Abstract
We have developed a sensitive and highly efficient whole-cell methodology for quantitative analysis and screening of protease activity in vivo. The method is based on the ability of a genetically encoded protease to rescue a coexpressed short-lived fluorescent substrate reporter from cytoplasmic degradation and thereby confer increased whole-cell fluorescence in proportion to the protease's apparent activity in the Escherichia coli cytoplasm. We demonstrated that this system can reveal differences in the efficiency with which tobacco etch virus (TEV) protease processes different substrate peptides. In addition, when analyzing E. coli cells expressing TEV protease variants that differed in terms of their in vivo solubility, cells containing the most-soluble protease variant exhibited the highest fluorescence intensity. Furthermore, flow cytometry screening allowed for enrichment and subsequent identification of an optimal substrate peptide and protease variant from a large excess of cells expressing suboptimal variants (1:100,000). Two rounds of cell sorting resulted in a 69,000-fold enrichment and a 22,000-fold enrichment of the superior substrate peptide and protease variant, respectively. Our approach presents a new promising path forward for high-throughput substrate profiling of proteases, engineering of novel protease variants with desired properties (e.g., altered substrate specificity and improved solubility and activity), and identification of protease inhibitors.
Proteases constitute a group of enzymes that irreversibly catalyze the cleavage of peptide bonds and represent approximately 2% of all protein-encoding genes in living organisms (39). Besides acting as virulence factors for many pathogens (16), proteases are crucial for the regulation of numerous biological processes that influence the life and death of a cell (4). These enzymes also underlie several pathological conditions, such as cancer (13) and neurodegenerative (20) and cardiovascular (8) diseases. A key issue for increasing our knowledge about such complex biological processes, and thereby hopefully also providing possibilities for new therapeutic strategies, is to deduce the proteases’ substrate repertoires. Consequently, a lot of efforts around the world are dedicated to the characterization of proteases and their substrates (2, 31). In addition to their biological importance, proteases have attracted much interest in several biotechnological and industrial applications, such as removal of “fusion tags” from recombinant target proteins (38), as supplements in dishwashing and laundry detergents, or for bating of hides and skin in the leather industry (41, 44). Sometimes, however, their use is hindered due to limitations inherent to a specific protease: for example, low solubility, poor enzyme stability and specificity, or limited activity. It would therefore be of great aid to have powerful and straightforward methods available that facilitate the engineering of novel protease variants not suffering from such limitations.
Traditionally, protease substrate specificity has been studied by comparison and alignment of naturally occurring substrate peptide sequences (7) or through biochemical analysis of cleavage products with synthetic peptides (47). More recent and powerful methods instead rely on the use of combinatorial substrate libraries, which can either be chemically or biologically generated (6, 15). Although all of these methods have proven useful in determining protease function, many suffer from being laborious and of limited throughput capacity, having an insufficient dynamic range, and resulting in limited information on the substrate profile. Moreover, only a small fraction of all proteases have been studied to date, and there is a need for novel approaches that allow for determination of protease specificity in a rapid, accurate, and quantitative manner.
Concerning the engineering of enzymes toward novel desired properties, like altered substrate specificity and improved activity, solubility, and stability; researchers have relied on the use of rational design and/or directed-evolution methods in combination with appropriate screening and selection procedures (1, 10, 11, 22). For instance, various mutagenesis procedures and subsequent screening via assays that report on the successful folding of a protein of interest (9, 32, 45) have been used to engineer protein variants exhibiting improved solubility (35, 37, 46). Despite the obvious success of using such folding reporters in solubility/folding engineering projects, there is a risk that the engineered protein may lose its inherent activity since these screening procedures in general do not select for retained activity but only improved solubility/folding. Therefore, as in the case of a protease, it would be advantageous to establish a screening or selection system that has the ability to simultaneously address traits such as improved folding/solubility without loss of proteolytic activity. However, directed evolution of desired catalytic properties has proven quite a challenge. A popular strategy has been to use phage display technologies, often in combination with transition state analogues (18) or mechanism-based suicide inhibitors, for selection (30). Although successful, the enrichment conferred by these methods is generally based on binding rather than catalysis. Georgiou and coworkers circumvented this potential problem by developing an interesting system that actually enables function-based isolation of novel protease variants from large libraries (34, 42, 43). However, their methodology is dependent on the use of cell surface-displayed proteases, which is not applicable to all proteases and therefore may limit its usefulness.
Herein, we present a novel, function-based, and highly efficient fluorescence-assisted whole-cell assay for characterization and engineering of proteases and their cognate substrate peptides. The method takes advantage of genetically encoded short-lived fluorescent substrates that upon coexpression of a substrate-specific protease result in a fluorescence signal, which can easily be monitored on a flow cytometer. Cells having a desired fluorescence profile can then be collected through sorting and sequenced to identify the protease-sensitive substrate peptide or protease capable of processing a particular peptide. Using this approach, we show that it is possible to analyze the efficiency with which the highly sequence-specific tobacco etch virus protease (TEVp) processes different substrate peptides and in model experiments also identify and enrich cells expressing the most favorable substrate peptide or protease from a large background of cells harboring less-efficient variants.
MATERIALS AND METHODS
Bacterial strains and reagents.
E. coli strain RR1ΔM15 [F′ lacIq Δ(lacZ)M15 supE44 lacY1 lacZ ara-14 galK2 xyl-5 mtl-1 leuB6 proA2 Δ(mcrC-mrr) recA+ rpsL20 thi-1 λ−] (40) was used as the host during construction of plasmids. E. coli strain DH5α [F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rk− mk+) phoA supE44 λ− thi-1 gyrA96 relA1] (Gibco) was used for flow cytometry analysis and cell sorting experiments. Culture media, chemicals, and DNA-modifying enzymes were purchased from Merck (Darmstadt, Germany), Sigma-Aldrich (St. Louis, MO), and New England Biolabs (Ipswich, MA), respectively.
Oligonucleotides.
The following oligonucleotides (Eurofins MWG Operon) were used (5′→3′): GEKO14 (GCAAACGACGAAAACTACAACTACGCTTTAGCAGCTTAAGCATGCAAG), GEKO15 (CTTGCATGCTTAAGCTGCTAAAGCGTAGTTGTAGTTTTCGTCGTTTGC), GEKO20 (CTCATCGATGGGCGCAACATGATAATTATTCGC), GEKO21 (GCGAATAATTATCATCAAGCGCCCATCGATGAG), PEAKfor (GGGGTACCCATCATCATCATCATCATCATGGAG), PEAKrev (GATGGGTACCCATAATCTATGGTCCTTGTTGGT), SAPA46 (CTCTCGAGCTCGAATTCTCTAGATTAAAGAGGAGAAAGGTACCCATGAGTAAAGGAGAAGAACTTTTC), SAPA47 (CTCTCAAGCTTGCATGCTTAAGCTGCTAAAGCGTAGTTTTCGTCGTTTGCTGCGTCGACTTTGTATGTTCATCCATGCCATG), SAPA60 (TCGATGAAGCCCTGAAAGACG), SAPA61 (GGCGATTAAGTTGGGTAACGC), SAPA62 (TCGACGAAAACCTGTACTTCCAGGGTG), SAPA63 (TCGACACCCTGGAAGTACAGGTTTTCG), SAPA66 (TCGACGAAAACCTGTACTTCCAGCCGG), SAPA67 (TCGACCGGCTGGAAGTACAGGTTTTCG), SAPA68 (TCGACGAAAACCTGTACTTCCAGGGTTAAG), SAPA69 (TCGACTTAACCCTGGAAGTACAGGTTTTCG), SAPA72 (GTTGGTATACACTCAGCATCG), and SAPA73 (CGATGCTGAGTGTATACCAAC).
