Abstract
The effect of eliminating d-lactate synthesis in poly(3-hydroxybutyrate) (PHB)-accumulating recombinant Escherichia coli (K24K) was analyzed using glycerol as a substrate. K24KL, an ldhA derivative, produced more biomass and had altered carbon partitioning among the metabolic products, probably due to the increased availability of carbon precursors and reducing power. This resulted in a significant increase of PHB and ethanol synthesis and a decrease in acetate production. Cofactor measurements revealed that cultures of K24K and K24KL had a high intracellular NADPH content and that the NADPH/NADP+ ratio was higher than the NADH/NAD+ ratio. The ldhA mutation affected cofactor distribution, resulting in a more reduced intracellular state, mainly due to a further increase in NADPH/NADP+. In 60-h fed-batch cultures, K24KL reached 41.9 g·liter−1 biomass and accumulated PHB up to 63% ± 1% (wt/wt), with a PHB yield on glycerol of 0.41 ± 0.03 g·g−1, the highest reported using this substrate.
Poly(3-hydroxybutyrate) (PHB) is the best-known and most common polyhydroxyalkanoate (PHA). PHAs are polymers with thermoplastic properties that are totally biodegradable by microorganisms present in most environments and that can be produced from different renewable carbon sources (38). Accumulated as intracellular granules by many bacteria under unfavorable conditions (1, 21), PHAs are carbon and energy reserves and also act as electron sinks, enhancing the fitness and stress resistance of bacteria and contributing to redox balance (12, 30). Escherichia coli offers a well-defined physiological environment for the construction and manipulation of various metabolic pathways to produce different bioproducts, such as PHB, from cost-effective carbon sources.
In recent years, a significant increase in the production of biodiesel has caused a sharp fall in the cost of glycerol, the main by-product of biodiesel synthesis. As a result, glycerol has become a very attractive substrate for bacterial fermentations (10), specially for reduced products, such as PHB (36). The E. coli strain used in this work, K24K, carries phaBAC, the structural genes responsible for PHB synthesis, from Azotobacter sp. strain FA8 (23) (Table 1). The pha genes in K24K are expressed from a chimeric promoter and consequently are not subject to the genetic regulatory systems present in natural PHA producers. Because of this, it can be assumed that regulation of PHA synthesis in the recombinants is restricted by enzyme activity levels, modulated principally by substrate availability. In most natural producers, and also in PHB-producing E. coli recombinants, PHB is synthesized through the condensation of two molecules of acetyl-coenzyme A (acetyl-CoA), catalyzed by an acetoacetyl-CoA transferase or 3-ketothiolase, resulting in acetoacetyl-CoA. This compound is subsequently reduced by an NAD(P)H-dependent acetoacetyl-CoA reductase to R-(−)-3-hydroxybutyryl-CoA, which is then polymerized by a specific PHA synthase (34).
TABLE 1.
E. coli strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant characteristicsb | Reference or source |
|---|---|---|
| E. coli strains | ||
| K1060a | F−fadE62 lacI60 tyrT58(AS) fabB5 mel-1 | 29 |
| K24 | Same as K1060, carrying pJP24; Apr | 23 |
| K24K | Same as K1060, carrying pJP24K; Apr Kmr | 23 |
| ALS786a | F− λ−rph-1 ΔldhA::kan; Kmr | 14 |
| K24LT | Same as K1060 but ΔldhA::kan by K1060 × P1(ALS786), carrying pJP24; Apr Kmr | This work |
| K24KL | Same as K1060 but ΔldhA by allelic replacement, carrying pJP24K; Kmr | This work |
| TA3522a | F− λ− Δ(his-gnd)861 hisJo-701 | 2 |
| TA3514a | Same as TA3522 but pta-200 | 19 |
| TA3522L | Same as TA3522 but ΔldhA::kan by TA3522 × P1(ALS786); Kmr | This work |
| TA3514L | Same as TA3514 but ΔldhA::kan by TA3514 × P1(ALS786); Kmr | This work |
| Plasmids | ||
| pQE32 | Expression vector, ColE1 ori; Apr | Qiagen GmbH, Hilden, Germany |
| pJP24 | pQE32 derivative expressing a 4.3-kb BamHI-HindIII insert containing the phaBAC genes from Azotobacter sp. strain FA8 under the control of a T5 promoter/lac operator element; Apr | 23 |
| pJP24K | pJP24 derivative; Apr Kmr | 23 |
| pCP20 | Helper plasmid used for kan excision; Saccharomyces cerevisiae FLP λ cI857 λ PRrepA(Ts); Apr Cmr | 7 |
| Oligonucleotides | ||
| ΔldhA-F | 5′-TAT TTT TAG TAG CTT AAA TGT GAT TCA ACA TCA CTG GAG AAA GTC TTA TGG TGT AGG CTG GAG CTG CTT C-3′ | This work |
| ΔldhA-R | 5′-CTC CCC TGG AAT GCA GGG GAG CGG CAA GAT TAA ACC AGT TCG TTC GGG CAC ATA TGA ATA TCC TCC TTA G-3′ | This work |
Strain obtained through the E. coli Genetic Stock Center, Yale University, New Haven, CT.
