Abstract
In order to introduce specificity for Mycobacterium avium subsp. paratuberculosis prior to a phage amplification assay, various magnetic-separation approaches, involving either antibodies or peptides, were evaluated in terms of the efficiency of capture (expressed as a percentage) of M. avium subsp. paratuberculosis cells and the percentage of nonspecific binding by other Mycobacterium spp. A 50:50 mixture of MyOne Tosylactivated Dynabeads coated with the chemically synthesized M. avium subsp. paratuberculosis-specific peptides biotinylated aMp3 and biotinylated aMptD (i.e., peptide-mediated magnetic separation [PMS]) proved to be the best magnetic-separation approach for achieving 85 to 100% capture of M. avium subsp. paratuberculosis and minimal (<1%) nonspecific recovery of other Mycobacterium spp. (particularly if beads were blocked with 1% skim milk before use) from broth samples containing 103 to 104 CFU/ml. When PMS was coupled with a recently optimized phage amplification assay and used to detect M. avium subsp. paratuberculosis in 50-ml volumes of spiked milk, the mean 50% limit of detection (LOD50) was 14.4 PFU/50 ml of milk (equivalent to 0.3 PFU/ml). This PMS-phage assay represents a novel, rapid method for the detection and enumeration of viable M. avium subsp. paratuberculosis organisms in milk, and potentially other sample matrices, with results available within 48 h.
The prospect of being able to detect viable Mycobacterium avium subsp. paratuberculosis organisms in food or veterinary samples within 48 h using a commercially available phage amplification assay (FASTPlaqueTB assay; Biotec Laboratories Limited, Ipswich, United Kingdom), rather than waiting weeks for conventional culture results, is an exciting recent development (7, 8, 26). However, the mycobacteriophage used in the phage amplification assay has a broader mycobacterial host range than M. avium subsp. paratuberculosis alone (23). Consequently, plaques obtained when naturally infected, rather than artificially spiked, samples are tested may not necessarily emanate from M. avium subsp. paratuberculosis alone if other Mycobacterium spp. are also present in the sample. Some additional selective step prior to phage infection, such as magnetic separation (12), is needed to introduce selectivity for M. avium subsp. paratuberculosis.
Magnetic separation (MS) has become a routine method in food and veterinary microbiology laboratories and is commonly used in combination with culture or molecular methods for the detection and isolation of pathogenic bacteria such as Listeria monocytogenes (13, 31), Salmonella spp. (22, 25), and Escherichia coli O157:H7 in both the food (15) and veterinary (20) clinical sample testing context. Magnetic-separation methods selectively separate the target bacterium from other, nontarget microorganisms and inhibitory sample components while concentrating the target bacterial cells into a smaller volume. Collectively, these properties of magnetic separation enhance the analytical specificity and sensitivity of the subsequent detection method, which can be culture, PCR, microscopy, an antigen detection immunoassay, or a phage assay. The latter is our proposed endpoint detection method. The combination of phage amplification and MS is not a new concept. Immunomagnetic (IMS)-phage assays for Salmonella enterica serovar Enteritidis and Escherichia coli O157:H7 have been described previously (5, 6).
The original IMS approach for M. avium subsp. paratuberculosis, employing a polyclonal anti-M. avium subsp. paratuberculosis antibody, was described by Grant et al. (9). This IMS approach showed good detection specificity for M. avium subsp. paratuberculosis as well as high detection sensitivity, because it was able to recover ≤10 CFU/ml directly from both spiked broth and milk. Its subsequent use in combination with IS900 PCR enhanced the speed of detection of M. avium subsp. paratuberculosis (10), and IMS-PCR was able to detect as few as 103 CFU/50 ml, 1 to 2 log10 units lower than the number detected by IS900 PCR applied directly to milk. However, our experience of using this and another polyclonal-antibody-based IMS method (Pathatrix PM-50 beads; Matrix Microscience, Newmarket, England) in conjunction with culture on Herrold's egg yolk medium for the isolation of M. avium subsp. paratuberculosis from mixed-broth cultures from milk (unpublished data) and from raw-milk cheeses (27) has been that these polyclonal-antibody-based IMS methods lack sufficient specificity for M. avium subsp. paratuberculosis, and that consequently, nontarget bacteria, which bind nonspecifically to the beads, often overgrow this bacterium in culture. With other food-borne pathogens, an appropriate selective culture medium can be employed after IMS to prevent the outgrowth of any nontarget bacteria. Unfortunately, no truly selective culture medium exists for M. avium subsp. paratuberculosis at present, so specificity for this bacterium via magnetic separation must be achieved by optimizing the types of bead and capture ligands used.
A monoclonal-antibody-based IMS method for M. avium subsp. paratuberculosis was reported by Metzger-Boddien et al. (17). Other groups have been attempting to produce monoclonal antibodies for application in IMS (3, 4). However, as an alternative to either polyclonal or monoclonal antibodies for the capture of M. avium subsp. paratuberculosis, new magnetic-separation approaches involving an M. avium subsp. paratuberculosis-specific peptide, aMp3 (30) or aMptD (28), have been described (i.e., peptide-mediated magnetic separation [PMS]). The first peptide (aMp3) was screened from nine recombinant bacteriophages specifically binding M. avium subsp. paratuberculosis that were produced using a commercially available phage-peptide display library (30). The second peptide, aMptD, was identified by biopanning of the M. avium subsp. paratuberculosis-specific ABC transponder operon (mpt) (29). The two chemically synthesized peptides, aMp3 and aMptD, were linked via carbodiimide to paramagnetic beads and were used in peptide-based capture PCR. Both PMS methods were reported to have high selectivity for M. avium subsp. paratuberculosis (i.e., no cross-reaction with other Mycobacterium spp.), and the analytical detection sensitivity, 5 ×102 CFU per ml (28), was comparable to the results previously reported by Grant et al. (10).