Construction of plasmids.
Heterologous expression of the catalytic domain of the tobacco etch virus protease (TEVp) in E. coli results in low yields of active protease due to the presence of several rare arginine codons in the TEVp gene (24) and self-inactivation caused by autoproteolysis between residues 218 and 219 (36). However, these negative effects can be relieved by exchanging the uncommon arginine codons for synonymous ones that are used more frequently in E. coli (24) and by creating a TEVp mutant (S219V) that is as active as but far more stable than the wild-type protease (26). Therefore, we combined relevant regions from the TEVp-encoding vectors pRK693 (24) and pRK793 (26) through gene splicing by overlap extension (23). First, two separate gene fragments were PCR amplified from pRK693 and pRK793, using primer pairs SAPA60/SAPA73 and SAPA72/SAPA61, respectively. Then, the two products were purified, mixed, and used as the template in an additional PCR, now with primers SAPA60 and SAPA61, to generate the PCR-spliced full-length product. The final amplicon was digested with HindIII and BamHI and ligated into the HindIII/BamHI-digested backbone of pRK693, resulting in pMal-TEV1. Based on this plasmid, we then created pMal-TEV2 by transferring the HindIII/MluI fragment (which encodes maltose-binding protein (MBP) fused to the TEVp substrate peptide, ENLYFQG) from pRK793 into the HindIII/MluI-digested backbone of pMal-TEV1. Thus, when TEVp is expressed from pMal-TEV2, the protease will only be transiently fused to the solubility-enhancing MBP moiety as this domain is removed from the fusion protein (MBP-ENLYFQG-TEVp) intracellularly, through TEVp-mediated substrate processing. We also constructed plasmid pTEV, which encodes TEVp without any solubility-enhancing MBP fusion. Thus, when the protease is expressed from this plasmid, it should exhibit very poor in vivo solubility (27). pTEV was constructed from pMal-TEV2, which served as the template in a PCR with primers PEAKfor and PEAKrev. The PCR product was treated with DpnI, purified on a Qiaquick PCR cleanup column (Qiagen), digested with KpnI, and then purified again. Finally, the digested amplicon was circularized by ligation, which resulted in pTEV. An expression vector for the catalytically inactive TEV protease variant (D81N) (26) was also created. pMal-TEV2 was used as the template in a QuikChange site-directed mutagenesis reaction (Stratagene) with primers GEKO20 and GEKO21 to introduce the D81N mutation, yielding pMal-InTEV. pGFP-ssrA, which encodes ssrA-tagged green fluorescent protein (GFP-ssrA; where ssrA is AANDENYALAA) that is destroyed by the cytoplasmic degradation complex, ClpXP (14, 17), was used as a parental vector for subsequent construction of various TEVp substrate reporter plasmids. For construction of pGFP-ssrA, GFPmut3 (12) was PCR amplified using primers SAPA46 and SAPA47. The amplicon was then digested with SacI and HindIII and ligated into the SacI/HindIII-digested backbone of pBAD33 (19), yielding pGFP-ssrA. A very similar plasmid, pGFP-ssrANY, that instead encodes a modified ssrA tag containing an additional pair of asparagine and tyrosine residues (in boldface) (AANDENYNYALAA), which improves ClpXP-mediated degradation efficiency (21), was also constructed. This was done by a QuikChange site-directed mutagenesis reaction (Stratagene) with primers GEKO14 and GEKO15 on the template pGFP-ssrA. Two TEVp substrate linkers, encoding either glycine (G) or proline (P) in the P1′ position of the wild-type substrate peptide (resulting in ENLYFQG/P), were created from primer pairs SAPA62/SAPA63 and SAPA66/SAPA67, respectively. In addition, a third linker, encoding the wild-type substrate peptide directly followed by a stop codon, was made of primers SAPA68 and SAPA69. The linkers were inserted between the GFP and the ssrA coding regions of SalI-digested pGFP-ssrANY (or pGFP-ssrA) to create pGFP-subG-ssrANY (or pGFP-subG-ssrA), pGFP-subP-ssrANY, and pGFP-subG, respectively (subG is ENLYFQG and subP is ENLYFQP). All plasmid constructs were verified by standard DNA sequencing. Vectors encoding the substrate reporters all contained a p15A origin of replication, a chloramphenicol acetyltransferase gene, and a PBAD promoter controlling the reporter expression. The plasmids constructed for TEVp expression all harbored the Ptac promoter (regulating protease expression), a ColE1 origin of replication, and an ampicillin resistance marker (bla gene, coding for β-lactamase).
Flow cytometry analysis and cell sorting.
