Abstract
Medium-chain fatty acids (octanoic and decanoic acids) are well known as fermentation inhibitors. During must fermentation, the toxicity of these fatty acids is enhanced by ethanol and low pH, which favors their entrance in the cell, resulting in a decrease of internal pH. We present here the characterization of the mechanisms involved in the establishment of the resistance to these fatty acids. The analysis of the transcriptome response to the exposure to octanoic and decanoic acids revealed that two partially overlapping mechanisms are activated; both responses share many genes with an oxidative stress response, but some key genes were activated differentially. The transcriptome response to octanoic acid stress can be described mainly as a weak acid response, and it involves Pdr12p as the main transporter. The phenotypic analysis of knocked-out strains confirmed the role of the Pdr12p transporter under the control of WAR1 but also revealed the involvement of the Tpo1p major facilitator superfamily proteins (MFS) transporter in octanoic acid expulsion. In contrast, the resistance to decanoic acid is composite. It also involves the transporter Tpo1p and includes the activation of several genes of the beta-oxidation pathway and ethyl ester synthesis. Indeed, the induction of FAA1 and EEB1, coding for a long-chain fatty acyl coenzyme A synthetase and an alcohol acyltransferase, respectively, suggests a detoxification pathway through the production of decanoate ethyl ester. These results are confirmed by the sensitivity of strains bearing deletions for the transcription factors encoded by PDR1, STB5, OAF1, and PIP2 genes.
The completion of alcoholic fermentation is one of the major objectives of most wine makers. Several factors can lead to stuck fermentation, such as insufficient nitrogen, the low lipidic content of the grape must, and high concentrations of medium-chain fatty acids (MCFA) in the fermenting must (3). The main MCFA, octanoic and decanoic acids, are produced by yeasts during alcoholic fermentation as by-products of lipid synthesis (40). The exposure of yeast cells to these acids in synthetic media leads to a fast decline of cell viability that is enhanced by low pH and the presence of ethanol (49). Medium-chain fatty acids as well as long-chain fatty acids penetrate inside the cell by passive diffusion in a nonionized form (21) and dissociate at the higher internal pH, leading to a decrease of the intracellular pH (50). The toxicity of short- to medium-chain fatty acids also is correlated with their lipophilic properties, indicating a probable effect on the cell membrane (45). Indeed, decanoic acid, the more toxic of the two compounds (49), has been shown to increase membrane permeability (3). However, the rapid exposure of yeast cells to sublethal concentrations of octanoic acid provokes an adaptive response, allowing the cells to resist larger amounts of inhibitor (7).
Two mechanisms have been hypothesized to explain yeast adaptation to MCFA, including their detoxification into ethyl esters (30) and the activation of a membrane transporter, which has not been identified (7). The WAR1-regulated Pdr12p transporter, which is responsible for the resistance to weak lipophilic organic acids such as sorbate or benzoate (24, 32), is a natural candidate for this function. Nevertheless, results concerning its ability to expulse octanoic acid are contradictory (18, 19), while it clearly is not involved in the decanoic acid response (19). So the question of the transporter involved in octanoic and/or decanoic acid expulsion has not been solved.
In this study, we present an investigation of the yeast response to the exposure to octanoic and decanoic acids. The transcriptome analysis of these two responses combined with the screening of deleted strains enabled us to identify the key transporters involved in yeast resistance to MCFA, and it reveals that ethyl ester synthesis is a possible detoxification pathway for these two acids. In addition, the comparison of these transcriptional responses to the responses caused by other lipophilic compounds enabled us to point out the role of several transcription factors.
MATERIALS AND METHODS
Strains and culture conditions.
The Saccharomyces cerevisiae strains used in this study are listed in Table S1 in the supplemental material. They were strains that were indigenous to wine and were collected in Alsatian wineries or were derivates of the By laboratory strain and eventually were deleted for one or two genes. All strains were cultivated aerobically at 30°C in YPD medium (1% [wt/vol] yeast extract [Difco], 2% Bacto peptone [Difco], 2% glucose [Euromedex]). Unless otherwise indicated, all yeast strains were grown routinely at 28°C.
Strains harboring multiple deletions have been obtained after crossing Euroscarf deletant strains of the opposite sexual type, followed by sporulation and spore dissection. The nonparental ditype (npdt) was verified by the amplification of the wild-type allele and control of kanamycin resistance.
MCFA susceptibility assay.
Tests for medium-chain fatty acid (MCFA) resistance phenotypes were performed with cells grown to the exponential growth phase (optical density at 660 nm [OD660] of 0.8) and diluted to an OD660 of 0.08. Identical volumes of yeast suspensions were spotted onto agar plates containing 0 to 0.25 mM decanoic acid or 0 to 0.8 mM octanoic acid. Growth was evaluated after 72 h of incubation. The resistance levels of strains indigenous to wine were ranked on a scale of 1 to 5 according to their resistance to octanoic or decanoic acids (Table 1).
TABLE 1.
Wine strain resistance level codification used for octanoic or decanoic acid resistance
| Resistance | Maximal MCFA concn allowing growth (mM) |
|
|---|---|---|
| Octanoic acid | Decanoic acid | |
| 1 | 0.4 | 0.2 |
| 2 | 0.6 (+)a | 0.25 (+) |
| 3 | 0.6 (+++)b | 0.25 (+++) |
| 4 | 0.8 (+) | 0.3 (+) |
| 5 | 0.8 (+++) | 0.3 (+++) |
Impaired growth.
Normal growth.
DNA microarray profiling experiment. (i) Medium-chain fatty acid exposure and cell harvest.
For the fatty acid treatment, the U13 wine strain was precultivated overnight in 10 ml agitated YPD media. Cells then were diluted in 300 ml fresh YPD (in 1-liter Erlenmeyer flasks) to obtain an OD660 of 0.05 and grown aerobically at 28°C until an OD660 of 1 to 1.1 was reached. Cultures were split in three, and 10 g/liter octanoic or decanoic acid (Sigma) dissolved in ethanol was added at a final concentration of 0.05 mM to one-third of the culture. The same volume of ethanol was added to the control culture. After 20 min, untreated and treated cultures were harvested by centrifugation at room temperature (2 min, 4,000 × g), and cells were immediately washed in ice-cold water, centrifuged at 4°C at 10,000 rpm/min, and frozen at −80°C. For each treatment, four independent samples were prepared and analyzed.
(ii) mRNA extraction and reverse transcription.
Total RNA was extracted using Trizol reagent (Gibco BRL, Life Technologies). For each sample, 109 cells were pelleted by centrifugation (5,000 × g for 5 min) in two microcentrifuge tubes, resuspended in 400 μl Trizol, and broken by being vortexed for 4 min with 300-μl glass beads. The two extracts were pooled, and the total volume was adjusted to 8 ml with Trizol reagent. After incubation for 5 min at room temperature, 1.6 ml chloroform was added to separate the aqueous and the organic phases with a brief agitation. After incubation for 3 min at room temperature, the mixture was centrifuged at 10,000 × g for 15 min, and the aqueous phase was recovered. The RNA was precipitated by the addition of an equal volume of cold (−20°C) isopropyl alcohol and centrifugation at 10,000 × g for 10 min. The precipitate was further dissolved in 150 μl of RNase-free water, and 100 μg of RNA was cleaned up with an RNeasy kit cartridge (Qiagen).