For oligonucleotides, the ATG codon of ldhA is underlined and the sequences with homology to FRT-kan-FRT in the template plasmid pKD4 (11) are shown in boldface.
Cells growing on glycerol are in a more reduced intracellular state than cells grown on glucose under similar conditions of oxygen availability. This has a significant effect on the intracellular redox state, which causes the cells to direct carbon flow toward the synthesis of more-reduced products when glycerol is used than when glucose is used in order to achieve redox balance (31). When metabolic product distribution was analyzed in bioreactor cultures of K24K using glucose or glycerol as the substrate, product distributions with the two substrates were found to be different, as glycerol-grown cultures produced smaller amounts of acetate, lactate, and formate and more ethanol than those grown on glucose. However, PHB production from glycerol was lower than that from glucose, except under conditions of low oxygen availability (13).
Manipulations to enhance the synthesis of a metabolic product include several approaches to increase the availability of the substrates needed for its formation or to inhibit competing pathways. The effect of eliminating competing pathways on PHB production from glucose has been investigated through the inactivation of different genes, such as those encoding enzymes participating in the synthesis of acetate (ackA, pta, and poxB) or d-lactate (ldhA). A pta mutant, which produces very little acetate (6), and an frdA ldhA double mutant (40) had increased PHB accumulation from glucose. A recent report using an ackA pta poxB ldhA adhE mutant under microaerobic conditions attained similar results (17). The inactivation of ldhA has also been shown to have an important effect on the metabolic product distribution in recombinant E. coli with glycerol as the carbon source, promoting ethanol synthesis (28). In the present work we analyzed the effect of ldhA inactivation in strain K24K using glycerol as the carbon source, with special emphasis on changes in carbon distribution and in the intracellular redox state, determined through cofactor levels.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides.
All Escherichia coli strains are listed in Table 1, along with plasmids and oligonucleotides used in this work.
DNA manipulations and mutant construction.
Unless otherwise indicated, all DNA procedures followed standard protocols and specific recommendations from manufacturers. Fermentative d-lactate dehydrogenase (ldhA) mutant derivatives were constructed by P1 transduction (37) of the ΔldhA::kan allele from E. coli ALS786 into different recipient strains. E. coli K24KL was constructed by allelic replacement (11) using a PCR fragment obtained with primers ΔldhA-F and ΔldhA-R. One Kmr isolate was selected, checked by PCR for kan insertion, and transformed with plasmid pCP20, encoding the Saccharomyces cerevisiae FLP recombinase (7). Kms transformants were selected at 42°C, and kan excision and ldhA deletion were confirmed by PCR and DNA sequencing (data not shown).
Growth media and culture conditions.
MYA medium contained 6.0 g·liter−1 Na2HPO4, 3.0 g·liter−1 KH2PO4, 1.4 g·liter−1 (NH4)2SO4, 0.5 g·liter−1 NaCl, 0.2 g·liter−1 MgSO4·7H2O, 10 g·liter−1 yeast extract, 5 g·liter−1 casein amino acids (Diagnostic Systems, Sparks, MD), and 30 g·liter−1 glycerol (Anedra, Buenos Aires, Argentina). Concentrations of antibiotics were 100 μg·ml−1 for ampicillin and 50 μg·ml−1 for kanamycin. Magnesium sulfate, antibiotics, and glycerol were added separately as filter-sterilized concentrated solutions after autoclaving and cooling the medium. Solid media also contained 30 g·liter−1 agar. Seed cultures for all experiments were prepared by dispersing a loopful of cells from a fresh LB plate into 50 ml of MYA medium in a 250-ml Erlenmeyer flask. Cultures were incubated in a rotary shaker overnight at 37°C and 250 rpm and used to inoculate the bioreactor at an initial cell dry weight (CDW) of approximately 0.05 g·liter−1. Aerobic shaken-flask cultures were grown at 37°C in 250-ml Erlenmeyer flasks containing 25 ml of MYA medium and shaken at 250 rpm.
Bioreactor cultivation.
Bioreactor cultivations were carried out in a 5.6-liter stirred-tank reactor equipped with six flat-bladed disk turbines (BioFlo 110; New Brunswick Scientific Co., Edison, NJ). Batch cultures were grown in a 3-liter working volume, with agitation set at 500 rpm. The fermentor was sparged with 3 liter·min−1 of filter-sterilized air, and the pH was controlled at 7.20 ± 0.05 by automatic addition of 3 M KOH or 1.5 M H2SO4. To prevent foam formation, 30 μl·liter−1 Antifoam 289 (Sigma-Aldrich, St. Louis, MO) was manually added at the onset of each run. Dissolved oxygen was measured using an Ag/AgCl polarometric oxygen probe (Mettler Toledo, Greifensee, Switzerland). Fed-batch cultures were developed under conditions similar to those described for batch cultures, except that the stirrer speed was automatically adjusted up to 1,000 rpm to keep the dissolved oxygen level above 40% of air saturation. Glycerol concentration was maintained above 5 g·liter−1 by periodic addition of a feeding solution consisting of 500 g·liter−1 glycerol, 40 g·liter−1 casein amino acids, and 4 g·liter−1 MgSO4.