As with other pathogenic bacteria that are likely to be present in raw milk, low numbers of viable M. avium subsp. paratuberculosis organisms are expected to be encountered in milk and dairy products (2, 11, 24). For other food-borne pathogens, such as Listeria monocytogenes (31), Salmonella spp. (22), and Escherichia coli O157:H7 (15), magnetic separation is generally applied after an enrichment culture step. This enrichment culture step aims to dilute food components known to be growth/PCR inhibitors, revive stressed or injured cells, and boost the numbers of the target bacterium (18, 21), so that magnetic separation and subsequent detection are likely to be more successful. Unfortunately, a prior enrichment culture step is impractical for M. avium subsp. paratuberculosis, since it would take too long, due to the slow-growing nature of this bacterium; thus, MS really needs to be applied directly to the sample. Consequently, any IMS or PMS method for M. avium subsp. paratuberculosis must achieve close to 100% capture efficiency, with minimal nonspecific binding by other mycobacteria, to limit false-negative or false-positive results. Capture efficiency is a measure of the completeness of capture of the original population of target cells present in the sample. Analytical specificity refers to the ability of an assay to measure one particular organism or substance, rather than others, in a sample (19). Therefore, the objectives of this study were (i) to identify the best magnetic-separation approach for the isolation of M. avium subsp. paratuberculosis from milk, in terms of capture efficiency and the percentage of nonspecific binding, by comparing as many paramagnetic-bead-coating antigen combinations as possible and (ii) to evaluate the potential use of the best magnetic-separation approach in conjunction with the previously optimized phage assay (7) as a novel IMS- or PMS-phage assay for the detection of M. avium subsp. paratuberculosis in milk.
MATERIALS AND METHODS
Mycobacterium spp.
Three strains of M. avium subsp. paratuberculosis, ATCC 19698, NCTC 8578, and 796PSS, were employed in the course of this study. They were grown in a shaker incubator for 4 to 6 weeks at 37°C, to stationary phase, in Middlebrook 7H9 broth containing a 10% oleic acid-albumin-dextrose-catalase (OADC) supplement (both from Becton Dickinson, United Kingdom) and 2 μg/ml mycobactin J (Synbiotics Europe SAS, Lyon, France). Other Mycobacterium spp. included Mycobacterium avium subsp. avium (field isolate provided by the Veterinary Sciences Division, Agri-Food and Biosciences Institute for Northern Ireland, and identified as such by the AccuProbe method), Mycobacterium fortuitum NCTC 10394, Mycobacterium kansasii NCTC 10268, Mycobacterium intracellulare NCTC 10425, Mycobacterium bovis BCG NCTC 5692, and Mycobacterium smegmatis mc2 155 (a gift from Ruth McNerney, London School of Hygiene and Tropical Medicine). These were cultivated in Middlebrook 7H9 broth containing 10% OADC for 3 to 14 days, depending on the Mycobacterium sp.
In-house coating of commercially available paramagnetic beads.
A range of different commercially available uncoated paramagnetic beads were coated with a polyclonal antibody (S624) and/or with nonbiotinylated or biotinylated M. avium subsp. paratuberculosis-specific peptides aMp3 (amino acid sequence, NYVIHDVPRHPA [30]) and aMptD (amino acid sequence, GHNHHHQHHRPQ [28]), essentially as described by the bead manufacturers in the accompanying instructions. The peptides used in this study were chemically synthesized by Cambridge Peptides Limited, Cambridge, United Kingdom. The paramagnetic bead-coating antigen combinations evaluated are summarized in Table 1. Coated beads were resuspended to the original volume and bead concentration in 0.05 M phosphate-buffered saline, pH 7.4 (PBS), and were stored at 4°C until required.
TABLE 1.
In-house-prepared paramagnetic-bead-coating-antigen combinations evaluated
| Paramagnetic beads | Coating antigena |
||||
|---|---|---|---|---|---|
| Polyclonal antibody S624 | aMp3 peptide | aMptD peptide | Biotinylated aMp3 peptide | Biotinylated aMptD peptide | |
| Dynabeads, M280 sheep anti-rabbit IgGb | + | − | − | − | − |
| Magnabind carboxyl derivatized beadsc | − | + | + | − | − |
| Amine-coated magnetic hollow glass microspheresd | + | + | + | − | − |
| Dynabeads, MyOne Carboxylic acidb | + | + | + | − | − |
| Dynabeads, MyOne Tosylactivatedb | + | + | + | + | + |
| Dynabeads, MyOne Streptavidin-T1b | − | − | − | + | + |
| Dynabeads, M280 Streptavidinb | − | − | − | + | + |
+, tested; −, not tested.
From Invitrogen, Life Technologies Corporation.
From Pierce Protein Research Products, Thermo Scientific.
From Microsphere Technology Limited, Adare, County Limerick, Republic of Ireland.
Magnetic separation.
Magnetic separation was carried out either manually or by using the Dynal BeadRetriever (Invitrogen) (so-called automated magnetic separation [AMS]). Manual magnetic separation was performed with 1-ml samples in Eppendorf tubes that were incubated for 30 min at room temperature on a Stuart rotator mixer operating at 8 rpm. In order to concentrate the bead-bacterium complexes, samples were placed in a magnetic separator rack (Dynal MPC-M) for 10 min. The sample supernatant was carefully aspirated off with a sterile Pasteur pipette and was discarded. Beads were washed twice with 1 ml PBS containing 0.05% Tween 20 (PBS-T) and were finally resuspended in 1 ml of Middlebrook 7H9-OADC broth. AMS was carried out with the BeadRetriever instrument (Invitrogen) using a customized program consisting of 30 min of capture with mixing, two washes in 1 ml PBS-T with mixing, and finally, resuspension of the beads in 1 ml Middlebrook 7H9-OADC broth. All protocol steps were carried out in disposable BeadRetriever tube strips (Invitrogen). Irrespective of the magnetic-separation method, 10 μl of in-house-coated or commercially available paramagnetic beads was added to 1 ml Middlebrook 7H9-OADC broth or ultra-heat-treated (UHT) whole milk (purchased from a local supermarket) containing 103 to 105 CFU M. avium subsp. paratuberculosis or another Mycobacterium sp.
Enumeration of M. avium subsp. paratuberculosis organisms before and after magnetic separation.
Samples were serially diluted, as necessary, in Middlebrook 7H9-OADC broth. M. avium subsp. paratuberculosis organisms were enumerated by spread plating 100 μl on Herrold's egg yolk medium containing 2 μg/ml mycobactin J (HEYM), followed by incubation for 4 to 6 weeks at 37°C. Other Mycobacterium spp. were enumerated by spread plating 100 μl on Middlebrook 7H9 agar plates enriched with 10% OADC, followed by incubation for appropriate periods (3 to 14 days), depending on the species. Petri dishes were sealed with DuraSeal laboratory sealing film (Diversified Biotech, Boston, MA) to prevent the desiccation of plates during incubation. Organisms in samples were also enumerated by using the optimized phage assay (as described below) before and after magnetic separation. The colony and plaque counts obtained were expressed as CFU or PFU per milliliter of broth or milk, respectively.