Fluorescent cDNAs were prepared using a ChipShot direct labeling and cleanup system (Promega Z4100) by direct labeling using dCTPs labeled with Cy3-Cy5 according to the manufacturer's instructions (Amersham).
The three modalities (octanoic acid, decanoic acid, and control) were compared in a triangular design in which each sample has been analyzed once, representing four biological replicates per modality.
Hybridization and microarray analysis.
Microarray slides were obtained from Eurogentec. They were washed with the buffers provided with a Pronto! universal hybridization kit for 25 slides (Corning 40026) according to manufacturer's instructions. Approximately 1,200 ng of labeled cDNA was deposited on the slide and hybridized overnight at 42°C. After being washed, the arrays were read with a Genepix 4000B scanner (Axon Instrument Inc.) and analyzed with Genepix Pro 3.0 (Axon Instrument Inc.). Artifactual or saturated spots were excluded from the analysis.
Statistical analysis.
Raw data were further analyzed using the LIMMA GUI R package (51). Data first were normalized for each slide according to a print-tip-group loess and then normalized between slides according to the quantile procedure. A linear analysis was further performed to detect the genes differentially expressed. Only genes for which a q value higher than 0.05 after Benjamini and Hochberg's (6) false-discovery rate (FDR) adjustment for multiple tests has been applied were retained for further analysis. The full data set has been deposited at GEO with accession number GSE18480 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=bzglpecmoiywudm&acc=GSE18480). For the different responses, the list of transcription factors involved in the regulation of each gene was obtained from the Yeastract database web site (http://www.yeastract.com) (43). The percentages of genes in each response regulated by every transcription factor obtained from Yeastract then were compared in a factorial correspondence analysis made with Statbox (Grimmersoft, Issy Les Moulineaux, France).
RESULTS
Screening of wine-indigenous strains reveals different responses to octanoic and decanoic acid stress.
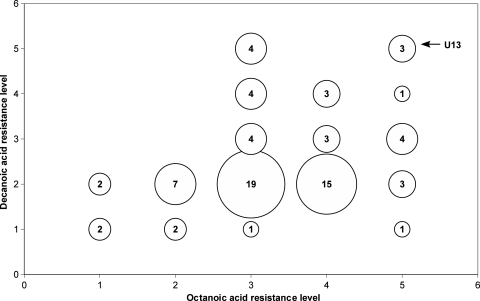
In an attempt to evaluate the variability of the resistance of wine strains to MCFA, we compared the sensitivities of 76 indigenous strains to these inhibitors in a drop test (Table 1). The strains were gathered in groups of similar sensitivities (see Table S1 in the supplemental material). Figure 1 shows that wine strains present a high variability in their ability to resist MCFA. Resistance to decanoic acid generally is associated with a medium to strong resistance to octanoic acid. In contrast, four of the strains showing the highest resistance to octanoic acid were sensitive to decanoic acid. These results suggest strongly that wine yeast activate two partially overlapping mechanisms to resist these MCFA, and that some mechanisms involved in decanoic acid resistance may contribute to octanoic acid resistance.
FIG. 1.
Variability of Saccharomyces cerevisiae wine strain resistance to octanoic acid (x axis) and decanoic acid (y axis) as revealed by drop test (resistance levels are given in Table 1). The dimension of the spots is related to the number of strains in each category indicated in the spots. The arrow indicates the group containing U13.
Transcriptome analysis reveals that octanoic and decanoic acids activate two partially overlapping sets of genes.
To get further insights into the genes involved in the S. cerevisiae response to octanoic and decanoic acids, we studied the transcriptome of wine yeast strain U13, chosen for its high resistance to both inhibitors in the former experiment. This strain was exposed for 20 min to 50 μM each acid. For both acids these conditions were found to be sufficient for the induction of high resistance in a preliminary experiment.
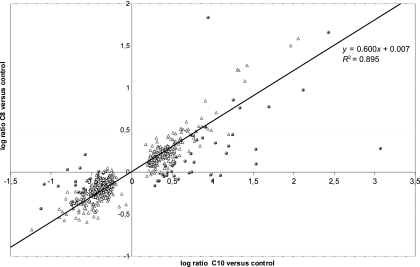
We carried out competitive hybridizations in a triangular design between cDNA obtained from nonexposed cells (T) and cDNA obtained from octanoic acid (C8)- or decanoic acid (C10)-treated cells, as well as between cDNAs from the two acid-exposed conditions. Considering all the genes whose expression was significantly altered between the two conditions, we found that exposure to octanoic or decanoic acid affected 81 and 620 genes, respectively, compared to control cells, with 76 being common to both responses. The comparative hybridization of C8 and C10 modalities revealed that 71 genes were differently affected by these organic acids. The imbalance between the numbers of genes involved in each response suggested that cells were exposed to different stress intensities. To determine if the two responses were correlated, we selected the genes induced by decanoic acid (i.e., genes with significant C10/T log ratios) but that were not detected in the C8/C10 comparison. The C10/T log ratio of these genes was plotted against the C8/T log ratio of the same genes (Fig. 2). The two ratios were highly correlated (R2 = 0.89), indicating that the two acids similarly affected this set of genes, and the slope of 0.6 reveals that the C8 response was weaker than the C10 response at the tested concentrations.
FIG. 2.
Correlation between the two responses. Triangles, expression ratio of genes whose expression level varied significantly compared to that of the control for one modality; spheres, expression ratio of genes whose expression level varied significantly for C8 compared to that of C10; these points were excluded for the estimation of the correlation between the two responses.
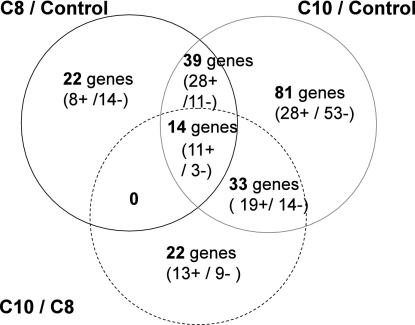
To minimize the biological noise, we restricted the analysis to the genes whose expression significantly differed by a minimum log ratio of 0.5 for C10 acid compared to that of the control. A ratio of 0.3 was chosen for C8 acid compared to that of the control to obtain similar cutoffs for the two acids. As a consequence, compared to the control, 75 genes were significantly modulated by octanoic acid and 165 by decanoic acid, with 53 genes being shared by the two responses (Fig. 3). The analysis of the functional categories through gene ontology (MIPS) (Table 2) shows that the yeast responses to these two organic acids share almost the same subset of genes involved in cell energy supply. Decanoic acid response also includes sets of genes involved in ribosome biogenesis and RNA processing. In agreement with this, GO biological processes for decanoic acid response include ribosome and large ribosomal subunit biogenesis (GO:0042254 and GO:0042273) and oxidation reduction (GO:0055114). The major facilitator superfamily proteins (MFS) also are especially well represented in this response. In addition, the analysis of these responses with Eu.Gene 1.2.1 (10) showed the significant activation of the fatty acid beta oxidation pathway.