Analytical procedures.
Biomass concentration was determined as the CDW of washed pellets dried in 15-ml polypropylene centrifuge tubes at 80°C for at least 36 h. Dried samples were allowed to cool and held in vacuo until weighed. PHB content was determined gravimetrically after alkaline treatment of the biomass with 0.2 N NaOH (8) or by gas chromatography after methyl esterification (5). Extracellular metabolic products were determined by high-pressure liquid chromatography (HPLC) (13), and residual glycerol concentration was obtained by using a colorimetric assay (27). Intracellular NADH, NADPH, NAD+, and NADP+ contents were estimated by using in vitro procedures based on rapid inactivation of the metabolism of growing cells followed by acid or alkaline nucleotide extraction. Nucleotide determination was conducted by means of spectrophotometric cycling assays with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide as the final electron acceptor (3, 27).
RESULTS AND DISCUSSION
ldhA mutants have increased PHB synthesis from glycerol.
Growth and polymer synthesis in several PHB-producing isogenic E. coli strains were analyzed using glycerol. For this purpose, strain TA3522 and its pta variant, TA3514, were used, and ldhA derivatives of these strains (TA3522L and TA3514L, respectively) were constructed by P1 transduction (Table 1). Plasmid pJP24, carrying the phaBAC genes from Azotobacter sp. strain FA8 under the control of a T5 promoter/lac operator element (23), was introduced into all the strains. Growth and PHB accumulation in 24-h aerobic shaken-flask cultures of all plasmid-bearing isogenic strains growing on MYA medium supplemented with 30 g·liter−1 glycerol were analyzed. All mutants grew slightly less but accumulated more PHB than the pta+ ldhA+ strain. While TA3522/pJP24 reached 6.0 g·liter−1 and accumulated PHB up to 30% ± 1% (wt/wt), strain TA3514/pJP24 (pta) grew to 4.8 g·liter−1 and accumulated PHB up to 37% ± 1% (wt/wt), TA3514L/pJP24 (pta ldhA double mutant) grew to 4.6 g·liter−1 and accumulated 39% ± 1% (wt/wt) of PHB, and strain TA3522L/pJP24 (ldhA) grew to 4.9 g·liter−1 and accumulated the largest amount of polymer, 44% ± 1% (wt/wt). In order to investigate the effect of ldhA in a genetic background more suitable for the synthesis of bioproducts, K24LT, a derivative of strain K24, a prototrophic strain that we have used in previous work, was constructed by P1 transduction (Table 1). In 24-h bioreactor batch cultures, strain K24LT grew to a 1.5-fold-higher level than K24 (17.9 and 11.6 g·liter−1, respectively) and produced more PHB (10.2 and 1.8 g·liter−1, respectively), reaching a PHB content of 57% ± 3% (wt/wt).
Previous experience indicates that strains obtained through transduction may contain unexpected mutations that can affect the phenotype of the mutants, masking the effect of the studied alleles (25). In order to have an appropriate ldhA derivative, K24KL, a precise deletion derivative of K24K, was constructed by allelic replacement. The construction of a markerless ldhA mutant enabled the use of pJP24K, a Kmr variant of plasmid pJP24 with enhanced segregational stability (23) (Table 1).
ldhA affects growth and distribution of metabolic products in the recombinants.