A 0.1 mM concentration of isopropyl-β-d-1-thiogalactopyranoside (IPTG) and 0.2% arabinose (final concentrations) were used for induction of protein expression in all cases, unless otherwise stated. For clone analysis and cell sorting, overnight cultures of DH5α cells harboring a TEVp expression vector (pMal-TEV2, pMal-InTEV, or pTEV) and a relevant TEVp substrate reporter plasmid (either pGFP-subG-ssrA, pGFP-ssrA, pGFP-subG, pGFP-subG-ssrANY, pGFP-subP-ssrANY, or pGFP-ssrANY) were subcultured by dilution (1:75 for clone analysis and 1:150 for the sorting experiments) into fresh LB broth, containing 100 μg/ml ampicillin and 20 μg/ml chloramphenicol, and incubated at 37°C in a rotary shaker set at 150 rpm. When the cultures reached a cell density (optical density at 600 nm [OD600]) of ∼0.5, IPTG was added to initiate TEVp expression and the cultures were transferred to a rotary shaker set at 30°C, 150 rpm. Thirty minutes later, l(+)-arabinose was added to induce the reporter expression. After 2 h, 1 ml of each culture was placed on ice and 5 to 10 μl from each sample was diluted with 1 ml ice-cold 1× PBS (11.68 g NaCl, 9.44 g Na2HPO4, 5.28 g NaH2PO4·2H2O, 1,000 ml MilliQ, pH 7.2) and kept on ice until analyzed on a FACSVantage SE flow cytometer (Becton Dickinson). The throughput rate for the flow cytometry analysis was 300 events/s, with an excitation wavelength of 488 nm (argon ion laser) and emission detection between 510 and 530 nm, and 10,000 events were recorded for each sample. For the experiments that aimed at finding induction conditions resulting in improved resolution between positive and negative cells with respect to the whole-cell fluorescence intensity, several different IPTG and arabinose concentrations were tested. In these instances, the final concentrations of IPTG and arabinose ranged from 0.1 mM to 0.00016 mM and 0.2% to 0.00032%, respectively, in 5-fold dilution steps. We also conducted several cell-sorting experiments. In one series, we tried to enrich cells expressing GFP-subG-ssrANY (DH5α/pMal-TEV2/pGFP-subG-ssrANY) from a large excess of cells expressing GFP-subP-ssrANY (DH5α/pMal-TEV2/pGFP-subP-ssrANY). In another set of experiments, the aim instead was to enrich cells expressing TEVp in a soluble format (DH5α/pMal-TEV2/pGFP-subG-ssrANY) from a large background of cells expressing “insoluble” TEVp (DH5α/pTEV/pGFP-subG-ssrANY). The samples were prepared for fluorescence-activated cell sorting in essentially the same way as for flow cytometry analysis of individual clones. However, right before the first round of sorting, the different cell types were mixed to achieve a 100,000-fold excess of “background” cells. The populations were then analyzed on the flow cytometer with a throughput rate of 300 to 500 events/s. The cells were sorted, according to desired fluorescence intensity criteria, directly into LB broth. Collected cells were then either plated on solid LB agar containing ampicillin and chloramphenicol or regrown overnight for additional rounds of analysis and cell sorting.
Western blot analysis.
Overnight cultures of DH5α cells containing pMal-TEV2 and a TEVp substrate reporter plasmid (either pGFP-subG, pGFP-subG-ssrANY, or pGFP-ssrANY) were diluted 1:75 into fresh LB broth supplemented with 100 μg/ml ampicillin and 20 μg/ml chloramphenicol. The cultures grew at 37°C in a rotary shaker set at 150 rpm until they reached a cell density (OD600) of ∼0.5. At this point, TEVp expression was initiated by adding IPTG to a final concentration of 0.1 mM, and then the cultures were incubated at 30°C, 150 rpm. Reporter expression commenced 30 min later by addition of l-arabinose to a final concentration of 0.2%. Two hours later, a volume corresponding to 1.75 OD600 equivalents was withdrawn from each culture, and the cells were harvested by centrifugation at 10,000 × g. The cell pellets were then treated with 400 μl BugBuster (Novagen) in order to release the protein content. After addition of 20 μl 3× SDS denaturing buffer (150 mM Tris-Base, 300 mM dithiothreitol [DTT], 6% SDS, 0.3% bromophenol blue, 30% glycerol) to 40 μl of each whole-cell lysate, the samples were heat denatured (96°C, 8 min) and run on an SDS-PAGE gel (Novex 4 to 12% Tris-glycine gradient gel; Invitrogen). The separated proteins were then transferred to a 0.45-μm-pore-size polyvinylidene difluoride (PVDF) membrane (Invitrogen) and treated overnight with blocking buffer (5% skim milk, 0.5% Tween 20) at 8°C. The blocked membrane was incubated with a rabbit anti-GFP polyclonal antibody (1:2,000 dilution; Dianova) at room temperature for 1 h. After extensive washing with TBST (150 mM NaCl, 10 mM Tris base, 0.05% Tween 20) the membrane was incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:1,000 dilution; Pierce) for 1 h at room temperature. Finally, the protein bands were visualized with a chromogenic HRP substrate (Moss).
RESULTS
Development of a fluorescence-assisted whole-cell assay for protease activity.
We sought to develop a label-free and sensitive high-throughput assay for in vivo monitoring of protease activity in E. coli by creating a genetically encoded protease-specific reporter system suitable for flow cytometry analysis and cell sorting. The reporter system was based on previous findings showing that a C-terminal fusion of the ssrA peptide (AANDENYALAA) to green fluorescent protein (GFP) renders the whole fusion protein susceptible to intracellular degradation by the cytoplasmic protease ClpXP, which effectively eliminates the GFP-mediated whole-cell fluorescence (14, 28).
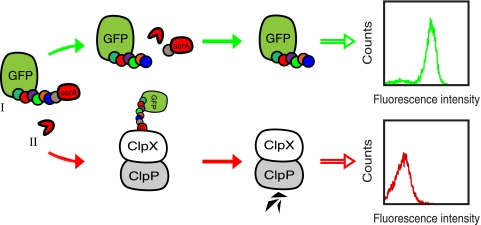
By including a protease substrate peptide between GFP and the ssrA moiety in the reporter construct, it should be possible to remove the ssrA degradation tag and thereby rescue GFP from destruction, given that the fusion protein is coexpressed with a substrate-specific protease. Consequently, there should be an increase in the whole-cell fluorescence intensity, reflecting the protease's apparent catalytic efficiency, which then can be monitored on a flow cytometer with the desired clones isolated through sorting (see Fig. 1 for an overview of the concept).
FIG. 1.
Schematic overview of the system. A reporter substrate, consisting of GFP fused to a protease recognition site with an ssrA degradation tag at the C terminus (I) and a target protease (II) are coexpressed in E. coli cells. (Upper section) A functional protease cleaves off the ssrA degradation tag and rescues GFP from ClpXP-mediated degradation, thus conferring increased whole-cell fluorescence. (Lower section) On the contrary, in the absence of a functional protease, the whole-cell fluorescence intensity is low due to the inevitable degradation of the reporter construct by ClpXP.
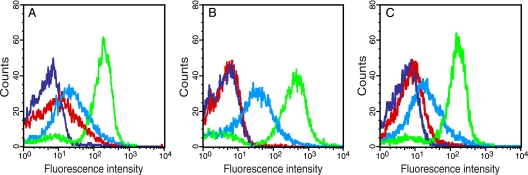
We started out by comparing the whole-cell fluorescence, using a flow cytometer, of E. coli DH5α cells that expressed tobacco etch virus protease (TEVp) together with either (i) a reporter construct containing TEVp's natural substrate peptide, ENLYFQG (GFP-subG-ssrA), or (ii) the negative control, ssrA-tagged GFP lacking any substrate peptide (GFP-ssrA). The reporter molecules were expressed from a low-copy-number plasmid (p15A based; pGFP-subG-ssrA or pGFP-ssrA) under the control of an inducible arabinose promoter, PBAD (0.2% arabinose), while TEVp was expressed from an IPTG-inducible tac promoter (0.1 mM IPTG) on a medium-copy-number plasmid (ColE1-based; pMal-TEV2). First, cells containing only the different reporter vectors (DH5α/pGFP-ssrA or DH5α/pGFP-subG-ssrA) exhibited very low whole-cell fluorescence intensities upon arabinose induction. The fluorescence intensities were in fact almost identical to that of noninduced cells, thereby demonstrating a highly efficient degradation of the reporter proteins by ClpXP (data not shown). Then, when the cells also harbored the TEVp expression vector pMal-TEV2 (DH5α/pMal-TEV2/pGFP-ssrA or DH5α/pMal-TEV2/pGFP-subG-ssrA) and expressed the reporter constructs alone (i.e., without TEVp expression being induced), they still appeared dark. However, cells expressing GFP-subG-ssrA displayed slightly elevated whole-cell fluorescence intensity (Fig. 2A)—most likely due to incomplete repression of the tac promoter controlling the protease expression.
