Abstract
Phosphorus was added as a nutrient to bench-scale and pilot-scale biologically active carbon (BAC) reactors operated for perchlorate and nitrate removal from contaminated groundwater. The two bioreactors responded similarly to phosphorus addition in terms of microbial community function (i.e., reactor performance), while drastically different responses in microbial community structure were detected. Improvement in reactor performance with respect to perchlorate and nitrate removal started within a few days after phosphorus addition for both reactors. Microbial community structures were evaluated using molecular techniques targeting 16S rRNA genes. Clone library results showed that the relative abundance of perchlorate-reducing bacteria (PRB) Dechloromonas and Azospira in the bench-scale reactor increased from 15.2% and 0.6% to 54.2% and 11.7% after phosphorus addition, respectively. Real-time quantitative PCR (qPCR) experiments revealed that these increases started within a few days after phosphorus addition. In contrast, after phosphorus addition, the relative abundance of Dechloromonas in the pilot-scale reactor decreased from 7.1 to 0.6%, while Zoogloea increased from 17.9 to 52.0%. The results of this study demonstrated that similar operating conditions for bench-scale and pilot-scale reactors resulted in similar contaminant removal performances, despite dramatically different responses from microbial communities. These findings suggest that it is important to evaluate the microbial community compositions inside bioreactors used for drinking water treatment, as they determine the microbial composition in the effluent and impact downstream treatment requirements for drinking water production. This information could be particularly relevant to drinking water safety, if pathogens or disinfectant-resistant bacteria are detected in the bioreactors.
Biological treatment of drinking water has been limited to date, especially in the United States, despite many obvious advantages, such as the ability of mixed microbial community bioreactors to simultaneously remove multiple contaminants from drinking water with limited additions of chemicals and little or no generation of by-products (6). Bench-scale and a few full-scale studies have demonstrated the effectiveness of biological processes in drinking water treatment in removing natural organic matter (11), taste/odor/color-causing compounds (35, 43), and several inorganic contaminants (3, 27). In some of these processes, preozonation is applied to improve the biodegradability of refractory organic matter (25, 35, 37, 43). In other processes, termed stimulated biological treatment processes in the current study, electron donors are added to support microbial activity for the removal of contaminants that can serve as electron acceptors, such as nitrate (20), perchlorate (7, 30), and other oxidized anions (34).
To allow wider application of biological treatment of drinking water, it will be necessary to address concerns, such as the possibility of microbial contamination of finished drinking water (8) and the generation of soluble microbial products, which may support biological growth in drinking water distribution systems (9). These concerns can be addressed only by expanding efforts to characterize the structures and activities of the microbial communities responsible for biological drinking water treatment and to link this information to the microbiological properties of finished drinking water. A few studies already have characterized microbial communities in drinking water treatment systems by using phospholipid fatty acids (32), culture-based methods (31, 38), and nucleic acid-based methods (18, 33, 41). More work is needed to elucidate how operational conditions, such as nutrient additions, influence microbial community composition and function to help drinking water utilities and regulators evaluate this emerging technology.
Phosphorus is an essential element in cell growth and function (28); thus, phosphorus addition to phosphorus-limited engineered bioreactor systems can increase total biomass density and improve bioreactor performance (36, 46). Furthermore, phosphorus limitations can have profound impacts on microbial behavior at cellular and community levels. For example, phosphorus limitation was found to activate a lethal phenotype in Pseudomonas aeruginosa, while phosphorus addition reversed this activation (56). In addition, studies of natural systems observed changes in the microbial community structure upon phosphorus addition (1, 12), and phosphorus limitation has been demonstrated to decrease the diversity of a microbial community by lowering the intensity of horizontal gene transfer (50). Moreover, nutrient limitations, including phosphorus limitations, promote the production of extracellular polymeric substances (EPS) in biofilms (51), which can have an impact on the effectiveness of backwashing and other operational parameters of fixed-bed biofilm systems (J. Brown and C. Lauderdale, presented at the Water Quality Technology Conference, Seattle, WA, 15 to 19 November 2009). These studies suggest that phosphorus addition to phosphorus-limited biological systems can be used to enhance microbial contaminant removal, other operational characteristics of bioreactor systems, the microbial community structure within such systems, and the microbiological properties of finished drinking water.
In the current study, efforts were made to elucidate how phosphorus addition affects microbial community function (i.e., reactor performance) and microbial community structure in biologically active carbon (BAC) reactors operated for drinking water treatment. Two BAC reactors, one bench scale and one pilot scale, were operated to remove perchlorate and nitrate from contaminated groundwater. Reactor performance and microbial community structure were monitored and compared before and after addition of phosphorus to these BAC reactors. Furthermore, the microbial results were used to suggest how stimulated biological treatment may affect downstream treatment such as disinfection.
MATERIALS AND METHODS
Reactor systems.
The bench-scale BAC reactor consisted of a glass column with an inner diameter of 4.9 cm and a height of 26.0 cm (see Fig. S1 in the supplemental material). Granular activated carbon (GAC) (bituminous F816; Galgon Carbon Corp., Pittsburgh, PA) was packed to a depth of 10.6 cm, resulting in an empty bed volume of 200 cm3. The remaining space in the glass column was reserved for bed expansion during backwashing. The system was seeded with biomass collected from a bench-scale BAC reactor previously operated for perchlorate removal (14) and with biomass collected from a GAC filter, which exhibited biological activity, in a full-scale drinking water treatment plant (Ann Arbor, MI). A synthetic groundwater was pumped into the bench-scale BAC reactor in a downflow mode at a flow rate of 10 ml/min, resulting in an empty bed contact time (EBCT) of 20 min. The synthetic groundwater composition was designed according to the composition determined for a real groundwater (Rialto, CA) and consisted of deionized water supplemented with 17.75 mg/liter Na2SO4, 6.90 mg/liter K2CO3, 289.18 mg/liter NaHCO3, 13.68 mg/liter NaCl, 2.81 mg/liter CaCl2, 3.88 mg/liter MgCl2, 34.27 mg/liter NaNO3, and 92.34 μg/liter NaClO4 (i.e., 75 μg/liter ClO4−). The dissolved oxygen (DO) level of the synthetic groundwater was adjusted to ∼7 mg/liter and maintained by utilizing a floating lid in the influent tank. Based on stoichiometric calculations with an assumed net yield value of 0.4 g biomass per g of chemical oxygen demand of acetate (CODacetate), 13 mg/liter of acetic acid as C would be needed to completely remove all three electron acceptors (i.e., DO, NO3−, and ClO4−). With a safety factor of 1.5, the influent acetic acid concentration was selected to be 20 mg/liter as C. Influent and effluent pH values varied between 7.5 and 7.9. On day 115 of operation, phosphoric acid was added to the synthetic groundwater at a concentration of 145 μg/liter as P, which was estimated through stoichiometric calculations (21). The bench-scale BAC reactor was operated in a temperature-controlled room set at 18°C and backwashed every 48 h with a mixture of water (i.e., 50 ml/min) and air for 4 min followed by high-flow-rate water flush (i.e., 500 ml/min) for 3 min. These operating conditions were designated the baseline operating conditions for the bench-scale BAC reactor.