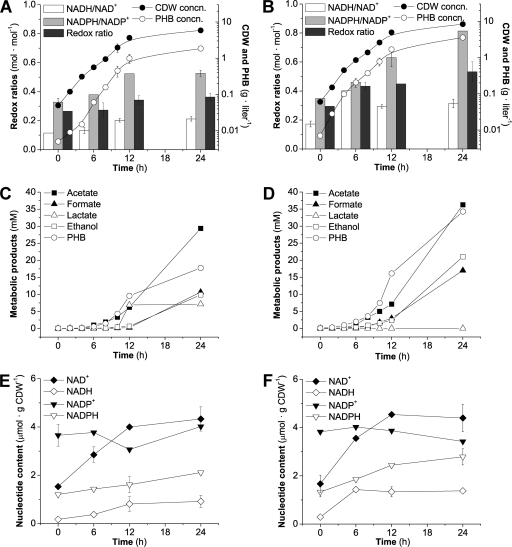
In shaken-flask cultures, strain K24KL accumulated 1.4-fold more polymer than K24K (44% ± 1% versus 32% ± 2% [wt/wt], respectively) and it also grew more (Fig. 1A and B). These results were similar to those obtained in microaerobic cultures grown on glucose using a multiple-fermentation-pathway mutant according to a recent report (17). In order to further analyze changes in carbon distribution, major metabolic products were measured in the cultures. Levels of acetate, formate, and ethanol production were higher for K24KL than for K24K (1.2-, 1.6-, and 2.1-fold, respectively) at 24 h (Fig. 1C and D). The presence of these metabolites reflects a limitation in oxygen availability that prompted the accumulation of fermentation products, also observed in bioreactor cultures. Ethanol and PHB production started earlier in the ldhA mutant. As expected, lactate was produced only by K24K, and its production peaked at 10 h. The increases in acetate and formate observed in the ldhA derivative compared to the parental strain were proportional to that of biomass, but the increases in ethanol and PHB accumulation were much higher. This can be due to the elimination of lactate synthesis, resulting in a higher availability of reducing power and of acetyl-CoA, which can be redirected toward the synthesis of biomass and other metabolites. The intracellular contents of NADH, NADPH, NAD+, and NADP+ were determined at several times. The redox ratio of the cells (measured as the ratio [NADH + NADPH]/[NAD+ + NADP+]) was, as expected, much more reduced in the ldhA strain (Fig. 1A and B) than in the wild-type strain, favoring the production of reduced metabolites, such as ethanol and PHB.
FIG. 1.
Physiological characterization of E. coli K24K (A, C, and E) and K24KL (its ldhA derivative) (B, D, and F) in shaken-flask cultures in MYA medium supplemented with 30 g·liter−1 glycerol under aerobic conditions. (A and B) Growth, polymer synthesis, and redox ratios; (C and D) time course plots for the synthesis of relevant bioproducts; (E and F) intracellular content of reduced and oxidized nucleotides. The redox ratio was defined as the ratio of reduced to oxidized cofactors ([NADH + NADPH]/[NAD+ + NADP+]). Symbols and bars represent the mean values for each parameter ± standard deviations of duplicate measurements from at least three independent cultures. CDW, cell dry weight; concn, concentration.
To obtain a higher biomass concentration and more-controlled conditions, both strains were grown in 24-h bioreactor batch cultures. The ldhA derivative grew more, as observed in the shaken-flask cultures, but the specific growth rates were similar (Table 2). K24KL had increased amounts of all measured metabolites, except formate, perhaps as a result of increased flux of carbon toward acetyl-CoA and higher oxygen consumption than K24K. The increased availability of both carbon and reducing equivalents had the greatest effect on ethanol and PHB. This was reflected in the high ethanol-to-acetate ratio obtained for K24KL, which was twice the value obtained for K24K, indicating a reduced internal state, as verified by the redox ratio. Taking into account the specific production rates, PHB synthesis was the most affected, as a 1.7-fold increase was observed for K24KL compared to K24K (Table 2). The final PHB concentration for K24KL in the 24-h bioreactor batch culture was 6.1 g·liter−1.
TABLE 2.
Fermentation parametersa for 24-h bioreactor batch cultures conducted under aerobic conditions in MYA medium with 30 g·liter−1 glycerol as the carbon source
| Parameter | Value for E. coli strain: |
|
|---|---|---|
| K24K | K24KL | |
| μmaxe (h−1) | 0.64 ± 0.03 | 0.67 ± 0.02 |
| Final CDWb concn (g·liter−1) | 10.7 ± 0.8 | 13.9 ± 0.7 |
| Final PHB content (% [wt/wt]) | 36 ± 2 | 44 ± 1 |
| Specific consumption rate (mmol·g CDW−1·h−1) for: | ||
| Glycerol | 1.8 ± 0.5 | 1.6 ± 0.3 |
| O2 | 0.085 ± 0.009 | 0.113 ± 0.006 |
| Specific production rate (μmol·g CDW−1·h−1) for: | ||
| Acetate | 52 ± 3 | 61 ± 2 |
| Formate | 77 ± 4 | 63 ± 5 |
| Lactate | 48 ± 5 | NDc |
| Ethanol | 149 ± 1 | 205 ± 1 |
| PHB | 103 ± 2 | 178 ± 3 |
| Ethanol-to-acetate ratio (mol·mol−1) | 0.36 ± 0.08 | 0.64 ± 0.02 |
| Redox ratiod ([NADH + NADPH]/[NAD+ + NADP+]) | 0.34 ± 0.03 | 0.62 ± 0.04 |
Maximum specific growth rates, as well as specific consumption and production rates, were calculated during exponential growth. Results represent the mean values ± standard deviations for duplicated measurements from at least two independent experiments.
CDW, cell dry weight.
ND, not detected.
Nucleotide content was enzymatically determined at 24 h.
μmax, maximum specific growth rate.
K24KL has high PHB and ethanol synthesis from glycerol in fed-batch cultures.