FIG. 3.
Venn diagram presenting the genes differentially expressed in these three conditions. C8, 20-min exposure to octanoic acid at 50 mM; C10, 20-min exposure to octanoic acid at 50 mM; C8/C10, comparison of exposure to C10 to exposure to C8. Repressed genes are followed by a minus sign; induced genes are followed by a plus sign.
TABLE 2.
Classification of genes involved in response to octanoic and decanoic acid according to MIPS functional categories
| MIPS category |
P |
|
|---|---|---|
| C8 | C10 | |
| Sugar, glucoside, polyol and carboxylate catabolism (01.05.02.07) | 2.3e-07 | 4.3e-06 |
| Electron transport (02.11) | 2.9e-07 | |
| Electron transport and membrane-associated energy conservation (02.11) | 3.2e-06 | 1.3e-06 |
| Ribosome biogenesis (12.01) | 2.5e-11 | |
| rRNA processing (11.04.01) | 7.7e-10 | |
| Purine nucleotide/nucleoside/nucleobase anabolism (01.03.01.03) | 1.9e-08 | |
| Tricarboxylic-acid pathway (02.10) | 2.1e-05 | |
Direct comparison of octanoic and decanoic acid-treated cells (with a log ratio of 0.3) allowed us to gain further indications on the analogies/differences between the two responses. We found that 68 genes presented significantly different expression profiles (Fig. 3). As the activation of genes was not similar for octanoic acid and decanoic acid, we divided the activated genes in three sets: genes activated by both C8 and C10 but not by the control were qualified as shared responses, the genes activated by C8 but not the control or C10 were called C8-specific responses, and the genes activated by C10 but not the control or C8 combined with genes differentially expressed after exposure to C10 were called C10-specific responses (Fig. 3).
Among the 53 genes whose expression was affected by both acids (C8 and C10 shared response) (see Table S2 in the supplemental material), 39 were upregulated and 14 repressed. Among them, C8 more efficiently induced PDR12 (3.5 times increase), whereas C10 more specifically induced ALD4, CWP1, TMA17, and HXK1.
The octanoic acid-specific response included 22 genes; 8 were upregulated (i.e., IDH2, ATP3, ALG12, TRX2, etc.), whereas 14 were repressed (EFT1, EFT2, ZRT1, FAS1, etc.).
The exposure to C10 resulted in the specific modulation of the expression of 114 genes (C10-specific response), among which EEB1, coding for an ethyl ester synthase, tops the list for its high induction (8.4 times increase). Two other genes (FAA1 and ELO1) involved in fatty acid metabolism also were induced (1.7 and 2.4 times increase), suggesting a potential metabolism of the fatty acid. However, two transporters involved in cell detoxification, TPO4 and PDR12 (two times higher expression), and, to a lesser extent, TPO1 (1.5 times increase), also were activated.
Comparison of octanoic and decanoic acid responses to stress responses already described.
The octanoic and decanoic acid responses were compared to those already described for different stresses (Table 3) after a similar incubation period: sorbic acid (a weak acid response has been described [36]), sodium dodecyl sulfate (39), octanol (14), fluphenazine (12), benomyl (27), 2,4-deoxyphenoxyacetic acid (44), polyoxyethylene-9-laurylether (POELE), 2,4-dichlorophenol (DCP) (37), oleic acid oxidative stress (23) and H2O2 (15). For each pair of stresses, we have counted the number of genes significantly induced or repressed by both stresses. When the significance information was not available, we selected genes induced (or repressed) by at least a factor of 2. The highest similarities were observed between the responses to octanoic and decanoic acid (71% of C8 responses are shared with C10 responses), but about half of the genes involved in both responses also are shared with H2O2 oxidative stress. Significant portions of these responses also are activated by detergent stresses: i.e., 45 and 35% of genes activated by octanoic and decanoic acids also are activated by SDS. Octanoic acid response presents 29% of genes in common with the sorbic acid response. In contrast with these different stresses, the oleic acid oxidative stress involved few of the genes activated by C8 or C10.
TABLE 3.
Comparison of different transcriptional responsesa
| Stressb | % Genes shared between the different responses | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Octanoic acid (0.05 mM) | 100 | |||||||||||
| Decanoic acid (0.05 mM) | 71 | 100 | ||||||||||
| Sorbic acid (8 mM) (36) | 29 | 19 | 100 | |||||||||
| POELE (1 mM) (37) | 43 | 31 | 43 | 100 | ||||||||
| DCP (3 mM) (37) | 19 | 22 | 20 | 28 | 100 | |||||||
| Octanol (1%) (14) | 0 | 23 | 18 | 14 | 19 | 100 | ||||||
| SDS (1%) (39) | 45 | 39 | 26 | 21 | 20 | 21 | 100 | |||||
| Fluphenazine (1 mM) (12) | 12 | 17 | 12 | 26 | 27 | 17 | 37 | 100 | ||||
| Benomyl (7 mM) (27) | 12 | 14 | 12 | 17 | 25 | 16 | 23 | 13 | 100 | |||
| 2,4-Dichlorophenoxyacetic acid (3 mM) (44) | 16 | 14 | 17 | 20 | 13 | 15 | 21 | 11 | 6 | 100 | ||
| Oleic acid (23) | 7 | 6 | 8 | 8 | 10 | 6 | 17 | 16 | 12 | 5 | 100 | |
| H2O2 (3 mM) (15) | 52 | 47 | 42 | 27 | 35 | 23 | 40 | 38 | 41 | 30 | 22 | 100 |
The percentage of genes shared between two different responses is given as the number of genes shared by the two responses divided by the number of genes of the smaller response.
The number of genes involved in each response was as follows: octanoic acid, 75; decanoic acid, 165; sorbic acid, 137; POELE, 548; DCP, 342; octanol, 621; SDS, 463; fluphenazine, 132; benomyl, 69; 2,4 dichlorophenoxyacetic acid, 172; oleic acid, 267; H2O2, 936.
As a consequence, our results suggest that the responses to octanoic and decanoic acid are composite responses involving the organic weak acid response and a detergent response, with both of them presenting many similarities with an oxidative stress.
Search for known transcription factors involved in MCFA response.