A pilot-scale fixed-bed BAC reactor was designed and constructed by Carollo Engineers, P.C. (Sarasota, FL), and Intuitech, Inc. (Salt Lake City, UT). Carollo Engineers operated the reactor at well 2 in Rialto, CA (see Fig. S1 in the supplemental material). The pilot-scale BAC reactor consisted of an epoxy-coated steel column with an inner diameter of 61.0 cm and a height of 243.8 cm and contained the same type of GAC as did the bench-scale BAC system. The height of the GAC bed was 152.4 cm (i.e., an empty bed volume of 0.445 m3), and the remaining volume was reserved for bed expansion during backwashing. The anion concentrations in the raw groundwater feed were the same as those in the bench-scale BAC system, except that the perchlorate concentration was 50 μg/liter. The influent DO of the pilot-scale BAC reactor was ∼7 mg/liter due to oxygen diffusion during pumping from a contaminated well and storage in a break tank before entering the reactor. The groundwater was pumped to the reactor at a flow rate of 24.7 liters/min, resulting in an EBCT of 18 min. The acetic acid requirement was determined according to the method described above, except that a safety factor of 1.13 was applied, resulting in an acetic acid addition of 15.1 mg/liter as C. The total organic carbon (TOC) level in the raw groundwater was generally below 2 mg/liter, and the biodegradable dissolved organic carbon (BDOC) concentration averaged 0.3 mg/liter, indicating that the contribution of naturally present organic compounds as electron donor sources was negligible compared to that for the acetic acid addition. On day 97 of operation, phosphoric acid was added to the groundwater with a final concentration of ∼180 μg/liter as P. The reactor was backwashed every 17 to 24 h according to the following procedure: fluidization (with surface wash) at 4.8 gallons per minute (gpm)/square foot of cross-sectional area (i.e., 195.3 liters per min per m2 of cross-sectional area [liters/min/m2]) for 69 s, 12.7 gpm/ft2 (i.e., 516.8 liters/min/m2) for 180 s, 3.2 gpm/ft2 (i.e., 130.2 liters/min/m2) for 120 s, 6.7 gpm/ft2 (i.e., 272.6 liters/min/m2) for 480 s, and 1.3 gpm/ft2 (i.e., 52.9 liters/min/m2) for 30 s and air scouring for a total of 24 s with a flow rate of 2 to 3.2 ft3 per min per ft2 of cross-sectional area (i.e., 610 to 980 liters/min/m2) during the entire fluidization step.
Chemical measurements.
In the bench-scale system, influent and effluent DO concentrations were measured using WTW multi340 m with CellOx325 sensors in WTW D201 flow cells (Weilheim, Germany) connected to the inlet and outlet of the reactor. For the pilot-scale system, DO concentrations were measured on-site using HACH sc100 LDO probes (Loveland, CO). Water samples from the pilot-scale system were shipped regularly to the University of Michigan by overnight service and stored according to standard methods until measurement (22). Water samples were filtered through 0.45-μm filters before measurement. Concentrations of acetic acid, nitrate, and nitrite were measured on an ion chromatography system with a conductivity detector (Dionex DX100; Sunnyvale, CA) according to standard methods (22). An AS-14 analytical column and an AG-14 guard column (Dionex, Sunnyvale, CA) were used with an eluent containing a mixture of 1.0 mM bicarbonate and 3.5 mM carbonate. Perchlorate concentrations were measured using another ion chromatography system (Agilent Chemistation; Santa Clara, CA) according to Environmental Protection Agency (EPA) standard method 314.0 (24). A Dionex AS-16 analytical column and an AG-16 guard column were used with an eluent containing 65 mM NaOH. The detection limits for DO, nitrate, acetic acid, and perchlorate were determined to be 0.01 mg/liter, 0.2 mg/liter, 0.2 mg/liter, and 2 μg/liter, respectively. Phosphorus concentrations were measured using an induced coupled plasma mass spectrometer (PerkinElmer ALEN DRC-e; Waltham, MA) with a detection limit of 5 μg/liter.
Clone library and microbial sequence analyses.
Composite BAC samples from the bench-scale reactor were collected after the reactor content was well mixed. Composite BAC samples from the pilot-scale reactor were collected by mixing the BAC content of a vertical core collected using a polyvinyl chloride (PVC) pipe, which was shipped overnight on dry ice to the University of Michigan. These biomass samples were stored at −80°C until analyses. Four BAC samples were collected for clone library analyses, two from the bench-scale BAC reactor (days 100 and 244) and two from the pilot-scale BAC reactor (days 84 and 210). DNA was extracted from the BAC samples using the FastDNA Spin kit (Qbiogene Inc., Irvine, CA) and quantified using a NanoDrop ND1000 spectrophotometer (NanoDrop Technology, Wilmington, DE). DNA quality was evaluated by electrophoresis on a 0.8% agarose gel. 16S rRNA genes of the bacterial domain were amplified in triplicate using the PCR on a Mastercycler thermocycler (Eppendorf International, Hamburg, Germany) with the forward primer 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) (17) and the reverse primer 1387R (5′-GGGCGG[A/T]GTGTACAAGGC-3′) (54). The composition of the PCR mixture was adopted from the work of Wobus et al. (54). Pooled PCR products were purified by electrophoresis on a 1% agarose gel and extracted using the MinElute gel extraction kit (Qiagen Inc., Valencia, CA). Purified PCR products were cloned using the pCR4-Topo cloning kit (Invitrogen Inc., Carlsbad, CA). Ninety-six-well microplates inoculated with randomly picked colonies were sent to the Genomic Center at Washington University (St. Louis, MO) for sequencing.