Based on results obtained in the batch bioreactor experiments, fed-batch fermentations were performed to further increase biomass and PHB content to evaluate E. coli K24KL as a potential biocatalyst for PHB synthesis. Biomass and PHB concentration increased almost linearly during the whole cultivation period (Fig. 2A), attaining 41.9 and 26.4 g·liter−1 at 60 h, respectively. PHB accumulation increased 1.4-fold compared with that for batch cultures (Table 2) and was similar to that obtained in fed-batch cultures with glucose of an E. coli strain carrying the phaCAB genes from Cupriavidus necator and overexpressing a NAD kinase (20). The final PHB content achieved was 63% ± 1% (wt/wt), with a volumetric productivity of 0.45 ± 0.08 g·liter−1·h−1. This polymer content ranks among the highest reported in the literature with glycerol as the carbon source, second only to that reached by Zobellella denitrificans MW1 in 50-h fed-batch cultures (66.9% ± 7.6% [wt/wt]) (16). The PHB-specific production rate (164 ± 9 μmol·g cell dry weight [CDW]−1·h−1) did not vary significantly compared to that for batch cultures (Table 2), and PHB yield on a carbon substrate at 60 h was 0.41 ± 0.03 g·g−1, the highest reported when using glycerol as the carbon source. This value comes close to that reported by Mothes et al. (22) for nitrogen-limited fed-batch cultures of C. necator DSMZ 4058 (0.37 g·g−1).
FIG. 2.
Fed-batch bioreactor culture of E. coli K24KL. (A) Time course plots for glycerol, as well as biomass and polymer concentrations; (B) metabolic profile (expressed as the PHB content [YPHB/X] and metabolic product yield on biomass [YP/X]) and intracellular redox state; (C) intracellular contents of reduced and oxidized nucleotides. The redox ratio was defined as the ratio of reduced to oxidized cofactors ([NADH + NADPH]/[NAD+ + NADP+]). Symbols and bars represent the mean values for each parameter ± standard deviations of duplicate measurements from at least two independent cultures.
Major metabolic products besides PHB were measured throughout the experiment, and their accumulation kinetics (expressed as g·g CDW−1) were analyzed (Fig. 2B). Interestingly, PHB content and ethanol accumulation followed similar linear patterns up to 36 h. This behavior can be explained by considering that both metabolites share acetyl-CoA as a precursor and both pathways use NAD(P)H as a cofactor. In line with this hypothesis, the redox ratio changed significantly over this period, increasing from 0.25 ± 0.02 to 0.64 ± 0.03 mol·mol−1 (Fig. 2B). Acetate accumulation rose sharply during the first 12 h, perhaps as a consequence of the aerobic activation of the Pta-AckA pathway (composed of phosphotransacetylase and acetate kinase), observed when excess carbon source is present (9), a phenomenon also detected in glycerol cultures of ethanologenic E. coli strains (28). After 36 h, ethanol and acetate accumulation decreased while PHB content kept increasing, albeit at a lower rate. As these three metabolites compete at the acetyl-CoA branching point, the observed distribution could be accounted for by an increased flux of acetyl-CoA toward the overexpressed reducing-power-consuming PHB synthesis pathway, limiting its availability for the native Pta-AckA and AdhE pathways. An analogous effect was observed in experiments performed using a recombinant E. coli redox mutant, in which the activity of a heterologous plasmid-encoded alcohol dehydrogenase was observed to increase in glycerol cultures, probably as a strategy to maintain redox balance (28). Furthermore, this metabolic effect was more evident in an ldhA mutant.
In accordance with the expected funneling of carbon skeletons into reduced metabolites in strain K24KL, we observed a final ethanol-to-acetate ratio of 1.31 ± 0.07 mol·mol−1, also reflected in the final redox ratio (0.76 ± 0.05 mol·mol−1) at 60 h. Formate accumulation was the lowest among the measured extracellular products, and its value remained almost constant during the cultivation, even though a slight decrease was observed after 36 h, similar to results for acetate and ethanol. As expected, lactate was not detected in supernatants throughout the experiment.
Cofactor distribution is altered in the ldhA mutant.
It has been proposed that PHB functions as an electron sink in natural producers (21) and promotes growth of E. coli redox mutants by consuming excess reducing power (30). Previous cofactor manipulations aimed at optimizing PHB synthesis from glucose in recombinant E. coli include the use of mutants that favor the use of the pentose phosphate pathway for catabolism, producing increased amounts of NADPH (18, 35). Transhydrogenases (32) and a NAD kinase (20) have also been manipulated to enhance PHB synthesis from glucose. On the other hand, increased PHB accumulation from glucose and glycerol has been observed in redox mutants defective in arcA (24, 26). In spite of the fact that PHB synthesis and the internal redox state of the cells are known to be associated, very few studies have analyzed the levels of the different cofactors in bacteria producing PHB (20, 32) and none used glycerol as a substrate. The distribution of reduced and oxidized cofactors in K24K and its ldhA derivative was studied to investigate possible relationships between cofactor distribution and metabolite production in PHB-synthesizing E. coli when grown on glycerol.