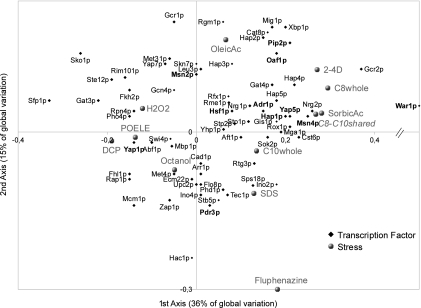
For both acid responses as well as for the other stresses cited in the former paragraph, we obtained from the Yeastract web site (43) the transcription factors involved in the regulation of each gene and scored the number of genes regulated per transcription factor. These scores were compared in a correspondence analysis. The result plotted in Fig. 4 revealed that the octanoic acid response presents, as expected, similarities to the weak acid response involving War1 and Msn4 (Fig. 4). In contrast, the C10 response appeared much closer to the SDS stress response. C8 and C10 responses also appeared as potentially regulated by transcription factors HAP1, HAP4, and HAP5. This could be interpreted as a sign of the activation of the fatty acid beta-oxidation pathway.
FIG. 4.
Factorial component analysis comparing the involvement of each transcription factor in the different stress responses. The proportion of each stress response explained by one transcription factor has been calculated from the Yeastract website. Codes for the different stresses: OleicAc, oleic acid; POELE, polyoxyethylene-9-laurylether; SorbicAc, sorbic acid; 2-4D, 2,4-dichlorophenoxyacetic acid; DCP, 2,4-dichlorophenol; C10whole, decanoic acid; C8whole, octanoic acid. The octanoic and decanoic shared response (C8-C10 shared) is analyzed as a supplementary individual and given in italic. Transcription factors with a cos2 lower than 0.1 are not drawn. Main transcription factors involved in the stress response are in boldface.
Phenotyping screening of Euroscarf deletion mutant strains allows us to identify genes involved in resistance. (i) Transporters.
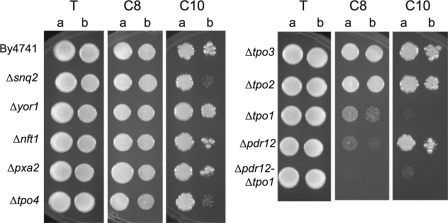
Looking for transporters involved in octanoic and decanoic acid expulsion, we screened a collection of haploid strains deleted for their PDR genes as well as for other transporters, including ADP1, AQR1, ATM1, AUS1, AZR1, BPT1, DIP5, FLR1, NFT1, PDR5, PDR10, PDR11, PDR12, PDR15, PDR18, PXA2, SNQ2, TPO1, TPO2, TPO3, TPO4, YBT1, and YOR1. Figure 5 shows the results obtained for the sensitivity test of some of these strains in the presence of 0 to 0.6 mM C8 and 0 to 0.25 mM C10.
FIG. 5.
Drop test presenting the sensitivity provoked by the deletion of different transporters on YPD (pH 4.5) medium containing 0.6 mM octanoic acid (C8) or 0.25 mM decanoic acid (C10) compared to that of the control (T). Cells used to prepare the spots were grown on liquid YPD (pH 4.5) medium until a standardized OD660 of 0.8 (a) and diluted to an OD660 of 0.08 (b). The growth observed for the nondiluted spot of the wild-type By laboratory strain corresponds to a rank of three in the first figure.
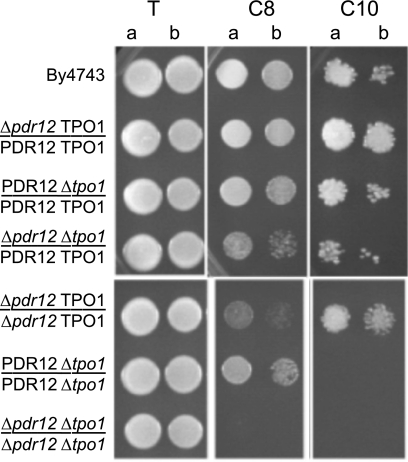
The highest sensitivity to octanoic acid was obtained for the Δpdr12 strain; however, the deletion of TPO1 also resulted in an increased sensitivity to this inhibitor. In addition, the Δpdr12-Δtpo1 double deletant strain was found to be more sensitive than the two single-deletion strains, showing a cumulative effect of the two transporters in the expulsion of this acid. In contrast to octanoic acid, the Δtpo1 strain exhibited the highest sensitivity to decanoic acid, while the deletion of PDR12 did not affect this phenotype. However, Δtpo4 and Δsnq2 strains also were slightly affected, indicating a possible ability of these transporters to expulse decanoic acid. None of the other strains tested showed modified resistance to octanoic or decanoid acid. Since Pdr12p and Tpo1p were the main transporters of octanoic and/or decanoic acid, we constructed diploid strains harboring one or two deleted copies of the genes (Fig. 6). In a background where two alleles of a given transporter were present, the presence of a single allele of the other transporter was sufficient to regenerate the wild-type phenotype. When the two tested genes were present as a single copy, the growth of yeast strains was injured on both octanoic and decanoic acids. Moreover, when a single copy of the TPO1 gene was present, decanoic acid resistance was correlated with the number of copies of the PDR12 gene, indicating that Pdr12p plays a part, though discrete, in resistance to decanoic acid. This observation was confirmed by analyzing the growth of the diploid on liquid medium complemented with inhibitors (Bioscreen analysis) (results not shown).
FIG. 6.
Drop test presenting the sensitivity of diploid strains deleted from one or two copies of PDR12 and TPO1 transporter genes on YPD (pH 4.5) medium containing 0.6 mM octanoic acid (C8) or 0.25 mM decanoic acid (C10) compared to that of the control (T). Cells used to prepare the spots were grown on liquid YPD (pH 4.5) medium until a standardized OD660 of 0.8 (a) and diluted to an OD660 of 0.08 (b).
(ii) Transcription factors.
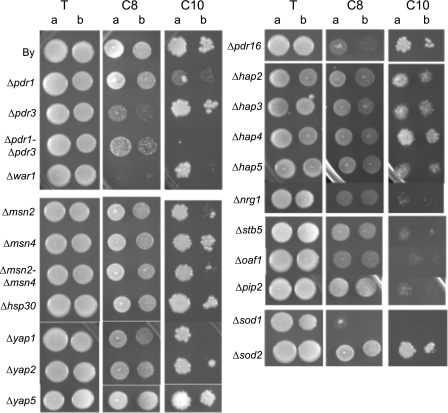
We also tested Euroscarf strains deleted for regulatory genes, including main regulators of stress responses (PDR1/PDR3, MSN2/MSN4, HSP30, etc.), transcription factors already described in weak organic acid stress or in acid stress (WAR1, HAA1, etc.), and other transcription factors suggested by the transcriptome analysis. Tested transcription factors were ADR1, AFT1, ARR1, CIN5, CRZ1, KFH2, FLO8, GCN4, GCR2, HAA1, HAC1, HAP2 to HAP5, HSF1, HSP30, MSN2, MSN4, NRG1, OAF1, PDR1, PDR3, PIP2, RPN4, SEF1, SKO1, SOK2, STB5, TOS8, WAR1, YAP1, YAP2, and YRR1. Figure 7 shows the results obtained for the drug sensitivity test of some of these strains in the presence of 0 to 0.6 mM C8 and 0 to 0.25 mM C10.