A total of 768 clones (two microplates for each library) were sequenced bidirectionally by using vector primers T3 and T7. Nucleotide sequences were analyzed and edited using BioEdit (23). Alignment of closely related sequences (i.e., Dechloromonas- and Azospira-like strains from all clone libraries) identified via the Ribosomal Database Project (RDP) (16) was conducted using ClustalW (13) for bacterial 16S rRNA genes. The genus-level changes between two clone libraries were evaluated using the Library Comparison function on RDP (16). Sequences identified as chimeras by Mallard (4) were excluded from further analyses. A phylogenetic tree was constructed using a 524-bp region in the 16S rRNA gene starting at the 8F primer region. Partial, instead of complete, 16S rRNA gene sequences were used for all phylogenetic analyses because the bidirectional sequence reads were shorter than anticipated and did not overlap. The tree was constructed using the software program MEGA (53). Clones sharing ≥95% identity in their 16S rRNA genes were considered one operational taxonomic unit (OTU) (19). OTUs, diversity statistics, and rarefaction curves were determined using DOTUR (47). The similarities of the bacterial 16S rRNA gene clone libraries were evaluated using ∫-LIBSHUFF (48). Sequences included in the phylogenetic tree are available at NCBI under accession numbers FJ525445 to FJ525546.
qPCR.
At every backwash event after phosphorus addition (day 115) for 20 days, BAC samples from the bench-scale reactor were collected in the same manner as were the BAC samples collected for the clone library analyses. Two real-time quantitative PCR (qPCR) primer sets (Table 1 ) were designed based on representative sequences of relevant clones by using the Primer3 program made available by Integrated DNA Technologies (44) and were synthesized by Invitrogen, Inc. (Carlsbad, CA). The primers were named according to published protocols (2, 5). The specificities of the designed primer sets were evaluated using the Probe Match function of RDP (16), while the coverage of the designed primer sets was evaluated against the clones of interest in relevant clone libraries by using the program OligoReport (http://www.cf.ac.uk) (Table 1). The annealing temperatures were optimized using the gradient function of a real-time PCR Mastercycler Realplex thermocycler (Eppendorf International, Hamburg, Germany) to distinguish between target and nontarget sequences (see Fig. S2 in the supplemental material). Target and nontarget templates used in this characterization were plasmid DNA extracted from relevant clones by using the QIAprep Miniprep kit (Qiagen, Inc., Valencia, CA). Plasmid concentrations were estimated after DNA quantification using a NanoDrop ND1000 spectrophotometer (42). The target templates contained the representative sequences based on which the primer sets were designed, while the nontarget templates contained sequences that were identified within the relevant clone libraries as containing the lowest number of mismatches with the designed primer sets (see Fig. S2). The chosen annealing temperatures for the two primer sets designed in this study (Table 1) could differentiate the fluorescence signals between equal amounts of target and nontarget templates (i.e., 106 copies/μl) by at least 15 cycle threshold units (see Fig. S2). The bacterial primer set (52) is reported in Table 1.
TABLE 1.
Sequences, coverage, specificity, and annealing temperatures of the qPCR primer sets used in this study
| Target (16S rRNA gene) | F or Ra | Name | Abbreviated name | Sequence (5′ to 3′) | Coverage in the clone library for the bench-scale BAC at dayb: |
Specificityc | Annealing temp (°C) | Source or reference | |
|---|---|---|---|---|---|---|---|---|---|
| 100 | 244 | ||||||||
| Dechloromonas | F | S-G-Dchm-0146-a-S-24 | Dchm0146F | TATCGGAACGTACCTTTCAGTGGG | 24/25 | 67/67 | 184/351 | 67.0 | This study |
| R | S-G-Dchm-0248-a-A-24 | Dchm0248R | GCTAATCTGATATCGGCCGCTCAA | 25/25 | 66/67 | 227/426 | This study | ||
| Azospira | F | S-G-Azsp-1009-a-S-24 | Azsp1009F | TACCCTTGACATGCCAGGAACTTT | 1/1 | 10/13 | 45/185 | 68.0 | This study |
| R | S-G-Azsp-1163-a-A-24 | Azsp1163R | CGGCAGTCTCATTAAAGTGCCCAA | 1/1 | 10/13 | 45/1046 | This study | ||
| Bacteria | F | S-D-Bact-1369-a-S-18 | Bact1369F | CGGTGAATACGTTCYCGG | 138/163 | 101/129 | —e | 56.0 | 48 |
| R | S-D-Bact-1492-a-A-19 | Bact1492R | GGWTACCTTGTTACGACTT | NAd | NA | — | 48 | ||
F, forward; R, reverse.
Number of target clones with perfect match to the corresponding primer/number of target clones in the clone library. The denominator for the Azospira primer set (i.e., 13) is different from the number calculated from Table 2 (i.e., 14), because Table 2 was based on the early half of the 16S rRNA gene sequence, whereas the Azospira primer set is targeting the late half of the 16S rRNA gene sequence. The denominator for the Dechloromonas primer set (i.e., 67) is slightly different from the number calculated from Table 2 (i.e., 65).
Number of target sequences in database with perfect match to the corresponding primer/number of sequences in database with perfect match to the corresponding primer. The nontarget sequences with perfect matches to the corresponding primer are not detected in the clone library results. Therefore, little interference is expected for the qPCR results. The numbers of sequences were obtained from the Probe Match function in RDP in August 2008.
NA, not available. The PCR primers used to construct the clone libraries were 8F and 1387R, which do not cover the region around 1492. Therefore, the coverage of the 1492R primer in the clone libraries is not available.
—, for the bacterial primer set, specificity was not measured.
A Mastercycler Realplex thermocycler was used for qPCR with the RealMasterMix SYBR green kit (Eppendorf International, Hamburg, Germany). The reaction mixtures in a 25-μl final volume contained 11.25 μl of 2.5× RealMasterMix SYBR green solution (including 0.05 U/μl HotMaster Taq DNA polymerase, 10 mM magnesium acetate, 1.0 mM deoxynucleoside triphosphates [dNTPs], and 2.5× SYBR green solution), 150 nM forward and reverse primers, pure water (Sigma-Aldrich, St. Louis, MO), and DNA templates of known concentrations or 10 ng of DNA template from environmental samples.