When the ratios of reduced to oxidized cofactors for K24K and K24KL were compared, it was found that values were higher for the ldhA derivative, as expected (Fig. 1A and B and 2B; Table 2). Furthermore, the highest ratio of reduced to oxidized cofactors observed for the ldhA derivative was due to differences in NADPH/NADP+ ratios, which were higher than those observed for K24K. For both strains, the NADPH/NADP+ ratios were higher than the NADH/NAD+ ratios throughout growth in the shaken-flask experiments (Fig. 1A and B). This trend was also observed in the fed-batch (Fig. 2C) and in the batch fermentation experiments, in which the final NADPH/NADP+ ratios were 0.55 and 1.13 for K24K and K24KL, respectively; while the NADH/NAD+ ratios were 0.17 and 0.28 for K24K and K24KL, respectively. This is surprising, as studies of the enzymes involved in PHB synthesis indicate that NADPH is the preferred cofactor for the reductase (34). However, previous work using shaken-flask cultures with glucose have reported similar results for other PHB-synthesizing E. coli strains (32).
The relative amounts of the oxidized forms of both cofactors varied, gradually changing from a higher proportion of NADP+ to a higher proportion of NAD+ as growth and metabolite synthesis proceeded. The change from a larger amount of NADP+ to a larger amount of NAD+ occurred between 5 and 12 h of growth in shaken-flask cultures, so that NAD+ was more abundant than NADP+ during the stationary phase (Fig. 1E and F). A similar trend was observed in fed-batch cultures, in which NADP+ levels continued decreasing, while the amounts of NAD+ increased (Fig. 2C). In shaken-flask cultures, the amounts of NADPH increased with time, while NADH increased during the first hours of growth and then remained approximately constant (Fig. 1E and F). In contrast, NADH increased throughout growth in fed-batch cultures of strain K24KL, probably as a consequence of repeated substrate addition (Fig. 2).
The amount of NADPH was higher than that of NADH at all times in shaken-flask experiments for both PHB-synthesizing recombinants (K24K and K24KL), and until the last hours of growth (50 h) for the K24KL fed-batch culture. Previous work with other PHB-accumulating recombinant E. coli strains reported a higher level of NADH than of NADPH in similar conditions using glucose (32). These dissimilarities could be due to differences in recombinant strain and/or growth conditions. A recent report showed that, under conditions in which NADH contents were reduced, E. coli produced significantly more NADPH (15). Accordingly, the high rate of NADH consumption due to increased production of reduced metabolites in K24K and K24KL could account for the augmented NADPH content observed. One of the mechanisms used by E. coli to control cofactor balance involves transhydrogenases, encoded by pntAB and udhA, that interconvert NADPH and NADH and are differentially activated in different growth conditions (33). In a previous study, cultures grown on glycerol were observed to have high expression levels of pntAB, encoding the enzyme that catalyzes the formation of NADPH from NADH (33). Glycerol as a substrate yields more reducing equivalents than glucose, reportedly in the form of NADH; NADPH in these cultures is believed to be produced mainly through the isocitrate dehydrogenase step of the tricarboxylic acid cycle (39), so cultures grown on glycerol are thought to have an NADPH deficiency. However, based on the ability of pntAB mutants to grow on glycerol, it has been suggested that there are metabolic adaptations that enable the cells to obtain sufficient NADPH under these conditions (33). Additionally, the synthesis of PHB introduces profound changes in the central carbon metabolic pathways of the recombinants that could lead to the cofactor distribution observed.
E. coli adjusts its metabolism to optimize cell growth in each environmental condition by using different combinations of metabolic pathways. There is an intimate association between carbon and electron flow, as carbon will be directed toward the synthesis of more-reduced or more-oxidized metabolic products according to intracellular redox conditions (4, 31, 41). In the ldhA mutant used in this work, the increased availability of carbon and reducing power affected the distribution of cofactors and carbon partitioning among the metabolic products and resulted in a significant increase of PHB and ethanol synthesis. This work illustrates how E. coli metabolism can be manipulated to enhance the production of biotechnologically relevant products from a cheap and readily available substrate.
Acknowledgments
We thank Beatriz S. Méndez and Miguel A. Galvagno for helpful discussions and M. Cecilia Ramirez for her help during construction of strain K24LT.
This work was partially supported by a grant from Universidad de Buenos Aires (project UBACyT X173). P.I.N. and M.J.P. are career investigators from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina). A.D.A. holds a postdoctoral fellowship and M.S.G. has a graduate student fellowship from CONICET.
Footnotes
Published ahead of print on 24 September 2010.
