Abstract
With the aim of obtaining new strategies to control plant diseases, we investigated the ability of antagonistic lipopolypeptides (paenimyxin) from Paenibacillus sp. strain B2 to elicit hydrogen peroxide (H2O2) production and several defense-related genes in the model legume Medicago truncatula. For this purpose, M. truncatula cell suspensions were used and a pathosystem between M. truncatula and Fusarium acuminatum was established. In M. truncatula cell cultures, the induction of H2O2 reached a maximum 20 min after elicitation with paenimyxin, whereas concentrations higher than 20 μM inhibited H2O2 induction and this was correlated with a lethal effect. In plant roots incubated with different concentrations of paenimyxin for 24 h before inoculation with F. acuminatum, paenimyxin at a low concentration (ca. 1 μM) had a protective effect and suppressed 95% of the necrotic symptoms, whereas a concentration higher than 10 μM had an inhibitory effect on plant growth. Gene responses were quantified in M. truncatula by semiquantitative reverse transcription-PCR (RT-PCR). Genes involved in the biosynthesis of phytoalexins (phenylalanine ammonia-lyase, chalcone synthase, chalcone reductase), antifungal activity (pathogenesis-related proteins, chitinase), or cell wall (invertase) were highly upregulated in roots or cells after paenimyxin treatment. The mechanisms potentially involved in plant protection are discussed.
In most plant-microbe interactions, the majority of the potential pathogens are prevented from causing damage by physical or chemical barriers or after the induction of a complex array of defense responses. The initiation of these inducible defense responses requires specific receptor-mediated recognition of pathogen- or plant cell wall-derived molecules, termed elicitors or PAMPs (pathogenesis-associated molecular patterns) (16). Following pathogen recognition, the activation of signal transduction pathways involving ion fluxes, protein kinase activation, and reactive oxygen species (ROS) production (5, 34) results in the expression of defense-related genes encoding, for example, pathogenesis-related (PR) proteins (17) and enzymes responsible for phytoalexin biosynthesis (45). Calcium influx, reactive oxygen and nitrogen species production, and mitogen-activated protein kinase (MAPK) activation are very early events occurring in response to elicitors (26, 39, 47), whereas phytoalexin synthesis occurs later (18, 22).
The use of plant growth-promoting rhizobacteria (PGPR) as inducers of plant resistance has been widely studied over the past 2 decades. The mechanisms involved include the production of antibiotics, which is a powerful mode of action in disease suppression that acts by directly inhibiting the development and/or the activity of the pathogen (19) or by indirectly inducing plant resistance (21, 44). However, the involvement of antibiotic compounds in the induction of plant resistance has not been investigated in detail (3, 30) and molecular mechanisms underlying early steps in the perception of rhizobacterium elicitors by the host plant are still poorly understood (42, 54). To study the possible involvement of antibiotics in resistance induction, strains producing specific antibiotics need to be applied separately from the pathogen to prevent direct interactions between them (55). Also, to confirm that the resistance induction is due to the antibiotic compounds, mutants lacking antibiotic production or the purified antibiotic compound should be tested (3).
Root and crown rot of alfalfa (Medicago sativa L.) is widespread throughout the world. This disease is characterized by the presence of dark-brown to black wedge-shaped necrotic lesions that spread from the center of affected crowns downward into vascular tissue. Infection reduces yield and longevity of the crop, and stands become unproductive after a few years. Several Fusarium species are associated with this disease, among which Fusarium acuminatum (Ellis & Everhart) is frequently isolated from diseased alfalfa roots (12, 52) and produces the T-2 toxin (38, 40). The size and complexity of legume genomes, the scarcity of genetic information, and difficulties encountered in transforming these plants are major limitations for reaching a better understanding of their defense mechanisms. The use of diploid model legumes, such as Medicago truncatula, which is a close relative of alfalfa that is self-fertile and easily transformed (11), with the large number of genomic tools available (53) could overcome these limitations.
Recently, some cyclic lipopeptide antibiotics from PGPR, such as massetolide A produced by Pseudomonas fluorescens SS101 (51) and surfactins and fengycins produced by Bacillus subtilis S499 (23, 36), were identified as elicitors of systemic resistance. Moreover, we have demonstrated that a mixture of cyclic lipopolypeptides (paenimyxin) isolated from Paenibacillus sp. strain B2 has an antagonistic activity against several plant-pathogenic bacteria and fungi (43). In this study, we investigated the capacity of paenimyxin to stimulate defense responses in M. truncatula, reduce disease symptoms from F. acuminatum, and upregulate transcripts from genes involved in defense mechanisms.
MATERIALS AND METHODS
Microorganism strains.
The Paenibacillus sp. strain B2 used in this study was isolated from the mycorrhizosphere of Sorghum bicolor inoculated with Glomus mosseae (Nicol & Gerd), Gerdemann & Trappe BEG 12 (6). The strain F. acuminatum Ellis & Ever (DSM 62148) was used as the pathogen.
Preparation of paenimyxin.
Bacterial growth conditions and purification of the Paenibacillus sp. strain B2 antagonistic factor (paenimyxin) were as described previously by Selim et al. (43) and referred to as Superdex purified material.
Plant materials and culture conditions. (i) Plantlets.
Seeds of M. truncatula Gaertn cv. Jemalong line J5 (provided by G. Duc, INRA, Dijon, France) were scarified for 6 min in 98% sulfuric acid, surface sterilized for 5 min in 96% ethanol followed by 10 min in 3% calcium hypochlorite, and rinsed in sterile distilled water. They were germinated on 0.7% Bacto agar at 25°C in the dark and transplanted after 48 h in to a hydroponic system in a sterile 12-well plate (flat-bottom multiwell [12-well] tissue culture plate with low-evaporation lid) containing 2 ml·well−1 filter-sterilized Long Ashton solution (20). Plants were grown in a growth chamber under constant conditions (16-h photoperiod, 19°C/22°C [night/day], 360 μmol m−2 s−1, and 70% relative humidity).
(ii) Cell suspension.
M. truncatula cv. Jemalong line J5 leaf cell suspension cultures were grown in 250-ml conical flasks in 100 ml of liquid callus-inducing medium with Murashige-Skoog basal medium (31) containing 30 g·liter−1 sucrose, 2 g·liter−1 casein, 250 mg·liter−1 Bacto tryptone, 3 mM morpholineethanesulfonic acid (MES), 5 mg·liter−1 nicotinic acid, 10 mg·liter−1 thiamine HCl, 10 mg·liter−1 pyridoxine HCl, 100 mg·liter−1 myoinositol, 2 mg·liter−1 glycine, 37.25 mg·liter−1 Na2EDTA, 27.85 mg·liter−1 FeSO4, 1 mg·liter−1 2,4-dichlorophenoxyacetic acid, and 2 mg·liter−1 trans-zeatin. Callus-inducing medium was adjusted to pH 5.6 with KOH, and suspensions were maintained at a constant temperature (25°C) under continuous illumination on a rotary shaker (125 rpm), with subcultures onto fresh medium every 14 days.
Elicitation of cell suspensions.
Plant cells harvested in the exponential phase of growth were filtered using a sintered glass filter (size 1), washed and resuspended at 0.1 g (fresh weight)·ml−1 in suspension buffer (175 mM mannitol, 0.5 mM K2SO4, 0.5 mM CaCl2, 2 mM MES [pH 5.75]). Samples of 10-ml suspensions were then aliquoted into 50-ml Erlenmeyer flasks. After a 2-h equilibration period, the cells were treated with paenimyxin.