As suggested in the manual of the RealMasterMix SYBR green kit, amplification involved one cycle of 95°C for 10 min for initial denaturation and then 40 cycles of denaturation at 95°C for 15 s followed by annealing at the temperatures presented in Table 1 for 20 s and extension at 68°C for 30 s. Melting profiles were collected after 40 cycles of amplification to check the specificity of the amplification (see Fig. S3 in the supplemental material). Purified Escherichia coli plasmid DNAs containing the 16S rRNA genes of Dechloromonas and Azospira were used to construct standard curves for primer sets Dchm0146F/Dchm0248R and Azsp1009F/Azsp1163R, respectively (see Fig. S4). Plasmid DNA containing the 16S rRNA genes of Dechloromonas from another study (X. Li, W. Yuen, E. Morgenroth, and L. Raskin, submitted for publication) was used to construct the standard curves for Bact1369F/Bact1492R.
RESULTS
Changes in microbial community function (reactor performance).
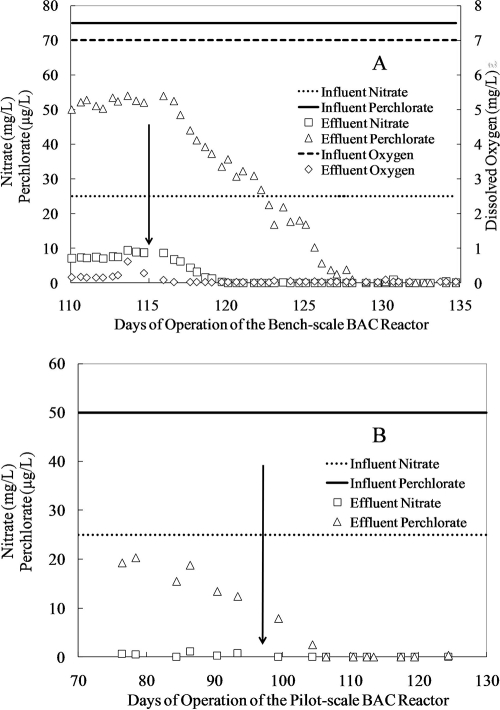
Because the bench-scale BAC reactor was designed for treatment of drinking water sources, the number of chemicals added to the simulated groundwater was minimized. Initially, only acetic acid was added to the synthetic groundwater, and during this initial operational period about 5.5 mg/liter of acetic acid was consumed. The bench-scale BAC reactor removed nearly all the DO, 17 mg/liter of nitrate (68% removal), and 25 μg/liter of perchlorate (33% removal) (Fig. 1A). The background phosphorus concentration in the synthetic groundwater was between 5 and 10 μg/liter as P. After the start of phosphorus addition on day 115 (145 μg/liter phosphoric acid as P), the effluent DO dropped below the detection limit within 2 days, and effluent nitrate and perchlorate started to decrease and fell below the detection limits within 5 and 15 days, respectively. Fifteen days after phosphorus addition, the amount of acetic acid consumed was 12.4 mg/liter.
FIG. 1.
Reactor performance of the bench-scale BAC reactor (A) and the pilot-scale BAC reactor (B). Arrows point to the dates when phosphorus started to be added to the BAC reactors. The influent concentrations are average values from routine measurements.
Similarly, before day 97 in the operation of the pilot-scale BAC reactor, acetic acid was the only chemical added to the contaminated groundwater. During this period, the phosphorus concentration in the groundwater ranged between 30 and 60 μg/liter as P. The reactor was able to completely remove DO (data not shown), nearly completely remove nitrate, and remove about 35 μg/liter perchlorate (∼70% removal) (Fig. 1B). The slightly decreasing trend in effluent perchlorate concentrations during the week prior to phosphorus addition (Fig. 1B) was believed to be due to operational variations but did not represent a real trend when data from the earlier operational period were evaluated (data not shown). After the start of phosphorus addition on day 97 (∼180 μg/liter P), the effluent nitrate concentration dropped below the detection limit within 2 days, and the effluent perchlorate concentrations fell below the detection limit within about 16 days.
Changes in the relative abundance of microbial populations.
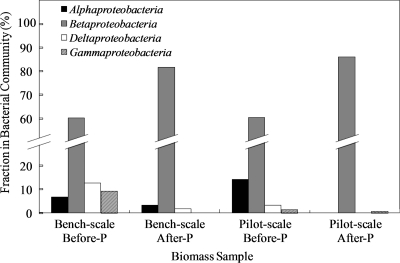
The class Betaproteobacteria was the most abundant microbial group in the bench-scale BAC reactor both before (day 100, 70.3%) and after (day 244, 91.7%) the start of phosphorus addition on day 115 (Table 2 and Fig. 2). In contrast, significant shifts in microbial community structure occurred at the genus level after phosphorus addition. The clone library results in Table 2 show that, after phosphorus addition, the relative abundance of Dechloromonas- and Azospira-like strains increased from 15.2% and 0.6% to 54.2% and 11.7%, respectively. Because most members of these two genera are perchlorate-reducing bacteria (PRB) and can use oxygen, nitrate, and perchlorate as electron acceptors (15), it is likely that phosphorus addition enhanced nitrate and perchlorate removal (Fig. 1A) by promoting and maintaining high numbers of Dechloromonas and Azospira bacteria. The third largest group identified at the genus level was Zoogloea. Zoogloea-like strains made up 7.3% and 7.5% of the total bacterial population before and after phosphorus addition, respectively. The changes in Dechloromonas and Azospira were considered significant (P < 0.01), whereas the change in Zoogloea was not significant (P = 0.54).
TABLE 2.