FIG. 2.
System development and optimizations. Fluorescence intensity of E. coli DH5α cells, harboring expression vectors for TEVp and different reporter constructs, subjected to various induction conditions. (A) DH5α/pMal-TEV2/pGFP-subG-ssrA (where subG is ENLYFQG) and DH5α/pMal-TEV2/pGFP-ssrA cells only induced with 0.2% arabinose (PBAD), and thus expressing the reporter constructs alone, are shown in cyan and blue, respectively. The same cell types, induced with both 0.2% arabinose and 0.1 mM IPTG (Ptac), now coexpressing TEVp and the reporter proteins, are shown in green and red, respectively. (B) Exactly the same experimental conditions as in panel A, except for the IPTG concentration, which was reduced to 0.02 mM. (C) Identical to panel A, except for the reporter constructs (pGFP-ssrANY and pGFP-subG-ssrANY), which now contained an ssrA variant (AANDENYNYALAA) engineered for improved ClpXP-mediated degradation.
On the contrary, when TEVp instead was expressed together with the different reporter constructs, there was a large shift toward higher fluorescence intensity, at least for cells expressing GFP-subG-ssrA (Fig. 2A). However, an unspecific gain of fluorescence could also be observed in cells expressing the negative control (i.e., GFP-ssrA). Although a limited increase, it still may interfere with the analysis of suboptimal protease substrates that only result in a minor shift in the whole-cell fluorescence intensity. Therefore, we wanted to minimize this unspecific gain of fluorescence.
System optimizations.
We reasoned that the proteolytic degradation machinery ClpXP may be overwhelmed when the cells, in addition to the reporter constructs, also express TEVp, whereby some GFP-ssrA evades degradation, thus resulting in increased whole-cell fluorescence. A potential solution to this problem may be to adjust the amount of TEVp and reporter molecules produced to a level that is compatible with (i) complete degradation of the negative control and (ii) efficient rescue of the GFP-subG-ssrA reporter.
It has been shown that adjusting the expression levels by titrating the inducer concentrations works fine at a whole-culture level. However, at the single-cell level, the promoters PBAD and Ptac are known to give rise to an all-or-none gene expression pattern at subsaturating inducer concentrations. But this only holds true in bacterial strains that are wild type with respect to the arabinose and lactose transporters, AraE and LacY, respectively (29, 33). Recently, Khlebnikov and Keasling showed that induction with IPTG gives rise to a single population of cells in both lacY-negative and lacY-positive strains regardless of the inducer concentrations used (29). Therefore, we thought it worthwhile to try different concentrations of IPTG (0.1 mM to 0.00016 mM) and arabinose (0.2% to 0.00032%) in 5-fold dilution steps.
In our hands, the only condition that proved productive was when the IPTG concentration was reduced to 0.02 mM. This resulted in an apparent complete degradation of the negative control, while the amount of TEVp produced still was large enough to allow for a substantial shift in the whole-cell fluorescence intensity for cells expressing GFP-subG-ssrA (Fig. 2B). However, a slightly wider distribution and increased fluorescence intensity of the GFP-subG-ssrA expressing population could also be observed (Fig. 2B). The reduction in IPTG (i.e., less TEVp being produced) probably shifted the cellular resources to allow for more reporter protein being produced and thereby yielding higher fluorescence intensity. Concerning the arabinose concentration, it had to be decreased to 0.008% before the signal from the negative control started to decline. However, at this low arabinose concentration, the overall production of reporter molecules also had dropped to such an extent that it was of no benefit to the resolution between positive and negative cells, as was revealed by the flow cytometry analysis (data not shown).
As an alternative approach for improving the apparent degradation efficiency of the negative control and the reporter molecules in general, we tried using an engineered ssrA tag containing an extra pair of asparagine and tyrosine residues (boldface) (AANDENYNYALAA), which has been reported to improve ClpXP-mediated degradation of ssrA-tagged proteins (21). When this modified tag was used, we actually observed an improved resolution between fully induced positive (DH5α/pMal-TEV2/pGFP-subG-ssrANY) and negative (DH5α/pMal-TEV2/pGFP-ssrANY) cells, compared to cells that instead employed the normal ssrA tag in the reporter constructs. This was mainly due to a more efficient degradation of GFP-ssrANY than GFP-ssrA (Fig. 2A and C). Encouraged by these positive results, we decided to continue using this modified degradation tag.
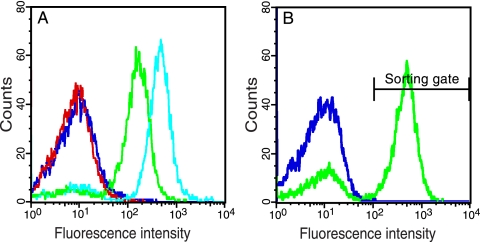
Finally, to rule out the possibility that the expression of TEVp enhances GFP fluorescence by some indirect mechanism other than the removal of the ssrA tag, we expressed the substrate reporters together with a catalytically inactive TEVp mutant (D81N) (26). This experiment indeed showed that catalytic activity was essential for the enhanced fluorescence since only the catalytically active TEVp variant yielded highly fluorescent cells (Fig. 3A). However, the inactive TEVp mutant actually conferred slightly elevated fluorescence intensity in cells expressing GFP-subG-ssrANY, but significantly less than the catalytically active protease did. We do not know the reasons behind the unspecific gain of fluorescence in this particular case but speculate that the inactive TEVp mutant can still bind but not cleave the subG substrate peptide, thereby interfering with the ClpXP-mediated degradation so that some GFP evades proteolysis.
FIG. 3.