Measurement of ROS concentration.
The concentration of ROS in cell suspensions was determined by chemiluminescence as described previously (26, 39, 47). Aliquots of 250 μl were collected from the batch of cell suspensions and mixed with 300 μl of 50 mM MES buffer, containing 175 mM mannitol, 0.5 mM CaCl2, 0.5 mM K2SO4, and 50 μl of 0.3 mM luminol. MES buffer (50 mM) with different pH values (5.6, 6.5, 7.5, and 8.5) was tested to determine the optimum pH for ROS production. Chemiluminescence was measured using a luminometer and expressed in micromoles of H2O2 per gram (fresh weight) of cells by calibrating the response of the luminometer using known concentrations of hydrogen peroxide (0.1 mg·ml−1) in elicitation buffer. The viability of paenimyxin-treated and control cells was evaluated by staining with fluorescein diacetate under UV light as described previously (57).
Inoculum preparation of F. acuminatum.
F. acuminatum was grown on petri plates containing potato dextrose agar (PDA). Following 10 days of incubation at 25°C, five mycelial discs (5-mm diameter) were placed in Erlenmeyer flasks (250 ml) containing 50 ml of PDA. These were incubated on a rotary shaker (125 rpm) at 25°C. After 7 days, the contents of the flasks were collected by centrifugation at 4,000 rpm for 15 min. The fungal biomass (mycelia and microconidia) was then washed twice with a sterile solution of 10 mM MgSO4, briefly homogenized in a blender, and filtered to obtain the microconidia. Spore concentration was determined by measurement with a hemocytometer, and the inoculum concentration was adjusted to 105 microconidia·ml−1.
Protection assay.
The roots of 9-day-old plantlets were soaked in different concentrations of paenimyxin (0 to 100 μM) in autoclaved Milli-Q water. After 24 h, the roots were washed three times with autoclaved Milli-Q water before inoculation with a spore suspension of F. acuminatum (105·ml−1). Plantlets were grown under the same conditions as those mentioned previously. The first symptoms were observed at 4 days postinoculation (dpi). At 4 dpi with F. acuminatum, the fresh weight of shoots and roots was determined and the plant height was measured. Roots were excised and incubated in 10% (wt/vol) KOH for 30 min at 90°C. Fungal structures were stained by dipping roots in trypan blue. Roots were then examined by light microscopy. Resistance or susceptibility was evaluated at 7 dpi, and necrotic spot number and necrosis on the root surface were measured.
To test for the presence of paenimyxin externally or internally of the roots treated with the bioactive molecule (1.23, 3.7, and 11.1 μM), a bioassay (top agar technique) based on Escherichia coli strain MRAP2 was developed and used as follows: 3 ml molten LB-0.7% agarose seeded with a 100-μl suspension of E. coli strain MRAP2 (optical density at 600 nm of 0.5) was placed in a petri dish (90 by 90 mm) containing 30 ml of LB medium. Also, PDA plates inoculated by spreading 500 μl (105 microconidia·ml−1) of the F. acuminatum spore suspension over the agar surface were used. Roots were washed three times in autoclaved Milli-Q water and placed on the surface of the medium to test for the presence of paenimyxin on the external surface of the roots. To test for the presence of paenimyxin inside the roots, three roots were ground and 5 μl of root extract was applied to the wells, punched out of LB or PDA medium with a sterile Pasteur pipette.
Total RNA extraction and DNase treatment.
For gene expression experiments, plant cells were incubated with paenimyxin at a concentration of 5 μM for 10, 20, and 30 min. Cells were centrifuged, harvested, immediately stored in liquid nitrogen, and subsequently used for RNA extraction.
Plantlet roots were treated for 1 day with 1.23 μM paenimyxin, inoculated with F. acuminatum, and harvested 1 and 3 dpi. Roots were stored immediately in liquid nitrogen and subsequently used for RNA extraction. Total RNA was isolated from M. truncatula roots and cells with the RNeasy plant mini kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's recommendations. RNA concentration was evaluated using absorption values at 260 and 280 nm, and RNA quality was checked by gel electrophoresis.
Reverse Northern analyses.
Sixty-four expressed sequence tags (ESTs) of defense-related genes of M. truncatula were selected essentially by their putative involvement in plant defense reactions, protein synthesis, and processing or primary metabolism, using the MENS database (http://medicago.toulouse.inra.fr/Mt/EST). The corresponding cloned cDNA was recovered from the M. truncatula EST libraries (24), multiplied, and purified with the NucleoSpin plasmid kit (Macherey-Nagel, Hoerdt, France). The PCRs were conducted with a PTC-200 thermocycler (MJ Research, Watertown, MA) in a 20-μl reaction volume containing 1 μl of a 1:10 dilution of miniprep materials in water, 0.5 U Taq polymerase (Gibco BRL, Invitrogen, Cergy-Pontoise, France), 125 μM concentration each deoxynucleoside triphosphate (dNTP), and 0.5 μM each primer (18.1for, 5′-GTC ACG ACG TTG TAA AAC G-3′, and 18.2rev, 5′-AGC TAT GAC CAT GAT TAC G-3′). Amplification cycles were as follows: 95°C for 5 min; 30 cycles of 93°C for 45 s, 56°C for 45 s, and 72°C for 2 min; and a final extension at 72°C for 5 min.
cDNA, synthesized from 2.5 μg total RNA, was 32P labeled by RT-PCR and purified on ProbeQuant G-50 microcolumns (Amersham Bioscience, Orsay, France) before denaturation for 5 min at 95°C. PCR products prepared from cloned DNA were separated on 1.2% agarose gels, transferred to Hybond-XL (Amersham Bioscience) by capillary blotting, and fixed under UV light (70,000 J·cm−2) (UV cross-linker; Hoefer Scientific Instruments, San Francisco, CA). Membranes were prehybridized for 1 h at 60°C and hybridized with probes overnight at 60°C in Church buffer (9) and then washed twice for 5 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at room temperature and twice for 20 min in 0.5× SSC-0.1% SDS at 60°C. Hybridization signals were quantified in a Molecular Dynamics Storm 860 phosphorimager with ImageQuant software (Amersham Bioscience) and normalized using the M. truncatula gap1 (Mtgap1) gene (encoding a glyceraldehyde phosphate dehydrogenase) (56).
Semiquantitative RT-PCR.
The expression patterns of genes that were upregulated at least 2-fold by paenimyxin in plant roots, according to reverse Northern blotting, were further analyzed by semiquantitative reverse transcription (RT)-PCR in plant roots incubated with 1.23 μM paenimyxin for 24 h and in cells treated with 5 μM paenimyxin with incubation periods of 10, 20, and 30 min. Semiquantitative RT-PCR was conducted as described previously (48). To prepare cDNA, 1 μg total RNA from paenimyxin-treated and control roots or cells was added to 1.5 μg oligo(dT)15-dNTP (2.5 mM each) and made up to a final volume of 11.5 μl with sterile distilled water. RNA was denatured for 5 min at 70°C and placed on ice, and 5 μl Moloney murine leukemia virus (MMLV) 5× reaction buffer, 300 U MMLV reverse transcriptase, and 80 U RNase inhibitor were added. First-strand cDNA was synthesized at 25°C for 15 min, followed by incubation for 50 min at 42°C and 2 min at 96°C. Gene-specific fragments were amplified by PCR using the specific primers, annealing temperature, and number of cycles described in Table 1 . Amplification was conducted at 95°C for 5 min, 93°C for 45 s, gene-specific annealing temperature for 45 s, 72°C for 2 min, and a final extension at 72°C for 5 min. Amplification products were analyzed by 2% agarose gel electrophoresis, stained by ethidium bromide, and quantified in a Molecular Dynamics Storm 860 phosphorimager with ImageQuant software (Amersham Bioscience).