Phylogenetic affiliations and estimated relative abundances of the clones in the four clone libraries
| Phylogenetic affiliation | % estimated relative abundance in BAC reactor |
|||
|---|---|---|---|---|
| Bench scale |
Pilot scale |
|||
| Before P addition (day 100) (total no. of clones = 165) | After P addition (day 244) (total no. of clones = 120) | Before P addition (day 84) (total no. of clones = 156) | After P addition (day 210) (total no. of clones = 177) | |
| Alphaproteobacteria | 6.7 | 3.3 | 14.1 | —a |
| Betaproteobacteria | 70.3 | 91.7 | 70.5 | 96.0 |
| Dechloromonas | 15.2 | 54.2 | 7.1 | 0.6 |
| Azospira | 0.6 | 11.7 | — | — |
| Zoogloea | 7.3 | 7.5 | 17.9 | 52.0 |
| Propionivibrio | — | — | — | 1.1 |
| Ferribacterium | 2.4 | 5.0 | 7.1 | 2.8 |
| Unclassified Rhodocyclaceae | 26.1 | 10.0 | 3.2 | 0.6 |
| Unclassified Oxalobacteraceae | 6.1 | — | — | — |
| Acidovorax | 7.9 | — | — | — |
| Hydrogenophaga | 1.2 | — | 0.6 | 2.3 |
| Simplicispira | — | — | — | 0.6 |
| Curvibacter | — | 1.7 | — | — |
| Unclassified Comamonadaceae | 1.2 | — | 5.1 | 25.4 |
| Aquabacterium | — | — | 9.6 | — |
| Pelomonas | — | 0.8 | — | — |
| Ideonella | — | — | 1.9 | — |
| Unclassified incertae sedis 5 | — | — | 15.4 | 2.3 |
| Unclassified Burkholderiales | 1.2 | — | 1.3 | 3.4 |
| Unclassified Betaproteobacteria | 1.2 | 0.8 | 1.3 | 5.1 |
| Deltaproteobacteria | 12.7 | 1.7 | 3.2 | — |
| Gammaproteobacteria | 9.1 | — | 1.3 | 0.6 |
| Unclassified Proteobacteria | — | — | 3.2 | 0.6 |
| Acidobacteria | — | 0.8 | 1.3 | — |
| Bacteroidetes | — | — | 1.9 | 1.1 |
| Chloroflexi | — | — | 0.6 | — |
| Firmicutes | — | — | 1.3 | 0.6 |
| Spirochaetes | 0.6 | — | — | — |
| Unclassified Bacteria | 0.6 | 2.5 | 2.6 | 1.1 |
—, not detected.
FIG. 2.
Fractions of the four classes of Proteobacteria in the bench- and pilot-scale BAC reactors before and after phosphorus addition.
The class Betaproteobacteria was also dominant in the pilot-scale BAC reactor (Table 2 and Fig. 2). Eight bacterial phyla/classes were identified before phosphorus addition, while only four remained after phosphorus addition, indicating a decrease in microbial richness at the phylum/class level (Table 2). The clone library results showed that Dechloromonas-like strains were the only known PRB in the pilot-scale BAC reactor, and the relative abundance of the Dechloromonas-like strains decreased from 7.1% to 0.6% after phosphorus addition. Differently from the bench-scale BAC reactor, no Azospira-like strains were detected in the pilot-scale BAC reactor. Opposite the trend for the Dechloromonas-like strains, the relative abundance of the Zoogloea-like strains increased from 17.9% to 52.0% after phosphorus addition. Both changes were statistically significant (P < 0.01).
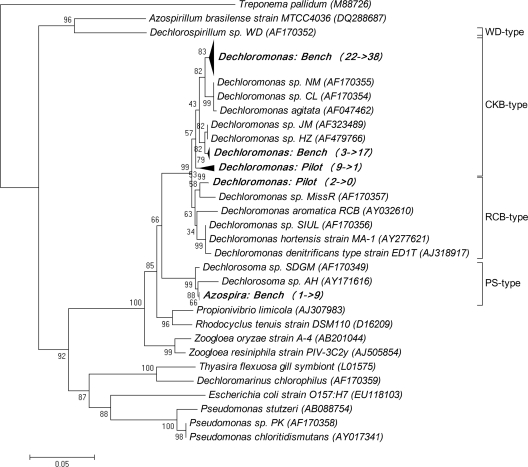
The Dechloromonas phylotypes were different in the two BAC reactors. All Dechloromonas-like strains detected in the bench-scale BAC reactor (i.e., before and after the start of phosphorus addition) belonged to the strain CKB type (Fig. 3 ) (15). In contrast, the Dechloromonas-like strains in the pilot-scale BAC reactor belonged to strain CKB and RCB types.
FIG. 3.
Phylogenetic tree of perchlorate-reducing bacteria in the two BAC reactors before and after phosphorus addition. The two numbers in parentheses represent the numbers of the clones before and after phosphorus addition. The numbers in the tree refer to percentage bootstrap values based on 1,000 replications. The bar represents 5% sequence divergence.
Changes in overall microbial community structure.
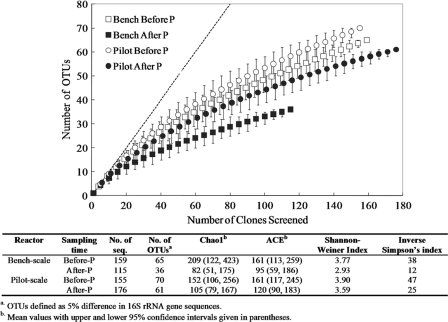
Phosphorus addition changed not only the relative abundance of specific bacterial populations but also the overall microbial community structure in the two BAC reactors. The Chao1 and abundance-based coverage estimate (ACE) richness indices for the bench-scale BAC reactor dropped from 209 and 161 to 82 and 95, respectively (Fig. 4). Although not statistically significant at the 95% confidence interval, the values suggest a decreasing trend in microbial richness after the start of phosphorus addition. The Shannon-Weiner and inverse Simpson indices also decreased from 3.77 and 38 to 2.93 and 12, respectively, indicating that the microbial diversity (including both richness and evenness) had decreased after phosphorus addition. Similar trends were observed for the pilot-scale BAC reactor, although the changes were less profound (Fig. 4). The rarefaction curves in Fig. 4 show that, for the level of sampling effort used, the microbial richnesses were similar in the bench- and the pilot-scale BAC reactors before phosphorus addition and had decreased in both reactors after the start of phosphorus addition. None of the four rarefaction curves leveled off completely, suggesting that further sequencing would have resulted in more OTUs. In addition, the changes of specific microbial populations reported in Table 2 were statistically confirmed by the results using ∫-LIBSHUFF (data not shown). All the comparisons had P values of less than 0.05, indicating that phosphorus addition caused significant microbial community shifts in both reactors.
FIG. 4.
Rarefaction curves indicating bacterial 16S rRNA gene richness within clone libraries from the bench- and the pilot-scale BAC reactors before and after phosphorus addition. The dashed line represents 1:1, indicating infinite diversity. The table lists the bacterial 16S rRNA gene sequence diversity indices. OTUs were defined as groups of sequences sharing 95% 16S rRNA gene sequence identity. The estimates of phylotype richness were calculated according to the abundance-based coverage estimate (ACE) and the bias-corrected Chao1 estimator. The Shannon-Weiner diversity index and the inverse Simpson diversity index, which take into account species richness and evenness, were also calculated.
Microbial population dynamics in the bench-scale BAC reactor.