Capture efficiency studies.
In an initial evaluation, the performances of two types of commercially available anti-M. avium subsp. paratuberculosis magnetic beads—Pathatrix-PM50 (coated with a polyclonal antibody; Matrix MicroScience, Newmarket, United Kingdom) and AnDiaTec ParaTub-S (coated with a monoclonal antibody; a gift from Johannes Kehle, AnDiaTec GmbH, Kornwestheim, Germany) beads—and three types of in-house-coated magnetic beads—Dynabeads M280 sheep anti-rabbit IgG beads coated with the polyclonal anti-M. avium subsp. paratuberculosis antibody S624 (the original antibody was produced at Queen's University Belfast in the late 1990s), Pierce MagnaBind carboxyl derivatized beads covalently linked to the M. avium subsp. paratuberculosis-specific peptide aMp3, and Pierce MagnaBind carboxyl derivatized beads covalently linked to aMptD—were compared. Subsequently, the capture efficiencies of five other types of surface-activated magnetic beads (amine-coated magnetic hollow glass beads [Microsphere Technology Limited, Adare, County Limerick, Ireland], Dynabeads MyOne carboxylic acid beads [Invitrogen Dynal AS, Oslo, Norway], Dynabeads MyOne Tosylactivated beads [Invitrogen], Dynabeads MyOne Streptavidin T1 beads [Invitrogen], and Dynabeads Streptavidin M280 beads [Invitrogen]) were evaluated. The beads were coated with a polyclonal antibody or with a peptide or biotinylated peptide as indicated in Table 1. In all evaluations, 10 μl of each type of coated bead was added to 1 ml Middlebrook 7H9-OADC broth containing 103 to 104 CFU M. avium subsp. paratuberculosis NCTC 8578, or one of the other Mycobacterium spp., and manual magnetic separation was performed. Beads were resuspended in 1 ml of Middlebrook 7H9-OADC broth after magnetic separation. Colony counts (CFU per milliliter) for each Mycobacterium sp. before and after magnetic separation were determined, and both the capture efficiency for M. avium subsp. paratuberculosis and the percentage of nonspecific recovery of other Mycobacterium spp. were calculated. Experiments were repeated in duplicate for each bead type and each Mycobacterium sp.
The abilities of four different blocking agents—PBS containing 1% bovine serum albumin (BSA) (both from Sigma, Poole, United Kingdom), PBS containing 2% BSA, sterile distilled water (SDW) containing 1% skim milk powder (Marvel, Chivers Ireland Ltd., Dublin, Ireland), and StabilCoat Immunoassay Stabilizer (SurModics, Inc., Eden Prairie, MN)—to potentially reduce the percentage of nonspecific binding were investigated. Beads were blocked overnight at 4°C with mixing. Blocked beads were resuspended to the original volume in fresh blocking buffer and were used in IMS experiments involving four types of uncoated paramagnetic beads: Dynabeads M280 sheep anti-rabbit IgG beads, Pierce MagnaBind carboxyl derivatized beads, amine-coated magnetic hollow glass beads (Microsphere Technology Ltd., Edinburgh, United Kingdom), and Dynabeads MyOne carboxylic acid beads (Invitrogen). The four types of blocked, uncoated beads were used in IMS experiments involving broth suspensions of each Mycobacterium sp. containing 103 to 105 CFU/ml. The recovery rates achieved using blocked beads were expressed as a percentage of the count of each Mycobacterium sp. in original broth samples. Experiments were repeated in triplicate for each Mycobacterium sp.
Evaluation of magnetic separation in combination with the optimized phage assay.
Middlebrook 7H9-OADC broth suspensions containing 103 to 104 CFU/ml M. avium subsp. paratuberculosis NCTC 8578 or Mycobacterium bovis BCG NCTC 5692 were subjected to manual magnetic separation using 10 μl of Dynabeads MyOne Tosylactivated beads coated with either polyclonal antibody S624, aMp3, biotinylated aMp3, or biotinylated aMptD. The use of a 50:50 mixture of beads coated with the two biotinylated peptides (i.e., 5 μl of each, added separately) was also evaluated. M. bovis BCG was studied as a surrogate for the containment level 3 (CL3) pathogen M. bovis, which may occur in raw milk from tuberculosis-affected herds. After magnetic separation and resuspension of the beads in 1 ml 7H9-OADC, 100 μl was plated on HEYM or 7H9-OADC agar (as appropriate for the Mycobacterium sp.) and was incubated at 37°C for as long as 6 weeks. The remaining 900 μl, after the addition of CaCl2 (to a final concentration of 2 mM), was incubated overnight at 37°C before being subjected to the optimized phage amplification assay (7). Briefly, 100 μl mycobacteriophage D29 (108 PFU/ml) was added to the sample before incubation at 37°C for 2 h. Samples were shaken occasionally to prevent bead sedimentation. A virucide (100 μl of 100 mM ferrous ammonium sulfate) was then added, and samples were mixed thoroughly (to wet all internal surfaces of the vial) and were then incubated for 5 min at room temperature in order to inactivate seed phage before the addition of 5 ml 7H9-OADC containing 2 mM CaCl2. The sample was returned to the incubator at 37°C for a further 1.5 h before being plated with 1 ml M. smegmatis mc2 155 culture (108 CFU/ml) and 5 ml molten Middlebrook 7H9 agar (cooled to 55°C). Plaques were counted after overnight incubation at 37°C and were expressed as PFU per milliliter. CFU and PFU counts were compared, and both the capture efficiency for M. avium subsp. paratuberculosis and the percentage of nonspecific binding by M. bovis BCG were calculated for each bead-coating-antigen combination.
Automated versus manual PMS.
Broth or UHT whole milk containing 103 to 104 CFU/ml M. avium subsp. paratuberculosis NCTC 8578 or M. bovis BCG NCTC 5692 was processed through manual and automated magnetic separation using 10 μl of mixed Dynabeads MyOne Tosylactivated beads coated with the two biotinylated peptides. After magnetic separation, samples were processed both by culture and by the optimized phage assay as described above. The experiment was repeated twice.