TABLE 1.
Oligonucleotide primer sequences, PCR conditions, and number of cycles used for analysis by semiquantitative RT-PCR
| Protein family or gene | Primers (forward and reverse; 5′-3′)a | MENS no.b | PCR melting temp (°C) | No. of cycles |
|---|---|---|---|---|
| Chitinase | ACC TCG TAG CCA CCG ACC | MtBC09D09 | 53 | 24 |
| ACT ACT ACC CTT GAC TGG | ||||
| Glutathione S-transferase | GGT ATG CTA CTG GAG ACG | MtBC35F10 | 56 | 22 |
| ATC AGT AGC TCC TGC TGC | ||||
| Pathogenesis-related protein (PR10) | GAA ATG GAG GAC CAG GAA | MtBC34D09 | 59 | 20 |
| ATG GAT CCA CCA TCA GAG CC | ||||
| Pathogenesis-related protein (PR10) | GTT ACG GAT GCT GAC ACC | MtBC53B02 | 53 | 24 |
| CTC ACA CTC ACA CTT CCA | ||||
| Pathogenesis-related protein (PR10-1) | GAT AAC CTT ATC CCG AAG G | MtBC10C12 | 53 | 20 |
| ATG CAT CAC AAC ACA CCG | ||||
| Peroxidase | TGC CCA AAC ATG TCC TAA CC | MtBC28H05 | 57 | 18 |
| AGT CCA TTC CCT CTT CCA CC | ||||
| Phenylalanine ammonia-lyase | CTG CAA GTA GAA ACC CAA G | MtBC46F12 | 54 | 24 |
| TGT CGC ACT CTT CTC CTG | ||||
| Chalcone synthases | GCA ACT AGA GAA GTG CTC | MtBA05H06 | 53 | 24 |
| GCA CAT CAA CAT GTT TGC TG | ||||
| Chalcon reductase | AGA TGA ACC TTG CAT GGC | MtBC34B01 | 53 | 24 |
| GGT CAT TGA TGC CTG GTT | ||||
| Cell wall invertase | ATC AGG GAA ACA GTT GGT GC | MtBC04D07 | 56 | 24 |
| ACT CTA GCT GTG ATG CAG G | ||||
| Cytochrome P450 | GTG AAA GGA GTC GTT TGG | MtBC26E08 | 53 | 22 |
| CTC TTC CTC CAA GAA CCG T | ||||
| Mtgap1 | TGA GGT TGG AGC TGA TTA CG | MtBC42E04 | 55 | 22 |
| AGC CTT GGC AGC TCC AGT GC |
The forward and reverse primers are given in that order.
MENS number according to the Medicago EST navigation system (http://medicago.toulouse.inra.fr/Mt/EST).
The Mtgap1 gene was used as an internal reference control for equivalent reverse transcription to cDNA and equivalent amplification in the PCR.
Statistical analyses.
Four independent biological repetitions were carried out. Comparisons between control and paenimyxin-treated samples were evaluated statistically using Student's t test.
RESULTS
ROS production.
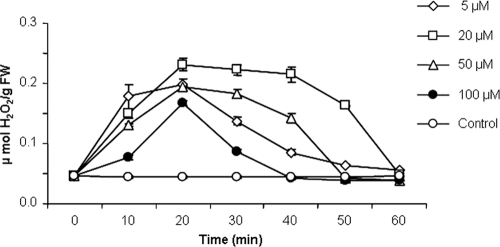
Hydrogen peroxide (H2O2) is commonly produced in plant and cell suspensions exposed to biotic and abiotic stressors, including elicitors of defense responses. The effect of paenimyxin on H2O2 production was tested in M. truncatula cv. Jemalong line J5 cell cultures. The optimum pH for the detection of chemiluminescence was determined to be 7.5 (data not shown). As shown in Fig. 1, paenimyxin at low concentrations induced significant H2O2 production in Medicago cells, which occurred after 5 min of treatment. Paenimyxin-induced H2O2 production reached a maximum after 20 min and returned to background levels within 60 min. At paenimyxin concentrations higher than 20 μM, the production of H2O2 was reduced; this reduction could be related to cell mortality. Indeed, the reduction in cell viability after 20 min in the presence of paenimyxin at 20, 50, and 100 μM reached 12.4, 60.6, and 79.3%, respectively. Because no significant increase in cell death, compared to that of the control, occurred with a concentration of 5 μM paenimyxin, this concentration was used subsequently for gene expression studies.
FIG. 1.
Time course of the production of reactive oxygen species by M. truncatula cells in a suspension treated with different concentrations of paenimyxin (i.e., cells were continually exposed for the indicated times) and control cells (not treated). FW, fresh weight.
Establishment of a paenimyxin-M. truncatula-F. acuminatum (elicitor-plant-pathogen) system.
To assess the protective effect of paenimyxin through the induction of plant defense mechanisms, direct contact between paenimyxin and F. acuminatum was avoided by extensive washing of the paenimyxin-treated roots before inoculation with F. acuminatum. Using the E. coli and F. acuminatum assays, no antagonistic activity associated with the plant roots, treated with 1.23, 3.70, or 11.10 μM paenimyxin, was detected after they were washed with water (data not shown).
Effect of paenimyxin on M. truncatula.
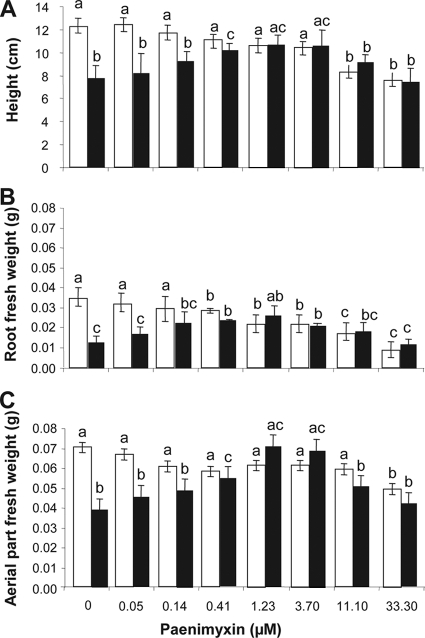
To test the effect of paenimyxin on plant growth, roots were incubated for 24 h with increasing concentrations (0 to 100 μM) of paenimyxin, and plant heights as well as fresh weights of roots and aerial parts were measured after 4 days. Plant height decreased significantly (P < 0.05) when the roots were incubated with a concentration of paenimyxin higher than 3.7 μM (Fig. 2A), whereas the fresh weights of root and aerial parts decreased after treatment with 0.41 and 33.3 μM paenimyxin, respectively (Fig. 2B and C). Plants had yellowish leaves at paenimyxin concentrations of 33.3 and 100 μM (data not shown).
FIG. 2.