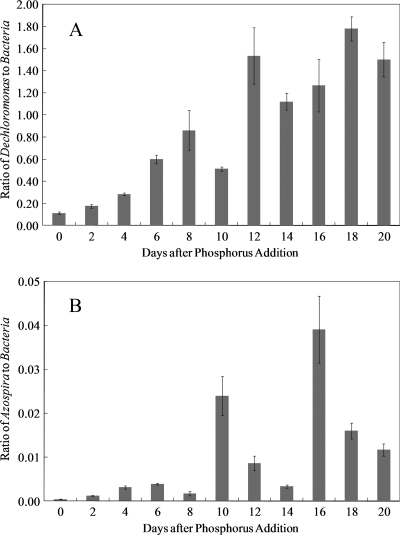
qPCR measurements showed that the relative abundance of Dechloromonas-like strains in the bench-scale BAC reactor started to increase within 2 days after the start of phosphorus addition and continued to increase for about 20 days (Fig. 5A). The relative abundance of Azospira-like strains also started to increase within 2 days after phosphorus addition and followed an increasing trend during the course of the experiment (Fig. 5B). On two occasions, 10 and 16 days after the start of phosphorus addition, the relative abundance of Azospira-like strains was substantially higher than the predominant trend derived from the other data points.
FIG. 5.
Relative abundance of Dechloromonas spp. (A) and Azospira spp. (B) in the bacterial community of the bench-scale BAC reactor after the start of phosphorus addition. The start of phosphorus addition occurred on day 115 of the operation of the bench-scale reactor. The error bars represent the standard deviations from triplicate measurements.
The total amount of biomass in the bench-scale BAC reactor increased after phosphorus addition. With the assumption of a net yield of 0.4 g biomass/g CODacetate, the total biomass produced inside the reactor during one backwash cycle (i.e., 48 h) was calculated to be 169.9 mg before phosphorus addition (average from days 72 to 115) and 380.4 mg after phosphorus addition (average from days 130 to 156). When phosphorus was provided, the measured amount of acetic acid consumed in the reactor was close to the value obtained through the stoichiometric calculation with the assumption of a net yield of 0.4 g biomass/g CODacetate (the measured average concentration of acetate consumed was 12.4 mg/liter versus the calculated concentration of 12.9 mg/liter).
DISCUSSION
The PRB that have been isolated so far are phylogenetically diverse and include species within the Alpha-, Beta-, and Epsilonproteobacteria (15). In this study, two genera of PRB within the Betaproteobacteria, Dechloromonas and Azospira, were detected in the bench-scale BAC reactor, while only Dechloromonas was detected in the pilot-scale BAC reactor. No other known PRB were found in either one of the BAC reactors. The dominance of these two genera in bioreactors fed acetic acid as the sole electron donor has also been reported in a previous study, in which approximately 23% and <1% of the bacterial domain belonged to the genera Dechloromonas and Azospira, respectively (57).
After the start of phosphorus addition, while the average perchlorate removal efficiency of the pilot-scale BAC reactor increased from ∼70% to 100%, the relative abundance of Dechloromonas-like strains, the only known PRB in the system, decreased from 7.1% to 0.6%. One possible explanation for this unanticipated result is that the pilot-scale BAC reactor did not need to sustain a large population of PRB in its microbial community, given that the fraction of perchlorate in the total concentration of electron acceptors was low (50 μg/liter perchlorate versus 7 mg/liter DO and 25 mg/liter nitrate). If so, then other bacterial populations in the microbial community could have been responsible for removing the competing electron acceptors (i.e., oxygen and nitrate), while the Dechloromonas-like strains, even at a low relative abundance, removed perchlorate completely. Zoogloea-like strains became the most abundant population (52.0%) in the pilot-scale BAC reactor after phosphorus addition. The genus Zoogloea can utilize oxygen and nitrate as electron acceptors (55). Therefore, the increase in Zoogloea abundance may be the result of the outcompetition of other bacteria by the Zoogloea-like strains and the increased role of the Zoogloea-like strains in removing competing electron acceptors in the pilot-scale BAC reactor.
Although the ability of Zoogloea to reduce perchlorate has not been demonstrated, the possibility of this genus being capable of doing so cannot be ruled out. A recent environmental genomic study revealed that chlorite dismutase, a key enzyme in the microbial pathway of perchlorate reduction, exists in a nitrite-oxidizing bacterium, which was never identified as a PRB species previously (29). To date, genomes of Zoogloea spp. have not been sequenced, and so it is not possible to check whether genes related to (per)chlorate reductase and chlorite dismutase genes are present in Zoogloea spp. without further experimentation, which is beyond the scope of the current study. If Zoogloea strains can utilize perchlorate as an electron acceptor, then the elevated relative abundance of Zoogloea-like strains could account for the improved perchlorate removal in the pilot-scale reactor, through a mechanism similar to those for the increases of Dechloromonas spp. and Azospira spp. in the bench-scale BAC reactor after phosphorus addition.
The bench-scale BAC reactor results obtained from the qPCR experiments did not quantitatively agree with those from the clone library experiments, although the same qualitative trend was observed for the two methods. The samples for qPCR were collected long before the sample for the second clone library was collected (Table 2 and Fig. 5), and so it may be difficult to compare the results directly. In addition, discrepancies between quantitative results obtained with two very different molecular methods are not uncommon (49). The relative abundances of Dechloromonas-like strains higher than 100% on days 12 to 20 were likely due to the coverage of the bacterial primer, which did not target a fraction of the clones (Table 1). The relative abundance of Azospira-like strains exhibited variations (i.e., days 10 and 16 after the start of phosphorus addition) from its predominant trend. Since the same DNA extracts were used for qPCR experiments with Dechloromonas and Azospira primer sets, and the trend for the relative abundance of Dechloromonas was smooth, the variations suggest that Azospira resided in the reactor in a less homogenous fashion than did Dechloromonas.
Similar to what was reported for natural systems (10, 45), a decrease in microbial diversity in response to nutrient addition was observed in the two BAC reactors. Furthermore, Gram-positive bacteria (i.e., Firmicutes) were not detected in the bench-scale reactor and were found only at very low levels in the pilot-scale reactor (Table 2). Low levels of Gram-positive bacteria may benefit downstream disinfection, since Gram-positive bacteria are usually more resistant to disinfectants than are Gram-negative bacteria (26). Since few studies have been performed to address disinfection kinetics of mixed microbial communities in biologically treated drinking water (40), further characterization of these effluents is necessary.