Detection sensitivity of the automated PMS-phage assay.
Twelve 1-ml aliquots of Middlebrook 7H9-OADC broth and UHT milk (purchased from a local supermarket), either without M. avium subsp. paratuberculosis or containing 102 to 103, 10 to 100, or 1 to 10 CFU/ml (four aliquots at each spiking level), were tested by the PMS-phage assay. Milk samples were centrifuged (14,000 rpm for 15 min) to remove milk fat and whey in preparation for magnetic separation, and the pelleted cells were resuspended in 1 ml Middlebrook 7H9-OADC broth. After PMS, a portion (100 μl) was plated on HEYM and was incubated at 37°C for 4 to 6 weeks; the remaining 900 μl after the addition of CaCl2 (final concentration, 2 mM) was incubated overnight at 37°C and was then processed by the optimized phage assay the next day. The colony count (expressed as CFU per milliliter) and plaque count (expressed as PFU per milliliter) obtained for each sample were compared with the corresponding values recorded for the original broth suspension. This detection sensitivity experiment was carried out for each of the three M. avium subsp. paratuberculosis strains in both broth and milk.
The detection sensitivity of the automated PMS-phage assay with a larger volume of milk (50 ml) was also determined, because this represents the volume of milk that would more commonly be tested for the presence of M. avium subsp. paratuberculosis. A blind trial was carried out with each of the three M. avium subsp. paratuberculosis strains. Each blind trial consisted of testing 12 50-ml aliquots of UHT whole milk containing different numbers of M. avium subsp. paratuberculosis organisms (3 with 102 to 103 CFU/50 ml, 3 with 101 to 102 CFU/50 ml, 3 with 1 to 10 CFU/50 ml, and 3 with no CFU/50 ml) prepared by a colleague. The operator was blinded to the quantity of cells present in each sample until the assay results were available. Each milk sample was centrifuged at 2,500 × g for 15 min, and the pellet was resuspended in 1 ml Middlebrook 7H9-OADC broth before being processed by the automated PMS-phage assay as described above, using the mixture of biotinylated-peptide-coated Dynabeads MyOne Tosylactivated beads.
Statistical analysis.
Plaque and colony counts were analyzed, where appropriate, by a paired t test (GraphPad Instat, version 3; GraphPad Prism, La Jolla, CA); differences with P values of <0.05 were considered significant. The 50% limit of detection (LOD50) and associated 95% confidence limits of the new PMS-phage assay were estimated using the generalized Spearman-Kärber LOD50 calculation for 4-level spiking protocols (1).
RESULTS
Capture efficiency studies.
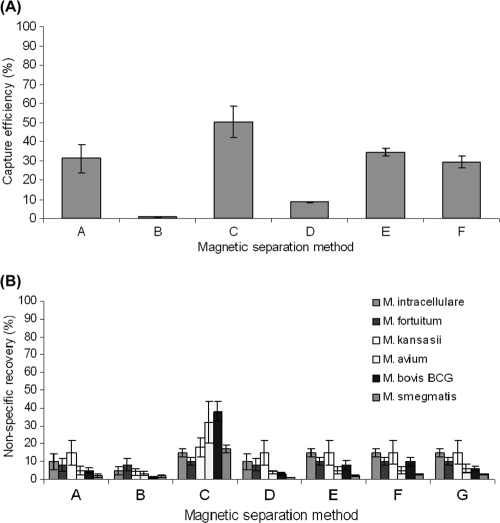
In the initial evaluation of different magnetic-separation approaches involving either antibody- or peptide-coated beads, the commercially available Pathatrix-PM50 beads, which are coated with a polyclonal antibody (method C) (Fig. 1A), consistently achieved the highest capture efficiency for M. avium subsp. paratuberculosis (mean, 50.3% ± 8.4%). Reasonable capture efficiencies (29.2% to 34.2%) were also recorded for M280 Dynabeads coated with the in-house polyclonal antibody S624 (method A) and Pierce MagnaBind carboxyl derivatized beads coated with either aMp3 or aMptD (methods E and F, respectively). AnDiaTec ParaTub-S beads coated with a monoclonal antibody and Dynabeads M280 sheep anti-rabbit IgG beads coated with a 1:10 dilution of polyclonal antibody S624 achieved much lower capture efficiencies (means, 8.54% ± 1.3% and 0.8% ± 0.6%, respectively). However, none of the magnetic-separation approaches in the initial evaluation achieved an efficiency of capture of M. avium subsp. paratuberculosis close to 100%.
FIG. 1.
Initial evaluation of the performance of six different in-house-coated or commercially available paramagnetic beads for magnetic separation applied to 1-ml aliquots of Middlebrook 7H9-OADC broth containing 103 to 104 CFU Mycobacterium sp./ml in terms of the mean efficiency of capture (expressed as a percentage) of M. avium subsp. paratuberculosis (A) and the mean percentage of nonspecific recovery of other Mycobacterium spp. (B). Methods A and B, M280 sheep anti-rabbit IgG Dynabeads coated with polyclonal antibody S624 and a 1:10 dilution of polyclonal antibody S624, respectively; method C, Pathatrix PM50 beads coated with a polyclonal antibody (Matrix Microscience, Newmarket, United Kingdom); method D, AnDiaTec beads coated with a monoclonal antibody (AnDiaTec GmbH, Kornwestheim, Germany); methods E and F, Pierce MagnaBind carboxyl derivatized beads carbodiimide linked with aMp3 and aMptD, respectively; method G, uncoated Pierce MagnaBind carboxyl derivatized beads.
Various degrees of nonspecific recovery of other Mycobacterium spp. were recorded for all the magnetic-separation methods. Pathatrix-PM50 beads (method C) (Fig. 1B) consistently achieved a mean percentage of nonspecific recovery (22% ± 11%) higher than that for all other coated beads (∼10% [Fig. 1B]) and uncoated Pierce MagnaBind carboxyl derivatized beads (Fig. 1B, method G).