Effect of different concentrations of paenimyxin on plant height (A), root fresh weight (B), and fresh weight of aerial parts (C) of M. truncatula at 4 dpi with 105 microconidia·ml−1 F. acuminatum (black) or without inoculation (white). Bars indicate means ± standard deviations. Bars with the same letters are not significantly different from the control grown in the absence of F. acuminatum and paenimyxin, as determined using Student's t test (P < 0.05).
Effect of paenimyxin-Fusarium interaction on growth of M. truncatula.
Pretreatment of the plant roots for 24 h with 0.41 to 3.7 μM paenimyxin resulted in a significant increase in the height of M. truncatula plants in the presence of F. acuminatum (Fig. 2A). Measurement of the fresh weights of roots and aerial plant parts (Fig. 2B and C) showed similar results. Concentrations of paenimyxin higher than 3.7 μM had a negative effect on plant height (Fig. 2A) and fresh weight (Fig. 2B and C).
Symptoms and microscope observations.
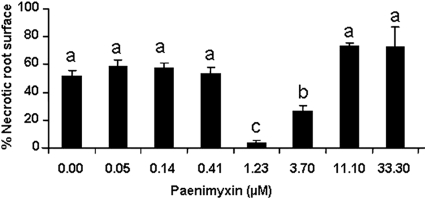
At 7 dpi with 105 microconidia of F. acuminatum, plant symptoms ranged from small brown spots of necrosis and necrotic surfaces (>2 cm) to wilting with symptoms of damping-off. In the absence of pretreatment with paenimyxin, 50% of the root surface, measured at 7 dpi, was necrotic, whereas after pretreatment of the root with 1.23 μM paenimyxin, only 5% of the root surface was necrotic (Fig. 3). Microscope observations after trypan blue staining showed that F. acuminatum hyphae had reached the root surface 3 days after inoculation of untreated plants. Two to 4 days later, extensive mycelium growth covered the root system and fungal hyphae proliferated in an inter- or intracellular fashion to colonize the root parenchymal cortex and the central cylinder (see Fig. S1 in the supplemental material).
FIG. 3.
Effect of different concentrations of paenimyxin on root necrotic surface at 7 dpi with 105 microconidia·ml−1 of F. acuminatum. Bars indicate means ± standard deviations. Bars with the same letters are not significantly different from the control inoculated with F. acuminatum but without paenimyxin pretreatment, as determined using Student's t test (P < 0.05).
Gene expression.
Modifications in plant gene expression in response to F. acuminatum-M. truncatula compatible interactions were first studied by analyzing the expression profiles of 64 ESTs by reverse Northern hybridization. cDNA produced from mRNA extracted from M. truncatula roots at 3 dpi with F. acuminatum without paenimyxin pretreatment was hybridized to selected ESTs. Downregulation or unchanged expression was the most frequent observation in this compatible interaction (Table 2). Twenty-three genes were upregulated at least 2-fold in roots inoculated with F. acuminatum compared to noninoculated roots (Table 2). These genes encode proteins related to plant response to abiotic stimuli, cell wall proteins, defense and cell rescue, lipid-related signaling, membrane transport, oxidative stress, phenylpropanoid and phytoalexin pathways, primary metabolism, protein synthesis and processing, and unknown functions.
TABLE 2.
Expression ratio of some M. truncatula defense-related genes encoding proteins from different classes, estimated by reverse Northern analysis
| Class and putative function | MENS no.a | Expression ratio after indicated treatmentb |
|||
|---|---|---|---|---|---|
| Pmx |
Pmx + Fusarium |
Fusarium at 3 dpi | |||
| 1 dat | 4 dat | 4 dat | |||
| I. Abiotic stimuli and development | |||||
| Germin-like protein (oxalate oxidase) (GLP) | MtBC09H07 | 1.01 | 1.44 | 0.21 | 0.27 |
| MtBC20D01 | 0.43 | 0.28 | 1.67 | 2.16 | |
| MtBC26G08 | 0.32 | 5.20 | 8.17 | 10.57 | |
| MtBC44D10 | 0.42 | 2.15 | 0.89 | 1.16 | |
| Prli-interacting factor G-like | MtBC17B07 | 0.84 | 0.78 | 0.76 | 0.98 |
| II. Cell wall proteins | |||||
| Cellulose synthase | MtBC23F02 | 0.89 | 0.59 | 0.97 | 1.25 |
| Exo-1,3-beta-glucanase precursor | MtBC34E10 | 0.92 | 0.77 | 0.85 | 1.10 |
| Extensin (HRGP) | MtBC14B06 | 0.62 | 8.93 | 6.04 | 7.81 |
| Repetitive proline-rich (RPR) | MtBC25E08 | 0.52 | 0.47 | 0.69 | 0.89 |
| III. Defense and cell rescue | |||||
| Chitinase | MtBB52A02 | 0.96 | 2.32 | 0.37 | 0.47 |
| MtBC09D09 | 2.33 | 0.24 | 0.96 | 1.24 | |
| Chitinase class V | MtBB53F12 | 0.94 | 4.63 | 4.35 | 5.63 |
| Glutathione S-transferase (GST) | MtBC36G01 | 0.61 | 0.31 | 0.06 | 0.08 |
| MtBC27E12 | 0.85 | 0.25 | 0.13 | 0.16 | |
| MtBC49F09 | 1.06 | 4.89 | 2.34 | 3.02 | |
| MtBC35F10 | 2.13 | 1.33 | 2.08 | 2.70 | |
| PR10 | MtBC36H09 | 0.93 | 0.58 | 0.49 | 0.63 |
| MtBC34D09 | 2.07 | 0.41 | 0.36 | 0.47 | |
| MtBB05D08 | 0.81 | 0.52 | 0.65 | 0.53 | |
| MtBC53B02 | 3.94 | 0.10 | 1.78 | 2.31 | |
| PR10-1 | MtBC10C12 | 2.30 | 2.34 | 2.50 | 3.23 |
| PR-1 | MtBC45F12 | 0.71 | 0.10 | 0.37 | 0.48 |
| IV. Gene expression and RNA metabolism | |||||
| Pre-mRNA splicing factor | MtBC09B05 | 0.88 | 1.58 | 1.00 | 1.29 |
| MtBC55B12 | 0.76 | 4.74 | 0.63 | 0.82 | |
| Small nuclear ribonucleoprotein G | MtBC54E06 | 0.88 | 0.11 | 0.38 | 0.50 |
| V. Lipid-related signaling | |||||
| Lipoxygenase | MtBC019C05 | 0.99 | 2.52 | 1.66 | 2.14 |
| VI. Membrane transport | |||||
| Vascular Atp synthase subunit G | MtBC01C05 | 0.82 | 1.06 | 1.66 | 2.15 |
| VII. Oxidative stress | |||||
| Peroxidase | MtBC28H05 | 1.52 | 0.44 | 0.69 | 0.90 |
| MtBC26H05 | 0.68 | 0.72 | 0.93 | 1.20 | |