In addition to disinfection efficiency, it is important to evaluate the viability and potential for regrowth of bacteria, which originate from the biological treatment step, in distribution systems. Pang and Liu compared bacteria that developed in medium-substrate environments (e.g., a bioreactor supplemented with an electron donor and nutrients) and low-substrate environments (e.g., a BAC reactor fed only groundwater) and found that the latter selected for microorganisms that are better adapted to survive in similar environments (e.g., distribution systems) (39). That study suggested that stimulated biological treatment with properly controlled addition of external electron donor and nutrients may help avoid selecting microorganisms that are adapted to low-substrate and low-nutrient environments. The current study investigated the microbial community of two BAC reactors representing medium-substrate environments. Further studies on microbial communities in low-substrate reactors are needed for comparison. Moreover, electron donor addition in stimulated biological treatment needs to be optimized to minimize substrate residual in finished water.
Supplementary Material
Acknowledgments
We thank the Department of Defense for financial support (ESTCP, ER-0544).
We thank Greg Estevao and Axel Ettori of Carollo Engineers for obtaining data from the pilot-scale BAC reactor.
Footnotes
Published ahead of print on 1 October 2010.
REFERENCES
- 1.Allers, E., L. Gomez-Consarnau, J. Pinhassi, J. M. Gasol, K. Simek, and J. Pernthaler. 2007. Response of Alteromonadaceae and Rhodobacteriaceae to glucose and phosphorus manipulation in marine mesocosms. Environ. Microbiol. 9:2417-2429. [DOI] [PubMed] [Google Scholar]
- 2.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, A., P. Laurent, A. Kihn, M. Prevost, and P. Servais. 2001. Impact of temperature on nitrification in biological activated carbon (BAC) filters used for drinking water treatment. Water Res. 35:2923-2934. [DOI] [PubMed] [Google Scholar]
- 4.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal-RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. 2007. Biological treatments of drinking water. Bridge 37(4):30-36. [Google Scholar]
- 7.Brown, J. C., V. L. Snoeyink, L. Raskin, and R. Lin. 2003. The sensitivity of fixed-bed biological perchlorate removal to changes in operating conditions and water quality characteristics. Water Res. 37:206-214. [DOI] [PubMed] [Google Scholar]
- 8.Camper, A. K., M. W. Lechevallier, S. C. Broadaway, and G. A. McFeters. 1986. Bacteria associated with granular activated carbon particles in drinking water. Appl. Environ. Microbiol. 52:434-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson, K. H., and G. L. Amy. 2000. The importance of soluble microbial products (SMPs) in biological drinking water treatment. Water Res. 34:1386-1396. [Google Scholar]
- 10.Cebron, A., L. Bodrossy, N. Stralis-Pavese, A. C. Singer, I. P. Thompson, J. I. Prosser, and J. C. Murrell. 2007. Nutrient amendments in soil DNA stable isotope probing experiments reduce the observed methanotroph diversity. Appl. Environ. Microbiol. 73:798-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charnock, C., and O. Kjonno. 2000. Assimilable organic carbon and biodegradable dissolved organic carbon in Norwegian raw and drinking waters. Water Res. 34:2629-2642. [Google Scholar]
- 12.Chenier, M. R., D. Beaumier, N. Fortin, R. Roy, B. T. Driscoll, J. R. Lawrence, and C. W. Greer. 2006. Influence of nutrient inputs, hexadecane and temporal variations on denitrification and community composition of river biofilms. Appl. Environ. Microbiol. 72:575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi, Y. C., X. Li, L. Raskin, and E. Morgenroth. 2008. Chemisorption of oxygen onto activated carbon can enhance the stability of biological perchlorate reduction in fixed bed biofilm reactors. Water Res. 42:3425-3434. [DOI] [PubMed] [Google Scholar]
- 15.Coates, J. D., and L. A. Achenbach. 2004. Microbial perchlorate reduction: rocket-fuelled metabolism. Nat. Rev. Microbiol. 2:569-580. [DOI] [PubMed] [Google Scholar]
- 16.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emtiazi, F., T. Schwartz, S. M. Marten, P. Krolla-Sidenstein, and U. Obst. 2004. Investigation of natural biofilms formed during the production of drinking water from surface water embankment filtration. Water Res. 38:1197-1206. [DOI] [PubMed] [Google Scholar]
- 19.Fields, M. W., T. F. Yan, S. K. Rhee, S. L. Carroll, P. M. Jardine, D. B. Watson, C. S. Criddle, and J. Z. Zhou. 2005. Impacts on microbial communities and cultivable isolates from groundwater contaminated with high levels of nitric acid-uranium waste. FEMS Microbiol. Ecol. 53:417-428. [DOI] [PubMed] [Google Scholar]
- 20.Fuchs, W., G. Schatzmayr, and R. Braun. 1997. Nitrate removal from drinking water using a membrane-fixed biofilm reactor. Appl. Microbiol. Biotechnol. 48:267-274. [DOI] [PubMed] [Google Scholar]
- 21.Grady, C. P. L., G. Daigger, and H. Lim. 1999. Biological wastewater treatment, 2nd ed. Marcel Dekker Inc., New York, NY.
- 22.Greenberg, A. E., L. S. Clesceri, and A. D. Eaton (ed.). 1992. Standard methods for the examination of water and wastewater, 18th ed. American Public Health Association, Washington, DC.
- 23.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 24.Hautman, D. P., D. J. Munch, A. D. Eaton, and A. W. Haghani. 1999. Method 314.0. Determination of perchlorate in drinking water using ion chromatography. National Exposure Research Laboratory, Office of Research and Development, U.S. Environmental Protection Agency, Cincinnati, OH.
- 25.Kim, W. H., W. Nishijima, E. Shoto, and M. Okada. 1997. Pilot plant study on ozonation and biological activated carbon process for drinking water treatment. Water Sci. Technol. 35:21-28. [Google Scholar]
- 26.Lechevallier, M. W., R. J. Seidler, and T. M. Evans. 1980. Enumeration and characterization of standard plate count bacteria in chlorinated and raw water supplies. Appl. Environ. Microbiol. 40:922-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, X. Y., and H. P. Chu. 2003. Membrane bioreactor for the drinking water treatment of polluted surface water supplies. Water Res. 37:4781-4791. [DOI] [PubMed] [Google Scholar]
- 28.Madigan, M. T., J. M. Martinko, and J. Parker. 2003. Brock biology of microorganisms, 10th ed. Pearson Education, Upper Saddle River, NJ.