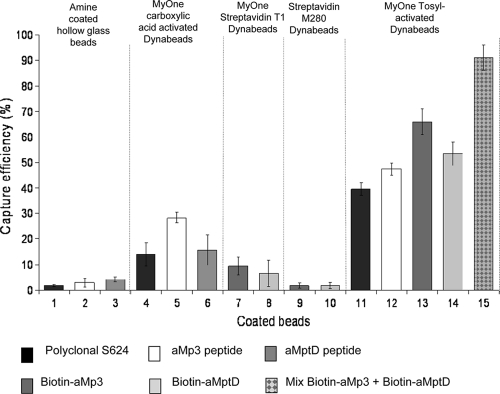
In a subsequent evaluation of other types of surface-activated magnetic beads, MyOne Tosylactivated Dynabeads coated with polyclonal antibody S624 or peptides achieved a much higher capture efficiency for M. avium subsp. paratuberculosis (Fig. 2, bars 11 to 15) than other surface-activated beads (with carboxylic acid or streptavidin). Peptide-coated beads (Fig. 2, bars 12 to 15) consistently achieved higher capture efficiencies than polyclonal-antibody-coated beads (Fig. 2, bar 11). The highest capture efficiency for M. avium subsp. paratuberculosis (91.5% ± 5.0%) was recorded by PMS with a 50:50 mixture of MyOne Tosylactivated Dynabeads coated with biotinylated aMp3 and biotinylated aMptD (Fig. 2, bar 15). The poorest capture efficiency was recorded for both types of streptavidin-coated beads and amine-coated hollow glass beads, for which mean capture efficiencies were consistently ≤10%.
FIG. 2.
Improved mean capture efficiency of M. avium subsp. paratuberculosis achieved by using additional paramagnetic bead-coating antigen combinations for magnetic separation applied to 1-ml aliquots of Middlebrook 7H9 broth containing 103 to 104 CFU/ml. Beads 1 to 3, amine-coated magnetic hollow glass beads coated with polyclonal antibody S624, aMp3, and aMptD, respectively; beads 4 to 6, MyOne carboxylic acid-activated Dynabeads coated with polyclonal antibody S624, aMp3, and aMptD, respectively; beads 7 and 8, MyOne streptavidin T1 Dynabeads coated with biotinylated aMp3 and biotinylated aMptD, respectively; beads 9 and 10, M280 streptavidin Dynabeads coated with biotinylated aMp3 and biotinylated aMptD, respectively; beads 11 to 14, MyOne Tosylactivated Dynabeads coated with polyclonal antibody S624, aMp3, biotinylated aMp3, and biotinylated aMptD, respectively; bead 15, a 50:50 mixture of beads 13 and 14.
The mean percentage of nonspecific recovery of Mycobacterium spp. other than M. avium subsp. paratuberculosis for all types of magnetic beads tested (with the exception of Pathatrix PM50 beads) was ∼10%, and moderate cross-reactivity of the coated MyOne Tosylactivated beads with four other Mycobacterium spp., ranging from 2.5% to 7.2% (mean, 5.1% ± 1.6%), was observed. The results of subsequent blocking experiments indicated that preblocking of peptide-coated MyOne Tosylactivated Dynabeads overnight with 1% skim milk significantly reduced nonspecific binding by other Mycobacterium spp. to <1% (P, 0.0008 by a paired t test) without adversely impacting the efficiency of capture of M. avium subsp. paratuberculosis. However, nonspecific binding by other Mycobacterium spp. was never completely eliminated. For this reason, beads were used unblocked in the remaining experiments during this study.
Combination of differently coated MyOne Tosylactivated beads and the optimized phage assay.
Unblocked MyOne Tosylactivated Dynabeads coated with either peptide recovered a higher percentage of M. avium subsp. paratuberculosis organisms than the same type of beads coated with polyclonal antibody S624, and the simultaneous use of beads coated with the two biotinylated peptides consequently achieved the highest mean efficiency of capture of M. avium subsp. paratuberculosis (98.5% ± 4.7%) and the lowest mean percentage of nonspecific recovery (5.5% ± 1.6%). The mean percentage of nonspecific recovery of M. bovis BCG NCTC 5692 by the other four types of coated Tosylactivated beads ranged from 11.0 to 17.4%. Mean capture efficiencies calculated from plaque counts were never significantly different from those calculated using colony counts (P, >0.05 by a paired t test), further substantiating the hypothesis that the PMS-phage assay is a promising method for enumerating viable M. avium subsp. paratuberculosis organisms (7).
Automated versus manual PMS.
The mean capture efficiencies for M. avium subsp. paratuberculosis achieved by automated and manual PMS using a 50:50 mixture of MyOne Tosylactivated Dynabeads coated with the two biotinylated peptides were not significantly different (P, 0.3178 by a paired t test). In contrast, the mean percentage of nonspecific recovery of M. bovis BCG by automated IMS (AIMS) was significantly less than that by manual IMS (P, 0.0312 by a paired t test), probably because AIMS moves the beads from tube to tube during processing, potentially leaving nontarget mycobacteria behind, adherent to tube surfaces, whereas in manual magnetic separation, the beads remain in the same tube throughout processing.
Detection sensitivity of the PMS-phage assay applied to spiked milk and broth.
The detection sensitivities (expressed as the mean LOD50s) of the combined assay involving PMS with a 50:50 mixture of biotinylated-aMp3- and biotinylated-aMptD-coated MyOne Tosylactivated beads, followed by the optimized phage assay, were 1.9 and 7.3 PFU M. avium subsp. paratuberculosis per ml of spiked broth and milk samples, respectively (Table 2). However, the number of PFU recovered from spiked milk after automated PMS was ∼10-fold lower than that recovered from spiked broth initially containing the same number of M. avium subsp. paratuberculosis organisms (Table 2). Corresponding colony counts, recorded in parallel with plaque counts, indicated a similar trend (data not shown). This result may represent lesser recovery of M. avium subsp. paratuberculosis from milk, but it may instead reflect greater clumping of cells in the fat-rich milk matrix, so that clumps of cells, or clumps of beads plus cells, rather than single cells, are recovered by magnetic beads. In either scenario, fewer PFU would result after the plaque assay.
TABLE 2.