| MtBC31D01 | 0.63 | 0.20 | 0.03 | 0.04 | |
| MtBC53C01 | 0.37 | 1.65 | 2.63 | 3.40 | |
| VIII. Phenylpropanoid and phytoalexin pathway | |||||
| Phenylalanine ammonia-lyase (PAL) | MtBC46F12 | 2.00 | 0.39 | 0.53 | 0.68 |
| Chalcone synthase (CHS) | MtBA05H06 | 4.17 | 5.75 | 6.25 | 1.62 |
| Isoflavone reductase | MtBC42C03 | 0.61 | 1.53 | 1.16 | 1.50 |
| Chalcone reductase (CHR) | MtBC34B01 | 6.45 | 10.08 | 11.56 | 14.96 |
| MtBC47F02 | 2.00 | 1.44 | 1.48 | 2.00 | |
| IX. Primary metabolism | |||||
| ADP-glucose synthase | MtBB03A11 | 0.66 | 2.23 | 0.98 | 1.27 |
| Cell wall invertase (CWIN) | MtBC04D07 | 3.66 | 4.28 | 3.63 | 4.70 |
| Geranylgeranyl hydrogenase (GGH) | MtBC27B11 | 0.45 | 2.62 | 1.55 | 2.01 |
| Geranylgeranyl pyrophosphate synthase (GGP) | MtBC41D06 | 0.91 | 0.54 | 0.30 | 0.38 |
| Glucose-1-phosphate adenylyl transferase | MtBB03A11 | 0.74 | 0.26 | 0.19 | 0.25 |
| Stearoyl-coenzyme A desaturase | MtBC09C08 | 0.66 | 0.09 | 1.08 | 1.40 |
| X. Protein synthesis and processing | |||||
| 60s Ribosomal protein L5 | MtBC27D07 | 0.62 | 0.43 | 3.90 | 5.04 |
| 60s Ribosomal protein L10 | MtBC45D06 | 0.85 | 0.39 | 0.84 | 1.09 |
| 60s Ribosomal protein L12 | MtBB29E07 | 0.73 | 1.24 | 1.39 | 1.79 |
| 60s Ribosomal protein L15 | MtBC02A12 | 0.58 | 1.45 | 2.09 | 2.70 |
| 60s Ribosomal protein L19 | MtBC25G02 | 0.84 | 1.06 | 0.61 | 0.79 |
| 60s Ribosomal protein L21 | MtBC14A09 | 0.64 | 0.62 | 1.49 | 2.00 |
| 60s Ribosomal protein L23 | MtBC54E01 | 0.80 | 0.13 | 0.28 | 0.37 |
| ATP-dependent Clp protease | MtBC51F07 | 0.97 | 2.06 | 1.59 | 2.06 |
| Elongation factor 1-alpha (EF1) | MtBC40A09 | 0.90 | 0.34 | 2.31 | 2.99 |
| Elongation factor 2 (EF2) | MtBC53E12 | 0.25 | 0.77 | 0.55 | 0.71 |
| Protein disulfide isomerase precursor | MtBC06G02 | 0.92 | 2.09 | 0.79 | 1.02 |
| XI. Secondary and hormone metabolism | |||||
| Chalcone-flavonone isomerase | MtBC02H11 | 0.98 | 0.52 | 1.24 | 1.60 |
| Cytochrome P450 | MtBC26E08 | 1.52 | 0.30 | 0.27 | 0.35 |
| Glutathione S-transferase (GST) | MtBC28B10 | 0.32 | 0.08 | 1.29 | 1.66 |
| MtBC33H02 | 1.04 | 0.74 | 1.11 | 1.43 | |
| MtBC05H10 | 0.85 | 0.93 | 3.43 | 4.44 | |
| Vestitone reductase | MtBC16B03 | 0.63 | 0.58 | 1.07 | 1.39 |
| Zeta-carotene desaturase | MtBB29F03 | 0.72 | 0.30 | 0.14 | 0.19 |
| XII. Unknown function | |||||
| Germin-like protein (oxalate oxidase) (GLP) | MtBC27H11 | 0.70 | 1.58 | 4.28 | 5.54 |
| Nodulin | MtBB01F04 | 0.71 | 4.79 | 0.19 | 0.25 |
| MtBB01C04 | 0.59 | 2.46 | 2.12 | 2.75 | |
| Tpr repeat-containing protein | MtBC03H04 | 0.66 | 2.08 | 0.73 | 0.95 |
MENS number according to Medicago EST navigation system (/medicago.toulouse.inra.fr/Mt/EST).
Expression ratios of ≥2 are in bold. Pmx, paenimyxin; dat, days after treatment; dpi, days postinoculation.
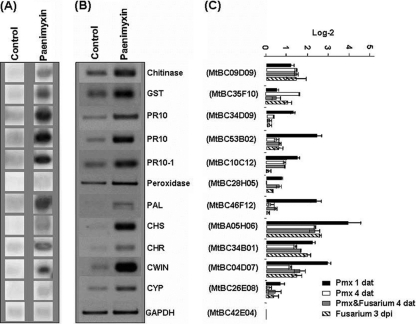
To characterize the mode of action of paenimyxin at a concentration of 1.23 μM in conferring protection of M. truncatula against infection by F. acuminatum, changes in plant gene expression in response to paenimyxin were studied. The results of reverse Northern hybridization showed that the mRNA levels of 10 genes were at least 2-fold higher in roots 1 day after treatment (dat) with paenimyxin than in untreated roots in four independent biological repetitions (Table 2 and Fig. 4). These genes are predicted to encode a cell wall invertase (CWIN; MtBC04D07 [MENS number according to the Medicago EST navigation system]), two chalcone reductases (CHR; MtBC34B01 and MtBC47F02), a chalcone synthase (CHS; MtBA05H06), a phenylalanine ammonia-lyase (PAL; MtBC46F12), a chitinase (MtBC09D09), a glutathione S-transferase (GST; MtBC35F10), and three PR proteins (PR10, MtBC34D09; PR10, MtBC53B02; PR10-1, MtBC10C12). The genes encoding CWIN, CHR, CHS, and PR proteins (PR10 and PR10-1) were upregulated more than 3-fold after 24 h of incubation with paenimyxin compared with genes in untreated roots. In contrast, 7 genes were repressed and 47 genes were not affected by paenimyxin treatment. The 7 genes inhibited more than 2-fold encoded elongation factor 2, geranylgeranyl hydrogenase, three germin-like proteins, GST, and a peroxidase. Among the 10 genes upregulated by paenimyxin 1 dat, 6 (encoding GST, PR10, PR10-1, two CHRs, and CWIN) were also upregulated in response to inoculation with F. acuminatum. At 4 dat with paenimyxin, only four genes of these six (encoding PR10-1, CHS, CHR and CWIN) were still upregulated in the roots inoculated with F. acuminatum or not, in addition to five genes (encoding germin-like protein [GLP; MtBC26G08], extensin[MtBC14B06], chitinase class V [MtBB53F12], GST [MtBC49F09], and nodulin [MtBB01C04]) which were activated later than 4 dat. All the other genes upregulated early (1 dat) or late (4 dat) were inhibited by the presence of F. acuminatum.
FIG. 4.
Expression ratios of 11 M. truncatula genes from paenimyxin-treated roots (Pmx), inoculated or not with F. acuminatum, obtained by reverse Northern hybridization (A) and semiquantitative reverse transcriptase-PCR (B and C). GST, glutathione S-transferase; PR10, pathogenesis-related protein 10; PAL, phenylalanine ammonia-lyase; CHS, chalcone synthase; CHR, chalcone reductase; CWIN, cell wall invertase; CYP, cytochrome P450; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; dat, days after treatment; dpi, days postinoculation. Bars indicate means ± standard deviations.