FIG. 7.
Drop test presenting the sensitivity of strains deleted for different transcription factors on YPD (pH 4.5) medium containing 0.6 mM octanoic acid (C8) or 0.25 mM decanoic acid (C10) compared to that of the control (T). Cells used to prepare the spots were grown on liquid YPD (pH 4.5) medium until a standardized OD660 of 0.8 (a) and diluted to an OD660 of 0.08 (b).
The octanoic acid response is clearly under the control of the WAR1 transcription factor. The deletion of PDR3 also has a great impact on this phenotype, while PDR1 deletion does not modify strain resistance to octanoic acid. None of the other transcription factors that were tested seemed to be involved in the modulation of the octanoic acid response, including HAA1 (not shown), which was described to be involved in a weak acid response (13).
The response to decanoic acid is clearly under the control of PDR1, while PDR3 deletion does not result in higher sensitivity. However, the Δpdr1-Δpdr3 double mutant is more sensitive to decanoic acid than the Δpdr1 mutant, indicating a slight role of PDR3. Moreover, the deletion of STB5 also lowers strain resistance to decanoic acid. This transcription factor is known to form a heterodimer with Pdr1p but not with Pdr3p and to regulate the pentose phosphate pathway and NADPH production in response to oxidative stress (25). Three other transcription factors are clearly involved in response to decanoic acid, namely, NRG1, OAF1, and PIP2.
Finally, the deletion of MSN2 (but not MSN4) or YAP1 seems to have little effect on decanoic acid resistance.
(iii) Other genes.
We also tested some other genes already described to be involved in weak acid resistance or in lipophilic compound resistance. We observed that PDR16 deletion had an impact on octanoic acid resistance, while the deletion of the homologous PDR17 gene had no impact.
Finally, SOD1 deletion had a drastic effect on both octanoic and decanoic acid sensitivity, while SOD2 deletion had no impact on these phenotypes.
DISCUSSION
Octanoic and decanoic acids are two compounds synthesized by yeast during alcoholic fermentation and are potent inhibitors, and they have been suggested to be involved in sluggish fermentation (4). The toxicity of these lipophilic molecules has been attributed to internal medium acidification (8). In the meantime, because toxicity is correlated with lipophilicity, an effect on membrane organization also has been suggested (3). Nevertheless, S. cerevisiae is able to adapt to these inhibitors through the induction of transporters (7) and possibly other mechanisms. The negative influence of these inhibitors on the fermentation process is offset by the fact that they are supposed to be the precursors of ethyl esters, which enrich wine flavor with fruit aromas (17), even though this has never been clearly shown (5). Our work was aimed at gaining insights into the mechanisms involved in the adaptation of S. cerevisiae to octanoic and decanoic acids. In a first approach we analyzed the resistance of a set of wild strains to both acids. The variability of the responses observed revealed unambiguously that two different and partially overlapping mechanisms were activated in response to the two inhibitors.
Transcriptome analysis reveals composite responses.
To get further information on the genes involved in the octanoic or decanoic acid responses, we used a transcriptomic analysis. Both responses appeared highly correlated, which we expected from the closely related structures of the two acids, but they also presented differences. We were able to distinguish a common response involving genes that were modulated by both acids and more restricted octanoic- or decanoic-specific responses. The responses to octanoic or decanoic acid also were compared to some already-described stress responses, including response to weak acids, to several detergents (SDS, POELE, etc.), to oxidative stress (H2O2), and to oleic acid as a unique carbon source. The octanoic and decanoic responses share more genes with each other than with any other stress response. They both present features in common with the oxidative stress response, but with an original component: the response triggered by octanoic acid exposure presents similarities to the weak acid response (36), whereas the decanoic acid response additionally presents some similarities to the oleic acid early stress response (23) and to the SDS stress response (39). This can be related to the chemical structures of the two inhibitors, and one can suggest that octanoic acid is perceived as an acid, while the hydrophobic part of decanoic acid is more prominent than its acidic part and induces a detergent-like response.
Phenotypic analysis of deletant strains highlights the main genes involved in resistance.
Transcriptome analysis reveals a number of genes induced or repressed as a response to an environmental change, turning the cell metabolism to the new conditions. However, many of these genes do not play a key role in the resistance to the stress agents (15). Several of the genes induced by MCFA according to transcriptome analysis did not participate significantly in MCFA resistance in the drop test phenotypic screening. As an example, the expression of TPO4 and PDR12 transporters was much more increased by exposure to decanoic acid (2.3 and 1.9 times increase, respectively) than the expression of TPO1 (1.5 times increase); however, the latter was the only transporter that was really efficient in the triggering of decanoic acid resistance. The weak induction of TPO1 is not related to a high basal expression but may be related to the mild intensity of the challenge. The higher activation of TPO4, which is not involved in the resistance, has been described likewise for 2,4-dichlorophenoxyacetic acid (44). In contrast, some of the genes involved in inhibitor resistance could be undetected in the transcriptome due to a delayed response or a posttranscriptional regulation. Drop tests of deleted strains submitted to octanoic and decanoic acids allowed us to identify genes involved in the resistance to both acids (common resistance mechanism) or to one of them (specific resistance mechanism).
Common resistance mechanism.
Among the numerous genes we tested, only TPO1, YAP1, and SOD1 seemed to be involved in both octanoic and decanoic acid resistance.
The MFS transporter Tpo1p is the key player in decanoic acid resistance and contributes significantly to octanoic acid resistance. Tpo1p was described to protect cells against a broad range of structurally unrelated molecules, including spermidine, cycloheximide, nystatin, artesunate, ibuprofen, 2,4-dichlorophenoxyacetic acid, etc. (for a review, see reference 33). To our knowledge, this work is the first evidence of Tpo1p being involved in the resistance to octanoic and decanoic acids. In our analysis of the resistance of wild strains to octanoic and decanoic acid, we observed that a high level of decanoic acid resistance always was associated with a high level of octanoic acid resistance. Strains with more efficient Tpo1p transport would exhibit such a phenotype. Interestingly, the TPO1 gene (as well as TPO4 and SNQ2) is activated progressively during the course of alcoholic fermentation (28), suggesting a key role during alcoholic fermentation. In analyzing the genomic differences between laboratory yeast strains and wine-making commercial strains by the CGH array, Dunn and coauthors (11) observed the amplification of TPO1 in all of the commercial wine strains tested, which could be the sign of an adaptive evolution, even if this amplification was not observed in a later study (9). The genome of the U13 wine strain that was used for transcriptomic analysis was compared to the S288c genome by a CGH array and did not reveal any amplification of the TPO1 gene (unpublished data), suggesting the gain of efficiency by another mechanism.