REFERENCES
- 1.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ardeshir, F., and G. F. L. Ames. 1980. Cloning of the histidine transport genes from Salmonella typhimurium and characterization of an analogous transport system in Escherichia coli. J. Supramol. Struct. 13:117-130. [DOI] [PubMed] [Google Scholar]
- 3.Bernofsky, C., and M. Swan. 1973. An improved cycling assay for nicotinamide adenine dinucleotide. Anal. Biochem. 53:452-458. [DOI] [PubMed] [Google Scholar]
- 4.Berríos-Rivera, S. J., G. N. Bennett, and K. Y. San. 2002. The effect of increasing NADH availability on the redistribution of metabolic fluxes in Escherichia coli chemostat cultures. Metab. Eng. 4:230-237. [DOI] [PubMed] [Google Scholar]
- 5.Braunegg, G., B. Sonnleitner, and R. M. Lafferty. 1978. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in bacterial biomass. Eur. J. Appl. Microbiol. Biotechnol. 6:29-37. [Google Scholar]
- 6.Chang, D. E., S. Shin, J. S. Rhee, and J. G. Pan. 1999. Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl coenzyme A flux for growth and survival. J. Bacteriol. 181:6656-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 8.Choi, J., and S. Y. Lee. 1999. Efficient and economical recovery of poly(3-hydroxybutyrate) from recombinant Escherichia coli by simple digestion with chemicals. Biotechnol. Bioeng. 62:546-553. [DOI] [PubMed] [Google Scholar]
- 9.Clark, D. P. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol. Rev. 5:223-234. [DOI] [PubMed] [Google Scholar]
- 10.da Silva, G. P., M. Mack, and J. Contiero. 2009. Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol. Adv. 27:30-39. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawes, E. A., and P. J. Senior. 1973. The role and regulation of energy reserve polymers in micro-organisms. Adv. Microb. Physiol. 10:135-266. [DOI] [PubMed] [Google Scholar]
- 13.de Almeida, A., A. M. Giordano, P. I. Nikel, and M. J. Pettinari. 2010. Effects of aeration on the synthesis of poly(3-hydroxybutyrate) from glycerol and glucose in recombinant Escherichia coli. Appl. Environ. Microbiol. 76:2036-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gokarn, R. R., J. D. Evans, J. R. Walker, S. A. Martin, M. A. Eiteman, and E. Altman. 2001. The physiological effects and metabolic alterations caused by the expression of Rhizobium etli pyruvate carboxylase in Escherichia coli. Appl. Microbiol. Biotechnol. 56:188-195. [DOI] [PubMed] [Google Scholar]
- 15.Holm, A. K., L. M. Blank, M. Oldiges, A. Schmid, C. Solem, P. R. Jensen, and G. N. Vemuri. 2010. Metabolic and transcriptional response to cofactor perturbations in Escherichia coli. J. Biol. Chem. 285:17498-17506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibrahim, M. H. A., and A. Steinbüchel. 2009. Poly(3-hydroxybutyrate) production from glycerol by Zobellella denitrificans MW1 via high-cell-density fed-batch fermentation and simplified solvent extraction. Appl. Environ. Microbiol. 75:6222-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jian, J., S. Q. Zhang, Z. Y. Shi, W. Wang, G. Q. Chen, and Q. Wu. 2010. Production of polyhydroxyalkanoates by Escherichia coli mutants with defected mixed acid fermentation pathways. Appl. Microbiol. Biotechnol. 87:2247-2256. [DOI] [PubMed] [Google Scholar]
- 18.Kabir, M. M., and K. Shimizu. 2003. Fermentation characteristics and protein expression patterns in a recombinant Escherichia coli mutant lacking phosphoglucose isomerase for poly(3-hydroxybutyrate) production. Appl. Microbiol. Biotechnol. 62:244-255. [DOI] [PubMed] [Google Scholar]
- 19.LeVine, S. M., F. Ardeshir, and G. F. L. Ames. 1980. Isolation and characterization of acetate kinase and phosphotransacetylase mutants of Escherichia coli and Salmonella typhimurium. J. Bacteriol. 143:1081-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Z. J., L. Cai, Q. Wu, and G. Q. Chen. 2009. Overexpression of NAD kinase in recombinant Escherichia coli harboring the phbCAB operon improves poly(3-hydroxybutyrate) production. Appl. Microbiol. Biotechnol. 83:939-947. [DOI] [PubMed] [Google Scholar]
- 21.Madison, L. L., and G. W. Huisman. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63:21-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mothes, G., C. Schnorpfeil, and J. U. Ackermann. 2007. Production of PHB from crude glycerol. Eng. Life Sci. 7:475-479. [Google Scholar]