Control experiments showing that substrate cleavage is needed for gain of fluorescence. (A) Comparison of the fluorescence intensity conferred by cells expressing either a catalytically active TEVp or an inactive TEVp variant (D81N). Shown are results for DH5α/pMal-TEV2/pGFP-ssrANY (red), DH5α/pMal-TEV2/pGFP-subG-ssrANY (green), DH5α/pMal-TEV2/pGFP-subG (cyan), and DH5α/pMal-InTEV2/pGFP-subG-ssrANY (lilac) 2.5 h after induction (0.2% arabinose, 0.1 mM IPTG). The catalytically active protease yielded significantly higher whole-cell fluorescence intensity (green) than the inactive variant (lilac) did. (B) Western blot analysis of DH5α cells upon coexpression of TEVp and different reporter constructs. The membrane was probed with GFP-specific antibodies to reveal the cytoplasmic GFP levels (boxed) in whole-cell lysates from cells that expressed TEVp together with GFP-ssrANY (lane 2), GFP-subG-ssrANY (lane 3), or GFP-subG (lane 4). Lysate from noninduced DH5α/pMal-TEV2/pGFP-ssrANY (lane 1) and a purified fusion protein, Z-GFP (lane 5), were used as controls. The theoretical molecular masses of the reporter proteins are as follows: GFP-ssrANY, 28 kDa; processed GFP-subG-ssrANY, 27 kDa; unprocessed GFP-subG-ssrANY, 29 kDa; and GFP-subG, 27 kDa. The GFP-reactive antibody exhibits cross-reactivity to two endogenous E. coli proteins of around 20 and 40 kDa, respectively.
In another control experiment, whole-cell lysates from cells expressing TEVp together with either GFP-ssrANY, GFP-subG-ssrANY, or GFP-subG (identical to GFP-subG-ssrANY, but which lacks the degradation tag, ssrANY) were first separated by SDS-PAGE and then subjected to Western blot analysis (using a GFP-specific antibody) to investigate the correlation between proteolysis and fluorescence. Cells that expressed GFP-subG-ssrANY yielded significantly more “rescued” GFP than cells expressing GFP-ssrANY, but less than the positive control, GFP-subG, did (Fig. 3B). This result was in line with the whole-cell fluorescence data (Fig. 3A). Moreover, increased fluorescence in cells expressing GFP-subG-ssrANY actually seemed to be the result of a specific removal of the ssrA tag, catalyzed by TEVp. First, GFP could only be detected at approximately 27 kDa and not 29 kDa, which correspond to the processed and unprocessed forms, respectively. Second, the molecular weight of the GFP band appeared to be identical to that of the positive control (GFP-subG), as expected (Fig. 3B).
Differences in the substrate processing efficiency can be revealed by the fluorescence-assisted whole-cell assay.
To test whether our system had the ability to detect differences in the efficiency with which TEVp processes different substrate peptides, we compared two reporter constructs that were expressed in DH5α cells together with the protease. More specifically, one construct (GFP-subG-ssrANY) contained the natural substrate peptide ENLYFQG, while another (GFP-subP-ssrANY) accommodated proline instead of glycine in position P1′ (ENLYFQP). We used these reporter constructs because they represent TEVp substrates of high and low efficiency of processing, respectively (25).
As expected, when the cells expressed the two different reporter substrates alone, the whole-cell fluorescence intensities were in parity with that of the negative control, GFP-ssrANY (data not shown). Thus, neither the inclusion of a substrate peptide nor its specific amino acid sequence seemed to influence the degradation properties of the reporter proteins to any significant extent.
When the cells then were allowed to coexpress TEVp and the different substrate reporters, there was a large increase in the whole-cell fluorescence intensity for cells expressing GFP-subG-ssrANY, whereas the GFP-subP-ssrANY-expressing cells still exhibited very low fluorescence intensity (Fig. 4A). In fact, the intensity was nearly identical to that of the negative control, GFP-ssrANY, indicating that this particular peptide (GFP-subP-ssrANY) cannot be processed by TEVp. These results reflected the substrates’ inherent efficiency of processing and were coherent with earlier observations obtained through alternative methods (25).
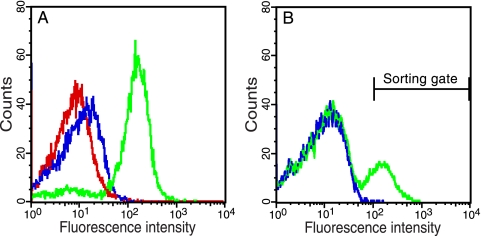
FIG. 4.
Discrimination between protease substrates and isolation of an efficiently processed peptide from a large background of suboptimal substrates. (A) Fluorescence intensity of DH5α/pMal-TEV2/pGFP-ssrANY, DH5α/pMal-TEV2/pGFP-subG-ssrANY, DH5α/pMal-TEV2/pGFP-subP-ssrANY, and DH5α/pMal-TEV2/pGFP-subG cells, 2.5 h after induction (0.2% arabinose, 0.1 mM IPTG). Cells expressing the negative control GFP-ssrANY, GFP-subG-ssrANY (where subG is ENLYFQG), the suboptimal reporter peptide GFP-subP-ssrANY (where subP is ENLYFQP), and, finally, the positive control GFP-subG are shown in red, green, blue, and cyan, respectively. (B) DH5α cells coexpressing TEVp and GFP-subG-ssrANY were mixed with cells coexpressing the protease and the suboptimal reporter peptide, GFP-subP-ssrANY, at a ratio of 1:100,000 (blue). The mixture of cells was then subjected to two rounds of flow cytometry analysis and cell sorting to collect highly fluorescent cells. This resulted in a 69,000-fold enrichment of cells coharboring pMal-TEV2 and pGFP-subG-ssrANY (green).
To estimate the dynamic range of our assay, we created a plasmid encoding a positive control (GFP-subG), GFP fused to the TEVp wild-type (wt) substrate peptide but lacking the ssrA tag. Thus, assuming that the TEVp-mediated liberation of the ssrA tag is quantitative and that all reporters are expressed to the same degree, the positive control should, theoretically, represent the largest possible signal one could expect from our system when using the current configuration. As can be seen in Fig. 4A, our system appears to have a theoretical dynamic range a little less than 2 orders of magnitude, which is based on the comparison of the whole-cell fluorescence intensity of the positive control (DH5α/pMal-TEV2/pGFP-subG) with that of the negative control (DH5α/pMal-TEV2/pGFP-ssrANY).
These initial experiments clearly demonstrated that the substrate processing was protease mediated, specific, and conferred increased whole-cell fluorescence intensity, which should allow for discrimination among different potential substrate peptides through the use of fluorescence-activated cell sorting.
Enrichment and isolation of an efficiently processed protease substrate from a large background of suboptimal substrates.
Since one of the ultimate goals with our system is to use it for identification of substrates for a given protease, we tried to isolate cells expressing a substrate reporter (GFP-subG-ssrANY) that is processed very efficiently by TEVp from a large background of cells containing a virtually noncleavable substrate peptide in the reporter construct (GFP-subP-ssrANY).