YAP1 is a transcription factor involved in oxidative stress adaptation (26) and the regulation of the expression of antioxidant genes such as thioredoxin, thioredoxin reductase, and glutathione reductase (reviewed in reference 20). YAP1 deletion hampers the cell resistance to octanoic and decanoic acid, suggesting that these inhibitors cause oxidative damage to the cell. Consistently with this hypothesis, MCFA resistance also is impaired in the SOD1-deleted strain but not in the mitochondrial SOD2-deleted strain. This indicates that, at least under conditions where respiration is glucose repressed, MCFA toxic action is due to superoxide anion production (42).
Octanoic acid resistance-specific mechanism.
The Pdr12p ABC transporter under the control of WAR1 is the main factor of octanoic acid resistance, which is enhanced by the activity of Tpo1p in a cumulative manner as revealed by the drop test (Fig. 6). Moreover, PDR16 deletion resulted in enhanced sensitivity to octanoic acid. The ABC transporter Pdr12p has been described to be the weak organic acid transporter (32) that is able to confer resistance to the C1 to C7 organic acids but not to the longer fatty acids (C8 to C10) (19), although the expression of PDR12 was induced by C3 to C8 acids (18). Our experiments show unambiguously that Pdr12p is indeed involved in octanoic acid resistance. The discrepancy observed between our results and previous ones could be due to the cumulative effects of the two transporters Pdr12p and Tpo1p or to the use of different genetic backgrounds.
Strikingly, the activity of these two transporters is based on two different energetic supplies. Pdr12p function is dependent on ATP consumption, while the activity of Tpo1p is linked to the proton gradient across the membrane. The second mechanism could be especially well adapted to the acidic pH of fermenting medium.
Among the other genes we tested, only PDR16 seemed to be involved in the octanoic acid response. This gene affects the lipid composition of the plasma membrane, limiting the passive uptake of the drug across the membrane (48). Curiously, the PDR16 homologous gene PDR17 has no effect on octanoic acid resistance, nor do other genes described to be involved in cell response to weak organic acids, such as SPI1 (38) or HAA1 (13).
Considering the regulation of the octanoic acid response, our results clearly highlight the essential role of WAR1, probably through the activation of Pdr12p. TPO1 and PDR16 genes are under the control of PDR1, but our phenotypic screening revealed a poor effect of PDR1 deletion on octanoic acid resistance, while the PDR3 deletion was more effective. Moreover, none of the other transcription factors tested had an effect on octanoic acid resistance. Octanoic acid seems to act essentially as a short-chain organic acid and to induce a relatively simple response, including expulsion through Pdr12p and Tpo1p and perhaps membrane adaptation through Pdr16p.
Decanoic acid resistance-specific mechanism.
The yeast response to decanoic acid is much more complex. Besides the key effect of TPO1 deletion, a slight effect of SNQ2 and TPO4 transporter deletion can be observed. It is noteworthy that the deletion of the PDR5 transporter, which often is associated with TPO1 in drug resistance (41, 2), had no impact on decanoic acid resistance. The deletion of the genes coding for the transcription factors PDR1, STB5, MSN2, NRG1, OAF1, and PIP2 resulted in impaired decanoic acid resistance.
PDR1, PDR3, and STB5 are three transcription factors that form homo- or heterodimers (1) and regulate many ABC transporters. The PDR1/PDR3 complex has been shown to be activated by membrane-active compounds (37) and, in this context, to act mainly through the Pdr5 transporter. In our case, neither PDR3 nor PDR5 deletions had any impact on decanoic acid resistance, suggesting another regulation network. The transcription factor Stb5p can form heterodimers with Pdr1p (1) but also is able to act without Pdr1p or Pdr3p to regulate the pentose phosphate pathway and NADPH production as an answer to oxidative agents (25). It is noteworthy that one of the Stb5p targets is the SNQ2 transporter, which has a (slight) effect on decanoic acid resistance. These results reinforce the idea of MCFA impairing the oxidative state of the cell, which is evidenced by the activation of PRX1 (also obtained during the screening of a multicopy expression library) as well as GPX2 (a phospholipid hydroperoxide glutathione peroxidase) that protects cells from phospholipid hydroperoxide during oxidative stress.
The MSN2/MSN4 complex is the mediator of the general stress response that is induced by different environmental changes. Moreover, Msn2p nucleus translocation and the activation of STRE genes also can be obtained by a range of membrane-disturbing agents (29). In our experiments, MSN2 deletion has more effect on decanoic acid resistance than MSN4 deletion, reinforcing the hypothesis of decanoic acid acting through membrane perturbation as well as internal medium acidification.
Finally, OAF1 or PIP2 deletion also resulted in impaired decanoic acid resistance. These transcription factors act as dimers to positively regulate genes encoding peroxisomal proteins in response to oleate induction in glucose-free medium. The induction of genes involved in fatty acid metabolism, such as EEB1, FAA1, and ELO1 (8.3, 1.7, and 2.4 times increase, respectively), is in agreement with the beta oxidation of fatty acids taking place in the peroxisome. One of the targets of these transcription factors is the gene EEB1 (22), which is the most induced by decanoic acid in our study. The high activation of EEB1 after exposure to decanoic acid as well as the slight (35%) but significant induction of YMR210w, coding for a putative acyltransferase with similarity to EEB1, suggests that ethyl ester synthesis plays a complementary role in the detoxification of culture media. Indeed, the deletion of EEB1 only or EEB1 and YMR210w in a laboratory strain causes 45 and 80% decrease in the production of ethyl ester during alcoholic fermentation, respectively (34). However, the same team (35) also found that the addition of octanoic acid during alcoholic fermentation resulted in an increase of EEB1 expression but not the addition of decanoic acid. The low sensitivity of defective EEB1 strains after MCFA exposure suggests that this metabolic route participates moderately in global resistance. However, we observed that during alcoholic fermentation strains possessing an inactivated copy of EEB1 presented a higher lag phase when exposed to decanoic acid (unpublished data), suggesting a more-significant role during alcoholic fermentation.
Activation mechanism of fatty acid resistance.
Fatty acids from C12 were shown to be able to trigger the transcriptional signal (47) through the direct activation of Oaf1p in a ligand-dependent manner (31). In a similar manner, various hydrophobic inhibitors are able to bind to a discrete xenobiotic-binding domain of Pdr1p and Pdr3p (46). In analogy with these recent results, we can hypothesize that these MCFA activate Pdr1p and Oaf1p. This activation of Oaf1p/Pip2p and Pdr1p/Stb5p is mediated by the complex Gal11p/MED15, and we also observed that the deletion of GAL11 results in a hypersensitivity of the Δgal11 mutant strain.
The mechanism activating the weak acid response is not understood. Gregori et al. (16) hypothesized the direct activation of War1p by the acid, but this has not been observed until now.