Limit of detection of the automated PMS-phage assay applied to Middlebrook 7H9 broth and whole milk spiked with low numbers of M. avium subsp. paratuberculosis
| M. avium subsp. paratuberculosis strain | Sample matrix | Mean plaque count (PFU/ml)a ± SD at the following spiking level (PFU/ml) before AMS: |
LOD50 (PFU/ml) (95% confidence limit)b | ||
|---|---|---|---|---|---|
| 100-1,000 | 10-100 | 1-10 | |||
| ATCC 19698 | Broth | 111.8 ± 22.1 (4/4)a | 10.2 ± 4.0 (4/4) | 0.3 ± 0.5 (1/4) | 2.9 (0.8-11.5) |
| Milk | 28.9 ± 18.2 (4/4) | 4.5 ± 3.7 (3/4) | 0.3 ± 0.5 (1/4) | 5.0 (0.8-31.0) | |
| NCTC 8578 | Broth | 241.0 ± 48.1 (4/4) | 27.3 ± 14.8 (4/4) | 1.5 ± 1.3 (3/4) | 1.7 (0.3-9.6) |
| Milk | 34.0 ± 15.9 (4/4) | 4.0 ± 3.0 (4/4) | 0.3 ± 0.5 (1/4) | 8.4 (1.5-46.9) | |
| 796PSS | Broth | 302.0 ± 29.3 (4/4) | 28.3 ± 15.2 (4/4) | 3.5 ± 1.7 (4/4) | 1.0 (0.3-2.8) |
| Milk | 27.2 ± 16.3 (4/4) | 2.3 ± 2.6 (2/4) | 0.3 ± 0.5 (1/4) | 8.6 (2.5-28.8) | |
For four replicate samples. The number of samples yielding plaques after AMS per number of samples tested at that spiking level is given in parentheses.
Calculated using the Limit of Detection Program for Qualitative Microbiology Methods (AOAC, 2010).
The detection sensitivity of the automated PMS-phage assay applied to 50-ml volumes of milk was estimated from the results of blind trials involving three different M. avium subsp. paratuberculosis strains. The mean LOD50 of the automated PMS-phage assay was 14.4 PFU M. avium subsp. paratuberculosis per 50 ml of milk (Table 3), which is equivalent to 0.3 PFU/ml and represents a higher detection sensitivity than the application of the automated PMS-phage assay to 1-ml aliquots of spiked milk (mean LOD50, 7.3 PFU/ml). This finding demonstrates that although some M. avium subsp. paratuberculosis cells may be lost during the centrifugation of larger volumes of milk (because an unknown proportion of the M. avium subsp. paratuberculosis cells present will be discarded with the whey and cream fractions), the detection sensitivity of the IMS-phage assay is increased by testing 50 ml of milk rather than 1 ml of milk.
TABLE 3.
Results of blind testing of 50-ml aliquots of whole milk spiked with three levels of three different M. avium subsp. paratuberculosis strains by automated PMS-phage assay
| Spiking level (PFU/50 ml) | ATCC 19698 |
NCTC 8578 |
796PSS |
||||||
|---|---|---|---|---|---|---|---|---|---|
| PFU spikeda | PFU recoveredb | LOD50 (PFU/50 ml) (95% CI)c | PFU spiked | PFU recovered | LOD50 (PFU/50 ml) (95% CI) | PFU spiked | PFU recovered | LOD50 (PFU/50 ml) (95% CI) | |
| 100-1,000 | 100 | 12.7 ± 2.1 (3/3) | 16.7 (2.9-96.2) | 206 | 123.0 ± 75.0 (3/3) | 9.4 (1.2-70.1) | 620 | 262.7 ± 48.4 (3/3) | 16.4 (0.8-318.2) |
| 10-100 | 13 | 3.0 ± 5.2 (1/3) | 15 | 7.3 ± 9.5 (2/3) | 62 | 7.0 ± 6.1 (2/3) | |||
| 1-10 | 1 | 0.0 ± 0.0 (0/3) | 1 | 0.0 ± 0.0 (0/3) | 9 | 5.7 ± 6.1 (1/3) | |||
| 0 | Unspiked | 0 (0/3) | Unspiked | 0 (0/3) | Unspiked | 0 (0/3) | |||
PFU added to 50 ml milk before PMS was determined by testing dilutions of broth culture used for spiking by an optimized phage assay.
PFU/50 ml of milk after PMS was determined by processing of beads recovered through an optimized phage assay. The number of samples yielding plaques after PMS per triplicate samples tested is given in parentheses.
Calculated using the Limit of Detection Program for Qualitative Microbiology Methods (AOAC, 2010). CI, confidence interval.
DISCUSSION
The objective of this study was to identify a magnetic-separation approach to be used in conjunction with the previously reported optimized phage assay (7), so as to provide a novel IMS- or PMS-phage assay for the detection and enumeration of viable M. avium subsp. paratuberculosis organisms in milk. The magnetic-separation approach needed to achieve a capture efficiency close to 100% and to maintain high specificity for M. avium subsp. paratuberculosis. An initial evaluation of six magnetic-separation approaches involving in-house-coated or commercially available anti-M. avium subsp. paratuberculosis paramagnetic beads clearly demonstrated that not all magnetic-separation approaches developed for the selective concentration of M. avium subsp. paratuberculosis perform equally well. It became apparent that bead characteristics (composition, size, concentration, and surface modification), as well as the nature of the coating antigen (polyclonal or monoclonal antibody, peptide or biotinylated peptide), impacted the selectivity of capture, assessed by determining the efficiency of capture (expressed as a percentage) of M. avium subsp. paratuberculosis and the percentage of nonspecific recovery of other Mycobacterium spp. Pathatrix PM-50 beads, which are latex beads coated with an affinity-purified polyclonal antibody (Adrian Parton, Matrix Microscience CEO, personal communication), achieved the highest efficiency of capture of M. avium subsp. paratuberculosis in an initial evaluation but also showed significant nonspecific recovery of other Mycobacterium spp., particularly M. avium subsp. avium and M. bovis BCG. Better results were obtained with magnetic-separation approaches involving beads coated with the polyclonal antibody S624 and the two M. avium subsp. paratuberculosis-specific peptides aMp3 (30) and aMptD (28), which consistently captured around 30% of M. avium subsp. paratuberculosis CFU from spiked broth and showed less cross-reactivity with other Mycobacterium spp. in the initial evaluation (Fig. 1). The worst performance was shown by AnDiaTec beads, which are coated with a monoclonal antibody (17) and which, in spite of achieving the lowest percentage of nonspecific recovery (7 and 4%, respectively, for M. avium and M. bovis BCG), had a low efficiency of capture of M. avium subsp. paratuberculosis (ca. 10%). It was surprising to find so much variability in capture efficiency and nonspecific binding for the six different bead-antigen combinations tested, and it was disappointing that none of the initial six IMS or PMS approaches had a capture efficiency close to 100%, as sought. Thus, none of the magnetic-separation methods included in the initial evaluation showed the desired capture efficiency or specificity for M. avium subsp. paratuberculosis.