To challenge our system, we mixed the two cell types DH5α/pMal-TEV2/GFP-subG-ssrANY and DH5α/pMal-TEV2/pGFP-subP-ssrANY in a ratio of 1:100,000 before the mixture was subjected to flow cytometry analysis and cell sorting for preferential enrichment of cells expressing the favorable substrate reporter: i.e., GFP-subG-ssrANY. In short, the cells were grown in liquid culture, and expression of TEVp and substrate reporters was induced in early log phase. Approximately 2 h later, the cultures were mixed and then analyzed on a flow cytometer, and highly fluorescent clones were collected through cell sorting. After two rounds, approximately 71% of the population seemed to express the better substrate reporter GFP-subG-ssrANY, as revealed by the flow cytometry analysis (Fig. 4B). This finding was also corroborated by DNA sequencing of the coselected plasmids from 192 randomly picked colonies from the final sorting round, which revealed that 69% of the clones harbored the plasmid encoding GFP-subG-ssrANY. This translates into an enrichment factor of 69,000 after two rounds of sorting.
Discrimination and enrichment among TEVp variants exhibiting different in vivo solubilities.
Another potentially very useful application of our system is to use it for engineering of novel protease variants that exhibit desired traits such as improved solubility and activity or altered substrate specificity. For instance, one of the major hurdles connected to the use of TEVp is its low in vivo solubility when expressed in E. coli. Therefore, we wanted to investigate if our system could be used to discriminate among and isolate E. coli cells expressing a soluble TEVp variant from an aggregation-prone protease variant.
In order to do so, we used two different TEVp expression plasmids. In one of the constructs (pMal-TEV2), TEVp was encoded as a transient C-terminal fusion to maltose-binding protein (MBP), which increases the protease solubility in E. coli (27). In the other construct (pTEV), the protease was expressed from an identical plasmid backbone (same promoter, ribosomal binding site, translational initiation region, etc), but now without being fused to the solubility-enhancing MBP moiety. Kapust and Waugh have previously shown that a majority (60 to 70%) of the TEVp is soluble and active when produced as a fusion to MBP, as opposed to the almost completely insoluble protease that accumulated in cells when expressed without MBP (27). Thus, the use of these two plasmids should ensure that any observed dissimilarities in the whole-cell fluorescence intensity (i.e., the substrate processing efficiency) are due to differences in the amounts of soluble and active protease produced.
When each of these two TEVp constructs were expressed in E. coli together with GFP-subG-ssrANY and analyzed on a flow cytometer, they both resulted in fluorescence intensities greater than that from cells coexpressing the negative control, GFP-ssrANY (Fig. 5A). Furthermore, cells expressing “soluble” TEVp displayed significantly higher whole-cell fluorescence intensity than cells containing the “insoluble” protease variant (Fig. 5A).
FIG. 5.
Discrimination of TEVp variants exhibiting different in vivo solubility and isolation of the most soluble one from a large background of a less-soluble TEVp variant. (A) Fluorescence intensity of DH5α/pMal-TEV2/pGFP-ssrANY, DH5α/pTEV/pGFP-subG-ssrANY, and DH5α/pMal-TEV2/pGFP-subG-ssrANY cells, 2.5 h after induction (0.1 mM IPTG, 0.2% arabinose), are shown in red, blue and green, respectively. The plasmid pTEV encodes a TEVp variant, referred to as iTEVp, with poor in vivo solubility due to expression without the solubility-enhancing MBP moiety. On the contrary, pMal-TEV2 encodes TEVp as a transient MBP fusion, which improves the protease's in vivo solubility. (B) DH5α cells coexpressing TEVp and GFP-subG-ssrANY were mixed with cells coexpressing iTEVp and GFP-subG-ssrANY at a ratio of 1:100,000 (blue). The cell mixture was then subjected to two rounds of flow cytometry analysis and cell sorting to preferentially collect cells expressing the more soluble TEVp variant, and resulted in a 22,000-fold enrichment of cells harboring pMal-TEV2.
These results indeed demonstrated that the gain of fluorescence was protease mediated and reflected the intrinsic solubility and/or activity of the investigated protease. Encouraged by the results, we then tried to enrich cells expressing soluble TEVp (DH5α/pMal-TEV2/pGFP-subG-ssrANY) from a large background of cells that expressed the protease in a solubility-compromised manner (DH5α/pTEV/pGFP-subG-ssrANY).
After the two cell types had been mixed in a ratio of 1:100,000, the mixture was analyzed on a flow cytometer and highly fluorescent clones were preferentially collected. After two sorting rounds, a part of the cell population had clearly shifted toward higher whole-cell fluorescence intensity (Fig. 5B), indicating an enrichment of cells expressing the soluble version of TEVp. This was later also confirmed by DNA sequencing analysis of the coselected plasmids, showing that 22% of the clones contained the plasmid encoding the more soluble TEVp variant, which was equivalent to an enrichment factor of 22,000 after two rounds of sorting.
DISCUSSION
We have developed a novel and very efficient fluorescence-assisted whole-cell method for characterization and engineering of proteases and their cognate substrate peptides. The cornerstones of the presented technology are (i) use of live bacterial cells that simultaneously produce the protease of interest and a substrate reporter intracellularly; (ii) the appearance of a fluorescence signal as a result of site-specific substrate proteolysis, which can easily be monitored using a flow cytometer; and (iii) a fluorescence signal that reports on the proteolytic efficiency.
During the development of this methodology, we tried two different approaches that both proved viable for improving the resolution, with respect to the whole-cell fluorescence intensity, between positive and negative cells. One strategy was to find a combination of inducer concentrations resulting in an apparent complete degradation of the negative control and still allowing for efficient rescue of true substrate-containing reporter molecules (Fig. 2B). In the second approach, the normal ssrA degradation tag was replaced with an engineered variant promoting improved degradation efficiency through the ClpXP machinery (Fig. 2C). The effect of these adjustments mainly appeared as a reduction in the unspecific gain of fluorescence from negative cells. We believe this improvement to be important for successful analysis and discrimination among different substrate variants, especially those of intermediate processing efficiency, which would otherwise be hard to discern from negative cells. It is also possible to envision several alternative strategies for achieving the same effect: for example, fine-tuning at the transcriptional and translation level through the use of alternative promoter sequences and/or translation initiation regions (3) or bringing the degradation capacity of ClpXP into line with the system's needs by applying directed-evolution schemes.
Nevertheless, the dynamic range (approximately 2 orders of magnitude) and signal/noise ratio provided by the present system's configuration allowed for identification and enrichment of scarce clones in library-like situations. For instance, in just two rounds of flow cytometry analysis and cell sorting, we managed to isolate clones expressing the best-performing substrate and protease from a large background of cells expressing less-efficient variants (Fig. 4B and 5B).
Nonetheless, in certain instances an improved dynamic range and sensitivity may be advantageous. This could perhaps be accomplished by increasing the expression level of the reporters (e.g., adjusting the promoter strength or number of gene copies) or using enhanced GFP variants in the reporters. However, one must keep in mind that such modifications may lead to an increased number of false-positive clones, unless the modifications are matched by necessary improvements in the ClpXP degradation capacity.