In conclusion, we have shown that the resistance to octanoic and decanoic acids, two potent fermentation inhibitors, results from the activation of three different mechanisms. As for many drugs, the expulsion of these two acids by the two transporters Tpo1p and Pdr12p represents the main part of the resistance. The adaptation to these acids also involves an oxidative stress response similar to what has been observed for other acids (i.e., sorbic acid) or detergents (SDS), but the activation of beta-oxidation can explain a part of this oxidative stress. However, contrary to former observations, we also could observe, unexpectedly for decanoic acid (and not octanoic acid), the activation of the genes involved in ethyl ester synthesis, which are key odorant compounds of wine aroma.
Supplementary Material
Acknowledgments
This work has been supported by a grant from the Alsace Region, France.
We are grateful to F. Devaux and C. Rodrigues-Pousada for kindly providing strains. We thank A. Alais Perot for her helpful technical assistance.
Footnotes
Published ahead of print on 17 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akache, B., S. MacPherson, M. Sylvain, and B. Turcotte. 2004. Complex interplay among regulators of drug resistance genes in Saccharomyces cerevisiae. J. Biol. Chem. 279:27855-27860. [DOI] [PubMed] [Google Scholar]
- 2.Alenquer, M., S. Tenreiro, and I. Sá-Correia. 2006. Adaptive response to the antimalarial drug artesunate in yeast involves Pdr1p/Pdr3p-mediated transcriptional activation of the resistance determinants TPO1 and PDR5. FEMS Yeast Res. 6:1130-1139. [DOI] [PubMed] [Google Scholar]
- 3.Alexandre, H., B. Mathieu, and C. Charpentier. 1996. Alteration in membrane fluidity and lipid composition, and modulation of H+-ATPase activity in Saccharomyces cerevisiae caused by decanoic acid. Microbiology 142:469-475. [DOI] [PubMed] [Google Scholar]
- 4.Alexandre, H., and C. Charpentier. 1998. Biochemical aspects of stuck and sluggish fermentation in grape must. J. Ind. Microbiol. Biotechnol. 20:20-27. [Google Scholar]
- 5.Bardi, L., C. Crivelli, and M. Marzona. 1998. Esterase activity and release of ethyl esters of medium-chain fatty acids by Saccharomyces cerevisiae during anaerobic growth. Can. J. Microbiol. 44:1171-1176. [PubMed] [Google Scholar]
- 6.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289-300. [Google Scholar]
- 7.Cabral, M. G., C. A. Viegas, and I. Sá-Correia. 2001. Mechanisms underlying the acquisition of resistance to octanoic-acid-induced-death following exposure of Saccharomyces cerevisiae to mild stress imposed by octanoic acid or ethanol. Arch. Microbiol. 175:301-307. [DOI] [PubMed] [Google Scholar]
- 8.Carmelo, V., H. Santos, and I. Sá-Correia. 1997. Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1325:63-70. [DOI] [PubMed] [Google Scholar]
- 9.Carreto, L., M. F. Eiriz, A. C. Gomes, P. M. Pereira, D. Schuller, and M. A. Santos. 2008. Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics 9:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavalieri, D., C. Castagnini, S. Toti, K. Maciag, T. Kelder, L. Gambineri, S. Angioli, and P. Dolara. 2007. Eu. Gene Analyzer a tool for integrating gene expression data with pathway databases. Bioinformatics 23:2631-2632. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, B., R. P. Levine, and G. Sherlock. 2005. Microarray karyotyping of commercial wine yeast strains reveals shared, as well as unique, genomic signatures. BMC Genomics 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fardeau, V., G. Lelandais, A. Oldfield, H. Salin, S. Lemoine, M. Garcia, V. Tanty, S. Le Crom, C. Jacq, and F. Devaux. 2007. The central role of PDR1 in the foundation of yeast drug resistance. J. Biol. Chem. 282:5063-5074. [DOI] [PubMed] [Google Scholar]
- 13.Fernandes, A. R., N. P. Mira, R. C. Vargas, I. Canelhas, and I. Sá-Correia. 2005. Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem. Biophys. Res. Commun. 337:95-103. [DOI] [PubMed] [Google Scholar]
- 14.Fujita, K., A. Matsuyama, Y. Kobayashi, and H. Iwahashi. 2004. Comprehensive gene expression analysis of the response to straight-chain alcohols in Saccharomyces cerevisiae using cDNA microarray. J. Appl. Microbiol. 97:57-67. [DOI] [PubMed] [Google Scholar]
- 15.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregori, C., C. Schüller, I. E. Frohner, G. Ammerer, and K. Kuchler. 2008. Weak organic acids trigger conformational changes of the yeast transcription factor War1 in vivo to elicit stress adaptation. J. Biol. Chem. 283:25752-25764. [DOI] [PubMed] [Google Scholar]
- 17.Guth, H. 1997. Quantitation and sensory studies of character impact odorants of different white wine varieties. J. Agric. Food Chem. 45:3027-3032. [Google Scholar]
- 18.Hatzixanthis, K., M. Mollapour, I. Seymour, B. E. Bauer, G. Krapf, C. Schüller, K. Kuchler, and P. W. Piper. 2003. Moderately lipophilic carboxylate compounds are the selective inducers of the Saccharomyces cerevisiae Pdr12p ATP-binding cassette transporter. Yeast 20:575-585. [DOI] [PubMed] [Google Scholar]
- 19.Holyoak, C. D., D. Bracey, P. W. Piper, K. Kuchler, and P. J. Coote. 1999. The Saccharomyces cerevisiae weak-acid-inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy-dependent mechanism. J. Bacteriol. 181:4644-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikner, A., and K. Shiozaki. 2005. Yeast signaling pathways in the oxidative stress response. Mutat. Res. 569:13-27. [DOI] [PubMed] [Google Scholar]
- 21.Kamp, F., W. Guo, R. Souto, P. F. Pilch, B. E. Corkey, and J. A. Hamilton. 2003. Rapid flip-flop of oleic acid across the plasma membrane of adipocytes. J. Biol. Chem. 278:7988-7995. [DOI] [PubMed] [Google Scholar]
- 22.Karpichev, I. V., and G. M. Small. 1998. Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6560-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koerkamp, M. G., M. Rep., H. J. Bussemaker, G. P. Hardy, A. Mul, K. Piekarska, A. Szigyarto, J. M. De Mattos, and H. F. Tabak. 2002. Dissection of transient oxidative stress response in Saccharomyces cerevisiae by using DNA microarrays. Mol. Biol. Cell 13:2783-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kren, A., Y. M. Mamnun, B. E. Bauer, C. Schüller, H. Wolfger, K. Hatzixanthis, M. Mollapour, C. Gregori, P. Piper, and K. Kuchler. 2003. War1p, a novel transcription factor controlling weak acid stress response in yeast. Mol. Cell. Biol. 23:1775-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larochelle, M., S. Drouin, F. Robert, and B. Turcotte. 2006. Oxidative stress-activated zinc cluster protein Stb5 has dual activator/repressor functions required for pentose phosphate pathway regulation and NADPH production. Mol. Cell. Biol. 26:6690-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274:16040-16046. [DOI] [PubMed] [Google Scholar]