To confirm the previous data, the transcript accumulation of the upregulated genes was monitored by semiquantitative RT-PCR. As for the reverse Northern analysis, Mtgap1 was used to normalize the samples. At 1 dat with paenimyxin, transcripts of all selected genes increased with paenimyxin treatment, but changes were less than 2-fold for genes encoding GST (Fig. 4). The transcript accumulation of genes encoding CWIN, CHR, CHS, PAL, and PR10 was more than 4-fold higher after paenimyxin incubation. At 4 dat, five genes, encoding chitinase, PR10-1, CHS, CHR, and CWIN, were still active in the roots inoculated with F. acuminatum or not. For the five genes encoding GLP, extensin, chitinase class V, GST, and nodulin which were upregulated later than 4 dat with paenimyxin, none of them was confirmed to be active in root samples 1 and 4 dat.
Transcript accumulation of 11 genes was evaluated in M. truncatula cell cultures treated with 5 μM paenimyxin. The expression of most of the genes was not significantly modified after incubation with paenimyxin for 10 or 20 min. The transcript accumulations of CHS, PR10-1, CHR, chitinase, PR10, and CWIN genes were high after 30 min of incubation, with an increase over those of nontreated cells of 38.6-, 19.9-, 18-, 4.3-, 3.8-, and 3.3-fold, respectively (see Fig. S2 in the supplemental material). Transcript accumulation of the five additional genes increased by a factor of 1.5 to 2.3.
DISCUSSION
The role of PGPR as biological control agents is well documented. However, the mechanism by which PGPR exert their beneficial effects at the molecular level remains poorly understood (50). Elicitation of defense mechanisms is assumed to be a powerful approach for controlling plant diseases and represents an alternative strategy to environmentally undesirable chemical-based methods (10). In the present study, we analyzed the possibility of eliciting plant defense responses by paenimyxin, an antagonistic compound isolated from the PGPR Paenibacillus sp. strain B2 (43). For this purpose, M. truncatula cell suspensions were used and a pathosystem between M. truncatula and F. acuminatum was established.
The ROS production observed in plant cells in response to pathogen infection was first reported by Doke in 1983 (15). In the following years, induction of the oxidative burst was recognized as a central mechanism in plant signaling pathways leading to defense activation (29). Paenimyxin induces a fast and transient H2O2 production in Medicago cell suspensions in a concentration-dependent manner. This oxidation burst is inhibited at paenimyxin concentrations greater than 20 μM. This observation might be related to the observation that cell viability in M. truncatula is reduced by 12.4, 60.6, and 79.2% in the presence of 20, 50, and 100 μM paenimyxin. In several pathosystems, H2O2 was reported to play an important role in the establishment of the hypersensitive response (49). In addition, the rapid generation of reactive oxygen species has been demonstrated to be typically involved in early events associated with the plant defense response following pathogen perception (1). Therefore, these data suggest that H2O2 produced in response to paenimyxin might act as a component of the plant cell machinery contributing to plant defense reactions.
In plantlets, disease symptoms in M. truncatula infected by F. acuminatum range from small brown spots of necrosis to wilting with symptoms of damping-off. In the experimental system used in this study, roots treated with paenimyxin were washed three times with autoclaved Milli-Q water to ensure the absence of any direct effects of the antagonistic factor on the pathogen. No remaining antagonistic activity associated with the roots was detected after the washing, strongly suggesting a plant-mediated defense mechanism. Although a direct effect of paenimyxin against F. acuminatum cannot be totally excluded, the induced resistance against F. acuminatum in M. truncatula roots was obtained with a concentration of paenimyxin of 1.23 μM, which is 6-fold lower than the minimal inhibitor concentration (8 μM) of this antagonistic factor that acts against F. acuminatum (43). This reinforces the idea that paenimyxin protects the plants by priming their defense system. This locally induced resistance is characterized by a large reduction in root damage and disease symptoms. The morphological observations showed large reductions in necrosis on the root surface and of fungal hyphae around the roots, indicating that paenimyxin elicits an inhibitory effect in the plant against fungal growth. Previous studies in other plants have shown induction of one or more plant proteins that have antifungal activity, such as chitinase (25, 27), or of phytoalexin biosynthetic enzymes, such as PAL, CHS, and CHR (10, 32).
To study the molecular basis of the priming activity of paenimyxin, reverse Northern blotting was used instead of the standard approach, as it provides the opportunity for the analysis of large numbers of transcripts. Sixty-four genes of M. truncatula were selected based on their putative involvement in plant defense, protein synthesis and processing, or primary metabolism. Twenty-three of these genes were upregulated in response to F. acuminatum inoculation. Analysis of the expression levels of the genes in M. truncatula roots treated with paenimyxin by reverse Northern blotting and semiquantitative RT-PCR showed that eight (encoding chitinase, three PR10 proteins, PAL, CHS, CHR, and CWIN) were induced earlier (at 1 dat) in the roots by 1.23 μM paenimyxin. Of these eight genes, five genes (encoding chitinase, PR10-1, CHS, CHR, and CWIN) were still upregulated at 4 dat and were also inhibited by F. acuminatum alone. Semiquantitative RT-PCR, using gene-specific primers, is well adapted to studying the expression levels of members of a multigenic family or when the biological material is limited (7). While F. acuminatum upregulated 5 of the 12 plant genes analyzed, paenimyxin more specifically affected those related to plant defense pathways (encoding chitinase, GST, and PR10) and antimicrobial phytoalexin synthesis (encoding PAL, CHS, CHR, and CWIN). Genes encoding CHS, CHR, CWIN, PAL, and PR10 showed the highest increase in transcript levels with paenimyxin treatment, with levels at least 4-fold higher in treated roots than in untreated roots. PAL, CHS, and CHR are key enzymes in phenylpropanoid metabolism leading to a large array of phenolics, including precursors for cell wall reinforcement, antifungal compounds, and salicylic acid, which play an important role in disease resistance (13). The genes encoding CHS, PAL, and CHR have already been reported to be the most rapidly upregulated after elicitation of the isoflavonoid biosynthetic pathway for the synthesis of phytoalexin from l-phenylalanine in alfalfa (32). Isoflavonoid phytoalexins are low-molecular-weight antimicrobial compounds synthesized by plants in response to attempted infection by fungal pathogens, exposure to elicitors, or other biotic and abiotic stresses (14). Phytoalexins have been implicated as important factors in the defense of alfalfa against a number of fungal pathogens (4, 35). There is also strong evidence that lignification is an important mechanism in disease resistance (28). The first step in the phenylpropanoid pathway to forming lignin is the deamination of phenylalanine to cinnamic acid, catalyzed by the PAL enzyme (46). Interestingly, treatment of other plants like potato, tomato, and tobacco with PGPR or their purified LPs also resulted in the accumulation of plant phenolics derived from the phenylpropanoid metabolism (2, 23, 37). The PR proteins PR10 and PR10-1 as well as chitinase have also been reported to have antifungal activity and the expression of their corresponding genes has been associated with defense responses (10, 27).
