes in microbial community composition and geochemistry during uranium and technetium bioimmobilization. Appl. Environ. Microbiol. 73:5885-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nedelkova, M., M. L. Merroun, A. Rossberg, C. Hennig, and S. Selenska-Pobell. 2007. Microbacterium isolates from the vicinity of a radioactive waste depository and their interactions with uranium. FEMS Microbiol. Ecol. 59:694-705. [DOI] [PubMed] [Google Scholar]
- 33.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.North, N. N., S. L. Dollhopf, L. Petrie, J. D. Istok, D. L. Balkwill, and J. E. Kostka. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prakash, O., T. M. Gihring, D. D. Dalton, K.-J. Chin, S. J. Green, D. M. Akob, G. Wanger, and J. E. Kostka. 2010. Geobacter daltonii sp. nov., an iron(III)- and uranium(VI)-reducing bacterium isolated from the shallow subsurface exposed to mixed heavy metal and hydrocarbon contamination. Int. J. Syst. Evol. Microbiol. 60:546-553. [DOI] [PubMed] [Google Scholar]
- 36.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 37.Reardon, C. L., D. E. Cummings, L. M. Petzke, B. L. Kinsall, D. B. Watson, B. M. Peyton, and G. G. Geesey. 2004. Composition and diversity of microbial communities recovered from surrogate minerals incubated in an acidic uranium-contaminated aquifer. Appl. Environ. Microbiol. 70:6037-6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 39.Sangwan, P., S. Kovac, K. E. Davis, M. Sait, and P. H. Janssen. 2005. Detection and cultivation of soil verrucomicrobia. Appl. Environ. Microbiol. 71:8402-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shelobolina, E. S., S. A. Sullivan, K. R. O'Neill, K. P. Nevin, and D. R. Lovley. 2004. Isolation, characterization, and U(VI)-reducing potential of a facultatively anaerobic, acid-resistant bacterium from low-pH, nitrate- and U(VI)-contaminated subsurface sediment and description of Salmonella subterranea sp. nov. Appl. Environ. Microbiol. 70:2959-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song, J., H. M. Oh, and J. C. Cho. 2009. Improved culturability of SAR11 strains in dilution-to-extinction culturing from the East Sea, West Pacific Ocean. FEMS Microbiol. Lett. 295:141-147. [DOI] [PubMed] [Google Scholar]
- 42.Spain, A. M., A. D. Peacock, J. D. Istok, M. S. Elshahed, F. Z. Najar, B. A. Roe, D. C. White, and L. R. Krumholz. 2007. Identification and isolation of a Castellaniella species important during biostimulation of an acidic nitrate- and uranium-contaminated aquifer. Appl. Environ. Microbiol. 73:4892-4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA:DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteria. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 44.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waldron, P. J., L. Wu, J. D. Van Nostrand, C. W. Schadt, Z. He, D. B. Watson, P. M. Jardine, A. V. Palumbo, T. C. Hazen, and J. Zhou. 2009. Functional gene array-based analysis of microbial community structure in groundwaters with a gradient of contaminant levels. Environ. Sci. Technol. 43:3529-3534. [DOI] [PubMed] [Google Scholar]
- 46.Wilkins, M. J., N. C. Verberkmoes, K. H. Williams, S. J. Callister, P. J. Mouser, H. Elifantz, A. L. N′Guessan, B. C. Thomas, C. D. Nicora, M. B. Shah, P. Abraham, M. S. Lipton, D. R. Lovley, R. L. Hettich, P. E. Long, and J. F. Banfield. 2009. Proteogenomic monitoring of Geobacter physiology during stimulated uranium bioremediation. Appl. Environ. Microbiol. 75:6591-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winslow, C.-E. A., and G. E. Willcomb. 1905. Tests of a method for the direct microscopic enumeration of bacteria. J. Infect. Dis. Suppl. 1:273-283. [Google Scholar]
- 48.Winterberg, H. 1898. Zur Methodik der Bakterienzahlung. Zeitschr. Hyg. 29:75-93. [Google Scholar]
- 49.ZoBell, C. E. 1946. Marine microbiology: a monograph on hydrobacteriology. Chronica Botanica Co., Waltham, MA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.