- 23.Nikel, P. I., A. de Almeida, E. C. Melillo, M. A. Galvagno, and M. J. Pettinari. 2006. New recombinant Escherichia coli strain tailored for the production of poly(3-hydroxybutyrate) from agroindustrial by-products. Appl. Environ. Microbiol. 72:3949-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikel, P. I., M. J. Pettinari, M. A. Galvagno, and B. S. Méndez. 2006. Poly(3-hydroxybutyrate) synthesis by recombinant Escherichia coli arcA mutants in microaerobiosis. Appl. Environ. Microbiol. 72:2614-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikel, P. I., A. de Almeida, M. J. Pettinari, and B. S. Méndez. 2008. The legacy of HfrH: mutations in the two-component system CreBC are responsible for the unusual phenotype of an Escherichia coli arcA mutant. J. Bacteriol. 190:3404-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikel, P. I., M. J. Pettinari, M. A. Galvagno, and B. S. Méndez. 2008. Poly(3-hydroxybutyrate) synthesis from glycerol by a recombinant Escherichia coli arcA mutant in fed-batch microaerobic cultures. Appl. Microbiol. Biotechnol. 77:1337-1343. [DOI] [PubMed] [Google Scholar]
- 27.Nikel, P. I., M. J. Pettinari, M. C. Ramirez, M. A. Galvagno, and B. S. Méndez. 2008. Escherichia coli arcA mutants: metabolic profile characterization of microaerobic cultures using glycerol as a carbon source. J. Mol. Microbiol. Biotechnol. 15:48-54. [DOI] [PubMed] [Google Scholar]
- 28.Nikel, P. I., M. C. Ramirez, M. J. Pettinari, B. S. Méndez, and M. A. Galvagno. 2010. Ethanol synthesis from glycerol by Escherichia coli redox mutants expressing adhE from Leuconostoc mesenteroides. J. Appl. Microbiol. 109:492-504. [DOI] [PubMed] [Google Scholar]
- 29.Overath, P., H. U. Schairer, and W. Stoffel. 1970. Correlation of in vivo and in vitro phase transitions of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 67:606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz, J. A., R. O. Fernández, P. I. Nikel, B. S. Méndez, and M. J. Pettinari. 2006. dye (arc) mutants: insights into an unexplained phenotype and its suppression by the synthesis of poly(3-hydroxybutyrate) in Escherichia coli recombinants. FEMS Microbiol. Lett. 258:55-60. [DOI] [PubMed] [Google Scholar]
- 31.San, K. Y., G. N. Bennett, S. J. Berríos-Rivera, R. V. Vadali, Y. T. Yang, R. E. Horton, F. B. Rudolph, B. Sariyar, and K. Blackwood. 2002. Metabolic engineering through cofactor manipulation and its effect on metabolic flux redistribution in Escherichia coli. Metab. Eng. 4:182-192. [DOI] [PubMed] [Google Scholar]
- 32.Sánchez, A. M., J. Andrews, I. Hussein, G. N. Bennett, and K. Y. San. 2006. Effect of overexpression of a soluble pyridine nucleotide transhydrogenase (UdhA) on the production of poly(3-hydroxybutyrate) in Escherichia coli. Biotechnol. Prog. 22:420-425. [DOI] [PubMed] [Google Scholar]
- 33.Sauer, U., F. Canonaco, S. Heri, A. Perrenoud, and E. Fischer. 2004. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279:6613-6619. [DOI] [PubMed] [Google Scholar]
- 34.Schubert, P., A. Steinbüchel, and H. G. Schlegel. 1988. Cloning of the Alcaligenes eutrophus genes for synthesis of poly(β-hydroxybutyric acid) (PHB) and synthesis of PHB in Escherichia coli. J. Bacteriol. 170:5837-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi, H., J. Nikawa, and K. Shimizu. 1999. Effect of modifying metabolic network on poly-3-hydroxybutyrate biosynthesis in recombinant Escherichia coli. J. Biosci. Bioeng. 87:666-677. [DOI] [PubMed] [Google Scholar]
- 36.Solaiman, D. K., R. D. Ashby, T. A. Foglia, and W. N. Marmer. 2006. Conversion of agricultural feedstock and coproducts into poly(hydroxyalkanoates). Appl. Microbiol. Biotechnol. 71:783-789. [DOI] [PubMed] [Google Scholar]
- 37.Sternberg, N. L., and R. Maurer. 1991. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 204:18-43. [DOI] [PubMed] [Google Scholar]
- 38.Verlinden, R. A. J., D. J. Hill, M. A. Kenward, C. D. Williams, and I. Radecka. 2007. Bacterial synthesis of biodegradable polyhydroxyalkanoates. J. Appl. Microbiol. 102:1437-1449. [DOI] [PubMed] [Google Scholar]
- 39.Walton, A. Z., and J. D. Stewart. 2004. Understanding and improving NADPH-dependent reactions by nongrowing Escherichia coli cells. Biotechnol. Prog. 20:403-411. [DOI] [PubMed] [Google Scholar]
- 40.Wlaschin, A. P., C. T. Trinh, R. Carlson, and F. Srienc. 2006. The fractional contributions of elementary modes to the metabolism of Escherichia coli and their estimation from reaction entropies. Metab. Eng. 8:338-352. [DOI] [PubMed] [Google Scholar]
- 41.Zhu, J., and K. Shimizu. 2005. Effect of a single-gene knockout on the metabolic regulation in Escherichia coli for D-lactate production under microaerobic condition. Metab. Eng. 7:104-115. [DOI] [PubMed] [Google Scholar]