The CWIN gene induced by paenimyxin belongs to a family encoding cell wall invertase, which is central to phloem unloading in some, but not all, sucrose-importing structures by hydrolyzing sucrose to monosaccharides, such as glucose and fructose, which are the central units for carbon metabolism, storage, and transport. The close relationship between apoplastic sucrose hydrolysis by cell wall invertase to monosaccharide and environmental stimuli or elicitors has been reviewed (8). Other studies with different pathosystems have revealed that the active response of plants to microbial ingress is associated with dramatic reprogramming of cellular metabolism and the rapid induction or strong upregulation in expression of a large array of genes whose products are involved in diverse primary and secondary metabolic pathways (41). In contrast, in M. truncatula cell suspensions, the expression of all of these genes increased in response to treatment with paenimyxin (5 μM) for 30 min.
Paenimyxin has a direct antagonistic activity against several plant-pathogenic bacteria and fungi (43). In contrast, the bacterium producing paenimyxin, Paenibacillus strain B2, stimulates the growth and promotes root colonization of the mycorrhizal fungus Glomus mosseae (6). The present work has shown that a high level of protection against F. acuminatum in M. truncatula can be obtained using paenimyxin at a concentration that is not phytotoxic and that a part of the mechanisms involved may also act through the plant. Multiple strains of PGPR or its cyclic lipopeptides were demonstrated to stimulate plant defense responses (23, 36, 51), and the amount of cyclic lipopeptide synthesized in situ has been shown, for Bacillus subtilis, to be in the same range as that used in this work (33). When taken into consideration, these bacteria and their produced molecules are potentially very interesting for the promotion of plant protection.
Supplementary Material
Acknowledgments
We are grateful to V. Gianinazzi-Pearson for critical reading of the manuscript and to Odile Chatagnier for her helpful technical assistance.
Footnotes
Published ahead of print on 24 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Apel, K., and H. Hirt. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55:373-399. [DOI] [PubMed] [Google Scholar]
- 2.Audenaert, K., T. Pattery, P. Cornelis, and M. Höfte. 2002. Induction of systemic resistance to Botrytis cinerea in tomato by Pseudomonas aeruginosa 7NSK2: role of salicylic acid, pyochelin, and pyocynamin. Mol. Plant Microbe Interact. 15:1147-1156. [DOI] [PubMed] [Google Scholar]
- 3.Bakker, P. A. H. M., L. X. Ran, C. M. J. Pieterse, and L. C. van Loon. 2003. Understanding the involvement of rhizobacteria-mediated induction of systemic resistance in biocontrol of plant diseases. Can. J. Plant Pathol. 25:5-9. [Google Scholar]
- 4.Blount, J. W., R. A. Dixon, and N. L. Paiva. 1992. Stress response in alfalfa (Medicago sativa L.). XVI. Antifungal activity of medicarpin and its biosynthesis precursors; implications for the genetic manipulation of stress metabolites. Physiol. Mol. Plant Pathol. 41:333-349. [Google Scholar]
- 5.Blumwald, E., G. S. Aharon, and B. C. H. Lam. 1998. Early signal transduction pathways in plant-pathogen interactions. Trends Plant Sci. 3:342-346. [Google Scholar]
- 6.Budi, S. W., D. van Tuinen, G. Martinotti, and S. Gianinazzi. 1999. Isolation from the Sorghum bicolor mycorrhizosphere of a bacterium compatible with arbuscular mycorrhiza development and antagonistic towards soilborne fungal pathogens. Appl. Environ. Microbiol. 65:5148-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burleigh, S. H. 2001. Relative quantitative RT-PCR to study the expression of plant nutrient transporters in arbuscular mycorrhizas. Plant Sci. 160:899-904. [DOI] [PubMed] [Google Scholar]
- 8.Büttner, M., and N. Sauer. 2000. Monosaccharide transporters in plants: structure, function and physiology. Biochim. Biophys. Acta 1465:263-274. [DOI] [PubMed] [Google Scholar]
- 9.Church, G., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. U. S. A. 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cluzet, S., C. Torregrosa, C. Jacquet, C. Lafitte, J. Fournier, L. Mercier, S. Salamagn, X. Briand, M.-T. Esquerré-Tugayé, and B. Dumas. 2004. Gene expression profiling and protection of Medicago truncatula against a fungal infection in response to an elicitor from green algae Ulva spp. Plant Cell Environ. 27:917-928. [Google Scholar]
- 11.Cook, D. R. 1999. Medicago truncatula—a model in the making! Curr. Opin. Plant Biol. 2:301-304. [DOI] [PubMed] [Google Scholar]
- 12.Couture, L., C. Dhont, F.-P. Chalifour, R. Drapeau, G. Tremblay, Y. Castonguay, G. Bélanger, and P. Nadeau. 2002. Fusarium root and crown rot in alfalfa subjected to autumn harvests. Can. J. Plant Sci. 82:621-624. [Google Scholar]
- 13.Dixon, R. A., L. Achnine, P. Kota, C. J. Liu, M. S. S. Reddy, and L. J. Wang. 2002. The phenylpropanoid pathway and plant defense—a genomics perspective. Mol. Plant Pathol. 3:371-390. [DOI] [PubMed] [Google Scholar]
- 14.Dixon, R. A., P. M. Dey, and C. J. Lamb. 1983. Phytoalexines: enzymology and molecular biology. Adv. Enzymol. 55:1-135. [DOI] [PubMed] [Google Scholar]
- 15.Doke, N. 1983. Involvment of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with an inocompatible race of Phytophthora infestans and to the hyphal wall components. Physiol. Plant Pathol. 23:345-357. [Google Scholar]
- 16.Ebel, J., and E. G. Cosio. 1994. Elicitors of plant defense responses. Int. Rev. Cytol. 148:1-36. [Google Scholar]
- 17.Friting, B., T. Heitz, and M. Legrand. 1998. Antimicrobial proteins in induced plant defense. Curr. Opin. Immunol. 10:16-22. [DOI] [PubMed] [Google Scholar]
- 18.Hahlbrock, K., D. Scheel, E. Logemann, T. Nurnberger, M. Parniske, S. Reinold, W. R. Sacks, and E. Schmelzer. 1995. Oligopeptide elicitor-mediated defense gene activation in cultured parsley cells. Proc. Natl. Acad. Sci. U. S. A. 92:4150-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handelsman, J., and E. V. Stabb. 1996. Biocontrol of soilborne plant pathogens. Plant Cell 8:1855-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewitt, E. J. 1966. Sand and water culture methods used in studies of plant nutrition. Commonwealth Agricultural Bureau, London, United Kingdom.