Compared to many other methods used for characterization and/or engineering of proteases, our system offers several attractive features. First, there is no need for synthetic substrates (which may be complicated and costly to produce). Second, the protease does not have to be expressed and/or purified for use in a subsequent in vitro assay. Third, the assay does not suffer from substrate transportation limitations usually associated with methods using intracellularly expressed enzymes; the protease and its corresponding substrate are genetically encoded and coexpressed within the analyzed cells. Fourth, direct quantitative measurement of the substrate processing for a given clone is possible. Fifth, clones exhibiting a desired catalytic efficiency can be enriched selectively by setting the appropriate sort gates on a flow cytometer. Although some of these features are not unique to our system, we believe that it will be a valuable complement or alternative to already existing methods in this growing research field (15), but obviously, future work is still needed to assess its full potential as a protease engineering/characterization platform.
In its current configuration, the protease and substrate have to be functional within the bacterial cytoplasm for the method to work properly. Potentially, this could exclude the use of proteases and substrates whose activity is dependent on structurally important disulfide bridges since such bonds, under normal conditions, cannot be formed there. However, this problem may be solved by using a mutant bacterial strain having an oxidative cytoplasmic environment, which enables efficient folding of recombinant proteins with multiple disulfide bonds in the cytoplasm (5). Obviously, other proteases that still might prove difficult are those that (i) for some other reason fail to fold [e.g., proteases synthesized as (pre)proform often have difficulties converting to the active form in the cytoplasm] or (ii) are toxic to E. coli (e.g., proteases that attack cellular proteins, resulting in cell death). It should be stressed, however, that these limitations are not unique to our system but apply to many host types to which the protease in question is not native.
As we see it, one of the truly strong assets of our method is its simplicity in terms of the genotype-phenotype link. The fact that an increase in the whole-cell fluorescence intensity is function based, related to the apparent catalytic efficiency, and dependent on coexpression of a plasmid-encoded protease and its corresponding substrate reporter should make (i) substrate profiling and (ii) engineering of novel protease variants with desired properties (e.g., improved catalytic activity, increased solubility, and altered substrate specificity) straightforward. For instance, genetically encoded substrate libraries could be constructed by combinatorial randomization of the substrate sequence. If such a library then is coexpressed with a protease of interest and analyzed on a flow cytometer to collect highly fluorescent clones, it should be possible to identify the protease's corresponding substrate profile by sequencing the coselected plasmids. Conversely, isolation and identification of a protease that can cleave a desired substrate peptide should be possible by applying a similar strategy, but in this instance, the library would instead consist of mutant protease variants or cDNA sequences. Although these two applications may demand screening of relatively big libraries, this could readily be accomplished by using a modern high-speed flow cytometer capable of analyzing up to 2.5 × 108 events/h.
We also believe that our system could be used as a cost-efficient platform for identification of protease inhibitors. In such a case, cells grown in microtiter plates and expressing a protease of interest and its corresponding substrate reporter can be used to screen for substances that are able to cross the membrane barrier and inactivate the protease, thereby obstructing the emergence of whole-cell fluorescence.
Considering what has been presented here, our methodology presents a new path forward for high-throughput substrate profiling of proteases, engineering of novel protease variants, and identification of protease inhibitors, which all are areas of great biological, biotechnical, and medical interest.
Acknowledgments
This work was supported by Swedish Research Council grant 621-2004-4647.
David S. Waugh is acknowledged for the kind gift of plasmids pRK693 and pRK793. Bahram Amini is acknowledged for help with DNA sequencing.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Antikainen, N. M., and S. F. Martin. 2005. Altering protein specificity: techniques and applications. Bioorg. Med. Chem. 13:2701-2716. [DOI] [PubMed] [Google Scholar]
- 2.auf dem Keller, U., A. Doucet, and C. M. Overall. 2007. Protease research in the era of systems biology. Biol. Chem. 388:1159-1162. [DOI] [PubMed] [Google Scholar]
- 3.Bandmann, N., and P.-A. Nygren. 2007. Combinatorial expression vector engineering for tuning of recombinant protein production in Escherichia coli. Nucleic Acids Res. 35:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett, A. J., N. D. Rawlings, and J. F. Woessner. 1998. Handbook of proteolytic enzymes. Academic Press, San Diego, CA.
- 5.Bessette, P. H., F. Aslund, J. Beckwith, and G. Georgiou. 1999. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc. Natl. Acad. Sci. U. S. A. 96:13703-13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulware, K. T., and P. S. Daugherty. 2006. Protease specificity determination by using cellular libraries of peptide substrates (CLiPS). Proc. Natl. Acad. Sci. U. S. A. 103:7583-7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bricteux-Grégoire, S., R. Schyns, and M. Florkin. 1972. Phylogeny of trypsinogen activation peptides. Comp. Biochem. Physiol. B 42:23-39. [DOI] [PubMed] [Google Scholar]
- 8.Butler, G. S., and C. M. Overall. 2009. Updated biological roles for matrix metalloproteinases and new “intracellular” substrates revealed by degradomics. Biochemistry 48:10830-10845. [DOI] [PubMed] [Google Scholar]
- 9.Cabantous, S., Y. Rogers, T. C. Terwilliger, and G. S. Waldo. 2008. New molecular reporters for rapid protein folding assays. PLoS One 3:e2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherry, J. R., and A. L. Fidantsef. 2003. Directed evolution of industrial enzymes: an update. Curr. Opin. Biotechnol. 14:438-443. [DOI] [PubMed] [Google Scholar]
- 11.Chica, R. A., N. Doucet, and J. N. Pelletier. 2005. Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design. Curr. Opin. Biotechnol. 16:378-384. [DOI] [PubMed] [Google Scholar]
- 12.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 13.Coussens, L. M., B. Fingleton, and L. M. Matrisian. 2002. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science 295:2387-2392. [DOI] [PubMed] [Google Scholar]