The major weakness of the magnetic-separation approaches initially evaluated was the nonspecific recovery of other Mycobacterium spp.; this would be a barrier to the use of these methods in conjunction with the phage assay, which was our objective. The issue of nonspecific binding between paramagnetic beads and nontarget bacteria has been reported in the literature previously (14, 31, 32). Vytřasová et al. (32) observed similar results during IMS experiments with anti-Listeria sp. Dynabeads, in particular under competitive capture conditions. These authors defined a moderate nonspecific reaction as a recovery percentage of <10% and nonspecific binding as a recovery percentage of >10%. However, it is not clear whether Vytřasová et al. (32) considered the nontarget bacteria to be reacting with the anti-Listeria antibody on the Dynabeads or with the surfaces of the beads by physical absorption. To investigate which was the case during M. avium subsp. paratuberculosis magnetic separation, a negative control consisting of uncoated beads was included (Fig. 1B, method G). The results clearly showed that uncoated beads recover a moderate percentage (around 10%) of nontarget mycobacteria and that nonspecific interaction between the surfaces of paramagnetic beads and bacteria does occur. Such interaction is most likely due to electrostatic bonds or van der Waals forces. This nonspecific recovery level of ∼10% was common to all six magnetic-separation approaches initially evaluated (Fig. 1B), and we attributed these results to physical interaction between bacterial cells and paramagnetic beads, whereas recovery levels exceeding 10% were attributed to suspected cross-reaction between the coating antigen or antibody and nontarget bacteria. Consequently, it was considered unsuitable to pursue method C (Pathatrix PM-50 beads). Methods A, E, and F, which showed the highest capture efficiency, were selected for further study.
A variety of other surface-activated magnetic beads coated with polyclonal antibody S624 or with either of the two specific peptides, aMp3 and aMptD, were evaluated in subsequent experiments. The beads studied included polystyrene or glass particles providing chemically active groups (amine, carboxylic, tosyl) that could be linked through covalent bonds with the first available carboxylic or amine group provided by the coating antigen (peptide or polyclonal antibody). Besides using primary-group-activated beads, we also evaluated the performance of two different streptavidin-coated paramagnetic beads coupled with biotinylated peptides aMp3 and aMptD. The results of this set of experiments (Fig. 2) once again showed that the capture efficiency of coated paramagnetic beads for M. avium subsp. paratuberculosis differed widely for different paramagnetic-bead-coating-antigen combinations. Dynabeads MyOne Tosylactivated beads coated with polyclonal antibody S624 or either peptide achieved the highest capture efficiencies for M. avium subsp. paratuberculosis overall. A key difference between this type of surface-activated bead and the other beads employed was the higher number of beads added per sample, 5 × 108 in 10 μl compared to 106 to 107 in 10 μl. A higher number of beads increases the surface area available for the binding of the target bacterium, thus enhancing the potential for bacterial capture.
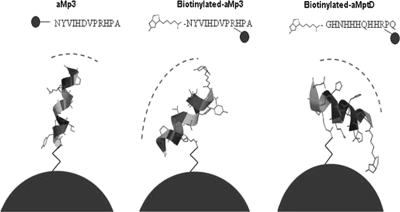
A higher efficiency of capture of M. avium subsp. paratuberculosis was consistently recorded with the biopanning-derived peptides reported by Stratmann et al. (28, 30) than with the original polyclonal-antibody-based IMS method (9). Surprisingly, biotinylated aMp3 and biotinylated aMptD peptides, which had originally been chemically synthesized to be applied to M280 and MyOne streptavidin beads, not to MyOne Tosylactivated beads, showed the highest capture efficiency and the lowest level of nonspecific binding by other Mycobacterium spp. Chemical modeling analysis carried out using Spartan '08 software (Wavefunction, Inc., Irvine, CA), which can predict the site of binding between peptides and paramagnetic-bead surfaces, provided some insight into the enhanced capture capability of the biotinylated peptides. It revealed that the presence of the biotin molecule linked to asparagine (N) in peptide aMp3 and to glycine (G) in peptide aMptD caused a change in the orientations of the two peptides (Fig. 3). Given that the two sites are no longer available for the amine bond with the paramagnetic beads, the most probable sites of linkage to the beads are the two arginines (R in the peptide sequences [Fig. 3]) located on the opposite parts of the two peptide chains. The linkage between beads and the two arginines, the fourth and third amino acids in the chains of aMp3 and aMptD, respectively, does not allow the peptide to bind perpendicularly to the surface of the bead and thus, due to steric hindrance, is likely to have produced a change in the angle of binding and the presentation of the peptide for capture purposes. It is possible that this condition enhances the surface available for binding with the target bacteria and makes interaction between the peptide and the cell wall receptor of the target bacteria more stable. This may explain the higher capture efficiency of M. avium subsp. paratuberculosis achieved using the two biotinylated phage-derived peptides, both separately and as a 50:50 mixture.
FIG. 3.
Schematic illustrating the predicted binding between the tosyl group on the surfaces of MyOne Tosylactivated Dynabeads and the M. avium subsp. paratuberculosis-specific peptides (aMp3, biotinylated aMp3, and biotinylated aMptD). Dashed lines indicate the supposed areas of interaction for binding with M. avium subsp. paratuberculosis in broth or milk samples.