- 21.Iavicoli, A., E. Boutet, A. Buchala, and J. P. Métraux. 2003. Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. Mol. Plant Microbe Interact. 16:851-858. [DOI] [PubMed] [Google Scholar]
- 22.Jabs, T., M. Tschöpe, C. Colling, K. Hahlbrock, and D. Scheel. 1997. Elicitor-stimulated ion fluxes and O2− from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. Proc. Natl. Acad. Sci. U. S. A. 94:4800-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jourdan, E., G. Henry, F. Duby, J. Dommes, J. P. Barthélemy, P. Thonart, and M. Ongena. 2009. Insight into the defense-related events occurring in plant cells following perception of surfactin-type lipopeptide from Bacillus subtilis. Mol. Plant Microbe Interact. 22:456-468. [DOI] [PubMed] [Google Scholar]
- 24.Journet, J. M., D. van Tuinen, J. Gouzy, et al. 2002. Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res. 30:5579-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klarzynski, O., and B. Fritig. 2001. Stimulation des défenses naturelles des plantes. C. R. Acad. Sci. III 324:953-963. [DOI] [PubMed] [Google Scholar]
- 26.Lebrun-Garcia, A., F. Ouaked, A. Chiltz, and A. Pugin. 1998. Activation of MAPK homologues by elicitors in tobacco cells. Plant J. 15:773-781. [DOI] [PubMed] [Google Scholar]
- 27.Mauch, F., B. Mauch-Mani, and T. Boller. 1988. Antifungal hydrolases in pea tissue. 2. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol. 88:936-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauch-Mani, B., and A. J. Slusarenko. 1996. Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehdy, M. C. 1994. Active oxygen species in plant defense against pathogens. Plant Physiol. 105:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meziane, H., I. Van Der Sluis, L. C. Van Loon, M. Höfte, and P. A. H. M. Bakker. 2005. Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Mol. Plant Pathol. 6:177-185. [DOI] [PubMed] [Google Scholar]
- 31.Murashige, T., and F. Skoog. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15:473-497. [Google Scholar]
- 32.Ni, W., T. Fahrendorf, G. M. Ballance, C. J. Lamb, and R. A. Dixon. 1996. Stress responses in alfalfa (Medicago sativa L.). XX. Transcriptional activation of phenylpropanoid pathway genes in elicitor-induced cell suspension cultures. Plant Mol. Biol. 30:427-438. [DOI] [PubMed] [Google Scholar]
- 33.Nihorimbere, V., P. Fickers, P. Thonart, and M. Ongena. 2009. Ecological fitness of Bacillus subtilis BGS3 regarding production of the surfactin lipopeptide in the rhizosphere. Environ. Microbiol. Rep. 1:124-130. [DOI] [PubMed] [Google Scholar]
- 34.Nurnberger, T., and D. Scheel. 2001. Signal transmission in the plant immune response. Trends Plant Sci. 6:372-379. [DOI] [PubMed] [Google Scholar]
- 35.O'Neill, N. R., and J. A. Saunders. 1994. Compatible and incompatible responses in alfalfa cotyledons to races 1 and 2 of Colletotrichum trifolii. Phytopathology 84:283-287. [Google Scholar]
- 36.Ongena, M., A. Adam, E. Jourdan, M. Paquot, A. Brans, B. Joris, J. L. Arpigny, and P. Thonart. 2007. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9:1084-1090. [DOI] [PubMed] [Google Scholar]
- 37.Ongena, M., P. Jacques, Y. Touré, J. Destain, A. Jabrane, and P. Thonart. 2005. Involvement of fengycin-type lipopeptides in the multifaceted biocontrol potential of Bacillus subtilis. Appl. Microbiol. Biotechnol. 69:29-38. [DOI] [PubMed] [Google Scholar]
- 38.Poppenberger, B., F. Berthiller, D. Lucyshyn, T. Sieberer, R. Schuhmacher, R. Krska, K. Kuchler, J. Glössl, C. Luschnig, and G. Adam. 2003. Detoxification of the fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 278:47905-47914. [DOI] [PubMed] [Google Scholar]
- 39.Pugin, A., J. Frachisse, E. Tavernier, R. Bligny, E. Gout, R. Douce, and J. Guern. 1997. Early events induced by the elicitor cryptogein in tobacco cells: involvement of a plasma membrane NADPH oxidase and activation of glycolysis and the pentose phosphate. Plant Cell 9:2077-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabie, C. J., E. W. Sydenham, P. G. Thiel, A. Lubben, and W. O. Marasas. 1986. T-2 toxin production by Fusarium acuminatum isolated from oats and barley. Appl. Environ. Microbiol. 52:594-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rushton, P. J., and I. E. Somssich. 1998. Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1:31l-315. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez, L., S. Weidmann, C. Arnould, A. R. Bernard, S. Gianinazzi, and V. Gianinazzi-Pearson. 2005. Pseudomonas fluorescens and Glomus mosseae trigger DMI3-dependent activation of genes related to a signal transduction pathway in roots of Medicago truncatula. Plant Physiol. 139:1065-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selim, S., J. Negrel, C. Govaerts, S. Gianinazzi, and D. van Tuinen. 2005. Isolation and partial characterization of antagonistic peptides produced by Paenibacillus sp. strain B2 isolated from the mycorrhizosphere of sorghum. Appl. Environ. Microbiol. 71:6501-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siddiqui, I. A., and S. S. Shaukat. 2003. Suppression of root-knot disease by Pseudomonas fluorescens CHA0 in tomato: importance of bacterial secondary metabolite, 2,4-diacetylpholoroglucinol. Soil Biol. Biochem. 35:1615-1623. [Google Scholar]
- 45.Somssich, I. E., and K. Hahlbrock. 1998. Pathogen defence in plants—a paradigm of biological complexity. Trends Plant Sci. 3:86-90. [Google Scholar]
- 46.Sticher, L., B. Mauch-Mani, and J. P. Métraux. 1997. Systemic acquired resistance. Annu. Rev. Phytopathol. 35:235-270. [DOI] [PubMed] [Google Scholar]
- 47.Tavernier, E., D. Wendehenne, J. P. Blein, and A. Pugin. 1995. Involvement of free calcium in action of cryptogein, a proteinaceous elicitor of hypersensitive reaction in tobacco cells. Plant Physiol. 109:1025-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor, J., and L. Harrier. 2003. Expression studies of plant genes differentially expressed in leaf and root tissue of tomato colonised by the arbuscular mycorrhizal fungus Glomus mosseae. Plant Mol. Biol. 51:619-629. [DOI] [PubMed] [Google Scholar]
- 49.Tenhaken, R., A. Levine, L. F. Brisson, R. A. Dixon, and C. Lamb. 1995. Function of the oxidative burst in hypersensitive disease resistance. Proc. Natl. Acad. Sci. U. S. A. 92:4158-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Timmusk, S., E. Gerhart, and H. Wagner. 1999. The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: a possible connection between biotic and abiotic stress responses. Mol. Plant Microbe Interact. 12:951-959. [DOI] [PubMed] [Google Scholar]
- 51.Tran, H., A. Ficke, T. Asiimwe, M. Höfte, and J. M. Raaijmakers. 2007. Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol. 175:888-901. [DOI] [PubMed] [Google Scholar]
- 52.Uddin, W., and T. R. Knous. 1991. Fusarium species associated with crown rot of alfalfa in Nevada. Plant Dis. 75:51-56. [Google Scholar]
- 53.VandenBosch, K. A., and G. Stacey. 2003. Summaries of legume genomic projects from around the globe. Community resources for crops and models. Plant Physiol. 131:840-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Loon, L. C. 2007. Plant responses to plant growth-promoting rhizobacteria. Eur. J. Plant Pathol. 119:243-254. [Google Scholar]
- 55.van Loon, L. C., P. A. H. M. Bakker, and C. M. J. Pieterse. 1998. Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36:453-483. [DOI] [PubMed] [Google Scholar]
- 56.Weidmann, S., L. Sanchez, J. Descombin, O. Chatagnier, S. Gianinazzi, and V. Gianinazzi-Pearson. 2004. Fungal elicitation of signal transduction-related plant genes precedes mycorrhiza establishment and requires the dmi3 gene in Medicago truncatula. Mol. Plant Microbe Interact. 17:1385-1393. [DOI] [PubMed] [Google Scholar]
- 57.Widholm, J. M. 1972. The use of fluorescein diacetate and phenosafranine for determining viability of cultured plant cells. Stain Technol. 47:189-194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.