- 27.Lucau-Danila, A., G. Lelandais, Z. Kozovska, V. Tanty, T. Delaveau, F. Devaux, and C. Jacq. 2005. Early expression of yeast genes affected by chemical stress. Mol. Cell. Biol. 25:1860-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marks, V. D., S. J. Ho Sui, D. Erasmus, G. K. van Der Merwe, J. Brumm, W. W. Wasserman, J. Bryan, and H. J. van Vuuren. 2008. Dynamics of the yeast transcriptome during wine fermentation reveals a novel fermentation stress response. FEMS Yeast Res. 8:35-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moskvina, E., E. M. Imre, and H. Ruis. 1999. Stress factors acting at the level of the plasma membrane induce transcription via the stress response element (STRE) of the yeast Saccharomyces cerevisiae. Mol. Microbiol. 32:1263-1272. [DOI] [PubMed] [Google Scholar]
- 30.Peddie, H. A. B. 1990. Ester formation in brewery fermentations. J. Inst. Brew. 96:327-331. [Google Scholar]
- 31.Phelps, C., V. Gburcik, E. Suslova, P. Dudek, F. Forafonov, N. Bot, M. MacLean, R. J. Fagan, and D. Picard. 2006. Fungi and animals may share a common ancestor to nuclear receptors. Proc. Natl. Acad. Sci. U. S. A. 103:7077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piper, P., Y. Mahé, S. Thompson, R. Pandjaitan, C. Holyoak, R. Egner, M. Mühlbauer, P. Coote, and K. Kuchler. 1998. The pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 17:4257-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sá-Correia, I., S. C. dos Santos, M. C. Teixeira, T. R. Cabrito, and N. P. Mira. 2009. Drug:H+ antiporters in chemical stress response in yeast. Trends Microbiol. 17:22-31. [DOI] [PubMed] [Google Scholar]
- 34.Saerens, S. M., K. J. Verstrepen, S. D. Van Laere, A. R. Voet, P. Van Dijck, F. R. Delvaux, and J. M. Thevelein. 2006. The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J. Biol. Chem. 281:4446-4456. [DOI] [PubMed] [Google Scholar]
- 35.Saerens, S. M., F. Delvaux, K. J. Verstrepen, P. Van Dijck, J. M. Thevelein, and F. R. Delvaux. 2008. Parameters affecting ethyl ester production by Saccharomyces cerevisiae during fermentation. Appl. Environ. Microbiol. 74:454-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schüller, C., Y. M. Mamnun, M. Mollapour, G. Krapf, M. Schuster, B. E. Bauer, P. W. Piper, and K. Kuchler. 2004. Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol. Biol. Cell 15:706-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schüller, C., Y. M. Mamnun, H. Wolfger, N. Rockwell, J. Thorner, and K. Kuchler. 2007. Membrane-active compounds activate the transcription factors Pdr1 and Pdr3 connecting pleiotropic drug resistance and membrane lipid homeostasis in Saccharomyces cerevisiae. Mol. Biol. Cell 18:4932-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simões, T., N. P. Mira, A. R. Fernandes, and I. Sá-Correia. 2006. The SPI1 gene, encoding a glycosylphosphatidylinositol-anchored cell wall protein, plays a prominent role in the development of yeast resistance to lipophilic weak-acid food preservatives. Appl. Environ. Microbiol. 72:7168-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sirisattha, S., Y. Momose, E. Kitagawa, and H. Iwahashi. 2004. Toxicity of anionic detergents determined by Saccharomyces cerevisiae microarray analysis. Water Res. 38:61-70. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, G. T., and B. H. Kirsop. 1977. The origin of medium chain length fatty acids present in beer. J. Inst. Brew. 83:241-243. [Google Scholar]
- 41.Teixeira, M. C., and I. Sá-Correia. 2002. Saccharomyces cerevisiae resistance to chlorinated phenoxyacetic acid herbicides involves Pdr1p-mediated transcriptional activation of TPO1 and PDR5 genes. Biochem. Biophys. Res. Commun. 292:530-537. [DOI] [PubMed] [Google Scholar]
- 42.Teixeira, M. C., J. P. Telo, N. F. Duarte, and I. Sá-Correia. 2004. The herbicide 2,4-dichlorophenoxyacetic acid induces the generation of free-radicals and associated oxidative stress responses in yeast. Biochem. Biophys. Res. Commun. 324:1101-1107. [DOI] [PubMed] [Google Scholar]
- 43.Teixeira, M. C., P. Monteiro, P. Jain, S. Tenreiro, A. R. Fernandes, N. P. Mira, M. Alenquer, A. T. Freitas, A. L. Oliveira, and I. Sá-Correia. 2006. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Database issue. Nucleic Acids Res. 34:D446-D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teixeira, M. C., A. R. Fernandes, N. P. Mira, J. D. Becker, and I. Sá-Correia. 2006. Early transcriptional response of Saccharomyces cerevisiae to stress imposed by the herbicide 2,4-dichlorophenoxyacetic acid. FEMS Yeast Res. 6:230-248. [DOI] [PubMed] [Google Scholar]
- 45.Tenreiro, S., P. A. Nunes, C. A. Viegas, M. S. Neves, M. C. Teixeira, M. G. Cabral, and I. Sá-Correia. 2002. AQR1 gene (ORF YNL065w) encodes a plasma membrane transporter of the major facilitator superfamily that confers resistance to short-chain monocarboxylic acids and quinidine in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 292:741-748. [DOI] [PubMed] [Google Scholar]
- 46.Thakur, J. K., S. Pan, W. S. Moye-Rowley, H. Arthanari, F. Yang, A. M. Näär, X. Fan, J. Breger, D. P. Frueh, K. Gulshan, D. K. Li, E. Mylonakis, K. Struhl, B. P. Cormack, and G. Wagner. 2008. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452:604-609. [DOI] [PubMed] [Google Scholar]
- 47.Thakur, J. K., H. Arthanari, F. Yang, K. H. Chau, G. Wagner, and A. M. Näär. 2009. Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J. Biol. Chem. 284:4422-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Hazel, H. B., H. Pichler, M. A. do Valle Matta, E. Leitner, A. Goffeau, and G. Daum. 1999. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J. Biol. Chem. 274:1934-1941. [DOI] [PubMed] [Google Scholar]
- 49.Viegas, C. A., M. F. Rosa, I. Sá-Correia, and J. M. Novais. 1989. Inhibition of yeast growth by octanoic and decanoic acids produced during ethanolic fermentation. Appl. Environ. Microbiol. 55:21-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viegas, C. A., and I. Sá-Correia. 1997. Effects of low temperatures (9-33 degrees C) and pH (3.3-5.7) in the loss of Saccharomyces cerevisiae viability by combining lethal concentrations of ethanol with octanoic and decanoic acids. Int. J. Food Microbiol. 34:267-277. [DOI] [PubMed] [Google Scholar]
- 51.Wettenhall, J. M., and G. K. Smyth. 2004. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 20:3705-3706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.