- 14.DeLisa, M. P., P. Samuelson, T. Palmer, and G. Georgiou. 2002. Genetic analysis of the twin arginine translocator secretion pathway in bacteria. J. Biol. Chem. 277:29825-29831. [DOI] [PubMed] [Google Scholar]
- 15.Diamond, S. L. 2007. Methods for mapping protease specificity. Curr. Opin. Chem. Biol. 11:46-51. [DOI] [PubMed] [Google Scholar]
- 16.Duesbery, N. S., C. P. Webb, S. H. Leppla, V. M. Gordon, K. R. Klimpel, T. D. Copeland, N. G. Ahn, M. K. Oskarsson, K. Fukasawa, K. D. Paull, and G. F. Vande Woude. 1998. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science 280:734-737. [DOI] [PubMed] [Google Scholar]
- 17.Farrell, C. M., A. D. Grossman, and R. T. Sauer. 2005. Cytoplasmic degradation of ssrA-tagged proteins. Mol. Microbiol. 57:1750-1761. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Gacio, A., M. Uguen, and J. Fastrez. 2003. Phage display as a tool for the directed evolution of enzymes. Trends Biotechnol. 21:408-414. [DOI] [PubMed] [Google Scholar]
- 19.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haass, C., and B. De Strooper. 1999. The presenilins in Alzheimer's disease—proteolysis holds the key. Science 286:916-919. [DOI] [PubMed] [Google Scholar]
- 21.Hersch, G. L., T. A. Baker, and R. T. Sauer. 2004. SspB delivery of substrates for ClpXP proteolysis probed by the design of improved degradation tags. Proc. Natl. Acad. Sci. U. S. A. 101:12136-12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hibbert, E. G., F. Baganz, H. C. Hailes, J. M. Ward, G. J. Lye, J. M. Woodley, and P. A. Dalby. 2005. Directed evolution of biocatalytic processes. Biomol. Eng. 22:11-19. [DOI] [PubMed] [Google Scholar]
- 23.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 24.Kapust, R. B., K. M. Routzahn, and D. S. Waugh. 2002. Processive degradation of nascent polypeptides, triggered by tandem AGA codons, limits the accumulation of recombinant tobacco etch virus protease in Escherichia coli BL21(DE3). Protein Expr. Purif. 24:61-70. [DOI] [PubMed] [Google Scholar]
- 25.Kapust, R. B., J. Tözsér, T. D. Copeland, and D. S. Waugh. 2002. The P1′ specificity of tobacco etch virus protease. Biochem. Biophys. Res. Commun. 294:949-955. [DOI] [PubMed] [Google Scholar]
- 26.Kapust, R. B., J. Tözsér, J. D. Fox, D. E. Anderson, S. Cherry, T. D. Copeland, and D. S. Waugh. 2001. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14:993-1000. [DOI] [PubMed] [Google Scholar]
- 27.Kapust, R. B., and D. S. Waugh. 1999. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 8:1668-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karzai, A. W., E. D. Roche, and R. T. Sauer. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7:449-455. [DOI] [PubMed] [Google Scholar]
- 29.Khlebnikov, A., and J. D. Keasling. 2002. Effect of lacY expression on homogeneity of induction from the P(tac) and P(trc) promoters by natural and synthetic inducers. Biotechnol. Prog. 18:672-674. [DOI] [PubMed] [Google Scholar]
- 30.Legendre, D., N. Laraki, T. Gräslund, M. E. Bjørnvad, M. Bouchet, P. A. Nygren, T. V. Borchert, and J. Fastrez. 2000. Display of active subtilisin 309 on phage: analysis of parameters influencing the selection of subtilisin variants with changed substrate specificity from libraries using phosphonylating inhibitors. J. Mol. Biol. 296:87-102. [DOI] [PubMed] [Google Scholar]
- 31.López-Otín, C., and C. M. Overall. 2002. Protease degradomics: a new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 3:509-519. [DOI] [PubMed] [Google Scholar]
- 32.Maxwell, K. L., A. K. Mittermaier, J. D. Forman-Kay, and A. R. Davidson. 1999. A simple in vivo assay for increased protein solubility. Protein Sci. 8:1908-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan-Kiss, R. M., C. Wadler, and J. E. Cronan, Jr. 2002. Long-term and homogeneous regulation of the Escherichia coli araBAD promoter by use of a lactose transporter of relaxed specificity. Proc. Natl. Acad. Sci. U. S. A. 99:7373-7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsen, M. J., D. Stephens, D. Griffiths, P. Daugherty, G. Georgiou, and B. L. Iverson. 2000. Function-based isolation of novel enzymes from a large library. Nat. Biotechnol. 18:1071-1074. [DOI] [PubMed] [Google Scholar]
- 35.Olson, C. A., and R. W. Roberts. 2007. Design, expression, and stability of a diverse protein library based on the human fibronectin type III domain. Protein Sci. 16:476-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parks, T. D., E. D. Howard, T. J. Wolpert, D. J. Arp, and W. G. Dougherty. 1995. Expression and purification of a recombinant tobacco etch virus NIa proteinase: biochemical analyses of the full-length and a naturally occurring truncated proteinase form. Virology 210:194-201. [DOI] [PubMed] [Google Scholar]
- 37.Pédelacq, J.-D., E. Piltch, E. C. Liong, J. Berendzen, C.-Y. Kim, B.-S. Rho, M. S. Park, T. C. Terwilliger, and G. S. Waldo. 2002. Engineering soluble proteins for structural genomics. Nat. Biotechnol. 20:927-932. [DOI] [PubMed] [Google Scholar]
- 38.Puhl, A. C., C. Giacomini, G. Irazoqui, F. Batista-Viera, A. Villarino, and H. Terenzi. 2009. Covalent immobilization of tobacco-etch-virus NIa protease: a useful tool for cleavage of the histidine tag of recombinant proteins. Biotechnol. Appl. Biochem. 53:165-174. [DOI] [PubMed] [Google Scholar]
- 39.Rawlings, N. D., E. O'Brien, and A. J. Barrett. 2002. MEROPS: the protease database. Nucleic Acids Res. 30:343-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rüther, U. 1982. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 10:5765-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saeki, K., K. Ozaki, T. Kobayashi, and S. Ito. 2007. Detergent alkaline proteases: enzymatic properties, genes, and crystal structures. J. Biosci. Bioeng. 103:501-508. [DOI] [PubMed] [Google Scholar]
- 42.Varadarajan, N., J. Gam, M. J. Olsen, G. Georgiou, and B. L. Iverson. 2005. Engineering of protease variants exhibiting high catalytic activity and exquisite substrate selectivity. Proc. Natl. Acad. Sci. U. S. A. 102:6855-6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varadarajan, N., S. Rodriguez, B.-Y. Hwang, G. Georgiou, and B. L. Iverson. 2008. Highly active and selective endopeptidases with programmed substrate specificities. Nat. Chem. Biol. 4:290-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, R., C. Min, C. Haiming, and Z. Li. 2009. Enzyme unhairing—an eco-friendly biotechnological process. J. Soc. Leath. Tech. Ch. 93:51-55. [Google Scholar]
- 45.Wigley, W. C., R. D. Stidham, N. M. Smith, J. F. Hunt, and P. J. Thomas. 2001. Protein solubility and folding monitored in vivo by structural complementation of a genetic marker protein. Nat. Biotechnol. 19:131-136. [DOI] [PubMed] [Google Scholar]
- 46.Yang, J. K., M. S. Park, G. S. Waldo, and S. W. Suh. 2003. Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 100:455-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yaron, A., A. Carmel, and E. Katchalski-Katzir. 1979. Intramolecularly quenched fluorogenic substrates for hydrolytic enzymes. Anal. Biochem. 95:228-235. [DOI] [PubMed] [Google Scholar]