- 29.Maixner, F., M. Wagner, S. Lucker, E. Pelletier, S. Schmitz-Esser, K. Hace, E. Spieck, R. Konrat, D. Le Paslier, and H. Daims. 2008. Environmental genomics reveals a functional chlorite dismutase in the nitrite-oxidizing bacterium ‘Candidatus Nitrospira defluvii’. Environ. Microbiol. 10:3043-3056. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. P., and B. E. Logan. 2000. Sustained perchlorate degradation in an autotrophic, gas-phase, packed-bed bioreactor. Environ. Sci. Technol. 34:3018-3022. [Google Scholar]
- 31.Moll, D. M., R. S. Summers, and A. Breen. 1998. Microbial characterization of biological filters used for drinking water treatment. Appl. Environ. Microbiol. 64:2755-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moll, D. M., R. S. Summers, A. C. Fonseca, and W. Matheis. 1999. Impact of temperature on drinking water biofilter performance and microbial community structure. Environ. Sci. Technol. 33:2377-2382. [Google Scholar]
- 33.Nerenberg, R., Y. Kawagoshi, and B. E. Rittmann. 2008. Microbial ecology of a perchlorate-reducing, hydrogen-based membrane biofilm reactor. Water Res. 42:1151-1159. [DOI] [PubMed] [Google Scholar]
- 34.Nerenberg, R., and B. E. Rittmann. 2004. Hydrogen-based, hollow-fiber membrane biofilm reactor for reduction of perchlorate and other oxidized contaminants. Water Sci. Technol. 49(11-12):223-230. [PubMed] [Google Scholar]
- 35.Nerenberg, R., B. E. Rittmann, and W. J. Soucie. 2000. Ozone/biofiltration for removing MIB and geosmin. J. Am. Water Works Assoc. 92:85-95. [Google Scholar]
- 36.Nishijima, W., E. Shoto, and M. Okada. 1997. Improvement of biodegradation of organic substance by addition of phosphorus in biological activated carbon. Water Sci. Technol. 36:251-257. [Google Scholar]
- 37.Nishijima, W., and G. E. Speitel. 2004. Fate of biodegradable dissolved organic carbon produced by ozonation on biological activated carbon. Chemosphere 56:113-119. [DOI] [PubMed] [Google Scholar]
- 38.Norton, C. D., and M. W. LeChevallier. 2000. A pilot study of bacteriological population changes through potable water treatment and distribution. Appl. Environ. Microbiol. 66:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pang, C. M., and W. T. Liu. 2006. Biological filtration limits carbon availability and affects downstream biofilm formation and community structure. Appl. Environ. Microbiol. 72:5702-5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pernitsky, D. J., G. R. Finch, and P. M. Huck. 1995. Disinfection kinetics of heterotrophic plate-count bacteria in biologically treated potable water. Water Res. 29:1235-1241. [Google Scholar]
- 41.Poitelon, J. B., M. Joyeux, B. Welte, J. P. Duguet, E. Prestel, O. Lespinet, and M. S. Dubow. 2009. Assessment of phylogenetic diversity of bacterial microflora in drinking water using serial analysis of ribosomal sequence tags. Water Res. 43:4197-4206. [DOI] [PubMed] [Google Scholar]
- 42.Ritalahti, K. M., B. K. Amos, Y. Sung, Q. Z. Wu, S. S. Koenigsberg, and F. E. Loffler. 2006. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72:2765-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rittmann, B. E., D. Stilwell, J. C. Garside, G. L. Amy, C. Spangenberg, A. Kalinsky, and E. Akiyoshi. 2002. Treatment of a colored groundwater by ozone-biofiltration: pilot studies and modeling interpretation. Water Res. 36:3387-3397. [DOI] [PubMed] [Google Scholar]
- 44.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 45.Salcher, M. M., J. Hofer, K. Hornak, J. Jezbera, B. Sonntag, J. Vrba, K. Simek, and T. Posch. 2007. Modulation of microbial predator-prey dynamics by phosphorus availability: growth patterns and survival strategies of bacterial phylogenetic clades. FEMS Microbiol. Ecol. 60:40-50. [DOI] [PubMed] [Google Scholar]
- 46.Sang, J. Q., X. H. Zhang, L. Z. Li, and Z. S. Wang. 2003. Improvement of organics removal by bio-ceramic filtration of raw water with addition of phosphorus. Water Res. 37:4711-4718. [DOI] [PubMed] [Google Scholar]
- 47.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Segawa, T., K. Miyamoto, K. Ushida, K. Agata, N. Okada, and S. Kohshima. 2005. Seasonal change in bacterial flora and biomass in mountain snow from the Tateyama Mountains, Japan, analyzed by 16S rRNA gene sequencing and real-time PCR. Appl. Environ. Microbiol. 71:123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Souza, V., L. E. Eguiarte, J. Siefert, and J. J. Elser. 2008. Microbial endemism: does phosphorus limitation enhance speciation? Nat. Rev. Microbiol. 6:559-564. [DOI] [PubMed] [Google Scholar]
- 51.Sutherland, I. W. 2001. Exopolysaccharides in biofilms, flocs and related structures. Water Sci. Technol. 43(6):77-86. [PubMed] [Google Scholar]
- 52.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 54.Wobus, A., C. Bleul, S. Maassen, C. Scheerer, M. Schuppler, E. Jacobs, and I. Roske. 2003. Microbial diversity and functional characterization of sediments from reservoirs of different trophic state. FEMS Microbiol. Ecol. 46:331-347. [DOI] [PubMed] [Google Scholar]
- 55.Xie, C. H., and A. Yokota. 2006. Zoogloea oryzae sp. nov., a nitrogen-fixing bacterium isolated from rice paddy soil, and reclassification of the strain ATCC 19623 as Crabtreella saccharophila gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 56:619-624. [DOI] [PubMed] [Google Scholar]
- 56.Zaborin, A., K. Romanowski, S. Gerdes, C. Holbrook, F. Lepine, J. Long, V. Poroyko, S. P. Diggle, A. Wilke, K. Righetti, I. Morozova, T. Babrowski, D. C. Liu, O. Zaborina, and J. C. Alverdy. 2009. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc. Natl. Acad. Sci. U. S. A. 106:6327-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, H., B. E. Logan, J. M. Regan, L. A. Achenbach, and M. A. Bruns. 2005. Molecular assessment of inoculated and indigenous bacteria in biofilms from a pilot-scale perchlorate-reducing bioreactor. Microb. Ecol. 49:388-398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.