Interestingly, the highest capture efficiency for M. avium subsp. paratuberculosis (determined either by colony counts or by plaque counts) was consistently achieved by PMS using a cocktail of MyOne Tosylactivated beads coated with biotinylated aMp3 and biotinylated aMptD (5 μl of each type of coated bead added separately per reaction rather than 10 μl of a mixture of the two types of beads). It should be noted that beads coated with the two types of peptide do not mix well, and a consistent 10-μl volume of mixed beads is difficult to achieve. This is why the two types of beads were added separately to the sample. The capture efficiency for M. avium subsp. paratuberculosis achieved by the 50:50 mixture was 91% ± 5% instead of 66% ± 5% or 53.5% ± 4.5%, achieved by biotin-aMp3- or biotin-aMptD-coated beads individually. The explanation for this finding is unclear, but the finding is in agreement with those of Kretzer et al. (16), who found that paramagnetic beads coated with phage-based cell wall-binding domain (CBD) molecules performed much better than commercially available antibody-based beads for IMS of L. monocytogenes, in terms of both sensitivity and the percentage of recovery. Furthermore, Kretzer et al. (16) found that a mixed preparation of two different types of CBD beads (CBD118 and CBD500) was required to capture all Listeria cells present in a sample. It is possible that broth cultures of M. avium subsp. paratuberculosis contain cells at different metabolic stages and that not all cells express the target proteins to which each peptide binds. It is reasonable to assume that the simultaneous use of two coating antigens would enhance the probability of recovering cells expressing either one or both of the two target proteins on the cell wall of M. avium subsp. paratuberculosis.
The best magnetic-separation approach (a 50:50 mixture of biotinylated-peptide-coated Dynabeads MyOne Tosylactivated beads) was then combined with the optimized phage assay into a novel automated PMS-phage assay, and experiments were carried out to determine the analytical sensitivity (LOD50) and specificity for M. avium subsp. paratuberculosis in broth and spiked milk. The automated PMS-phage assay consistently achieved a high capture efficiency for M. avium subsp. paratuberculosis (85 to 100%) and showed minimal cross-reaction with other Mycobacterium spp. Blocking reduced (to a recovery level of <1%), but did not completely eliminate, nonspecific binding between other mycobacteria and the paramagnetic beads, which means that a small percentage of plaques produced when a raw-milk sample is tested could be generated by the D29 phage infecting other Mycobacterium spp. Therefore, the method may overestimate the number of viable M. avium subsp. paratuberculosis organisms when it is employed to test raw milk, where other pathogenic or environmental mycobacteria may also be present. However, this issue can potentially be resolved by testing a proportion of the plaques obtained by IS900 PCR, so called plaque-PCR, according to the method described by Stanley et al. (26).
The present study found that automated PMS carried out using the BeadRetriever system, which enables the processing of 15 samples at once, required less plastic consumables, speeded up the magnetic-separation process (approximately 40 min for 15 samples), achieved a capture efficiency for M. avium subsp. paratuberculosis comparable to that of manual PMS, and lowered the percentage of nonspecific recovery of other Mycobacterium spp. The automated PMS-phage assay was able to detect <10 PFU or CFU of viable M. avium subsp. paratuberculosis organisms per ml of spiked broth or milk. However, a “matrix effect” was observed when milk rather than broth was processed, resulting in the recovery of ca. 10-fold fewer CFU or PFU from spiked milk than from spiked broth containing the same number of M. avium subsp. paratuberculosis organisms before testing (Table 2). The same trend was observed for blind trial results, when a larger volume (50 ml) of UHT milk spiked with M. avium subsp. paratuberculosis was tested. In this case, the automated PMS-phage assay was able to detect <370 CFU M. avium subsp. paratuberculosis per 50 ml of milk (the lowest spiking concentration tested), but the CFU and PFU were once again about 10-fold lower than the corresponding counts recorded for the original M. avium subsp. paratuberculosis-spiked samples (Table 3). This may be a real or artifactual observation in light of the fact that M. avium subsp. paratuberculosis cells have a strong propensity to clump, and milk contains agglutinins that may promote the clumping of cells once the milk has been spiked. Consequently, more clumps, rather than single cells, could have been captured by the beads from milk during magnetic separation than from broth, with a resultant artifactual decrease in CFU or PFU/ml. Alternatively, milk proteins may have blocked some binding sites on the coated beads, preventing the attachment of some of the M. avium subsp. paratuberculosis cells present. Notwithstanding the matrix effect observed, the detection sensitivity of the novel automated PMS-phage assay is comparable to or better than those of IMS-PCR methods used previously to test naturally infected milk samples (500 CFU/ml [28] and 103 CFU/50 ml [10]). Of course, the main advantage of the novel assay over IMS-PCR is that it detects the presence of viable M. avium subsp. paratuberculosis cells, and not just M. avium subsp. paratuberculosis DNA.
In conclusion, of the magnetic-separation approaches tested during this study, the best approach for the specific capture of M. avium subsp. paratuberculosis cells from milk was a 50:50 mixture of MyOne Tosylactivated Dynabeads coated with biotinylated M. avium subsp. paratuberculosis-specific peptides aMp3 and aMptD. This PMS approach in combination with the optimized phage assay consistently achieved a capture efficiency for M. avium subsp. paratuberculosis of 85 to 100% from spiked broth and minimal nonspecific binding by other Mycobacterium spp., particularly if coated beads were preblocked with 1% skim milk before use. Despite an apparent reduction in capture efficiency when milk was tested instead of broth, the detection sensitivity of the automated PMS-phage assay was 14.4 PFU/50 ml of milk (equivalent to 0.3 PFU/ml of milk), which, in light of our previous experience and in light of the numbers of M. avium subsp. paratuberculosis organisms reported likely to be present in raw milk (11, 24), is considered sufficient sensitivity for the testing of naturally infected milk samples. The novel PMS-phage assay represents a more rapid method of detecting and enumerating viable M. avium subsp. paratuberculosis organisms in milk, and potentially other food matrices, than chemical decontamination followed by culture, which is the current reference method for the detection of viable M. avium subsp. paratuberculosis but is slow (requiring weeks, not hours, of incubation) and more costly.
Acknowledgments
Antonio Foddai was the recipient of a Master and Back Studentship from the Sardinian government during this study.
Thanks are due to Francesco Ravalico of the Department of Chemistry and Chemical Engineering, Queen's University Belfast, for assistance in using the Spartan '08 software to visualize magnetic-bead-peptide linkages and to Anthony Hitchins, CFSAN, FDA, for providing the Excel version of the Limit of Detection Program for Qualitative Microbiology Methods to enable LOD50 calculations.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.AOAC International Presidential Task Force on Best Practices in Microbiological Methodology. 2006. Final report and executive summaries. Appendix K. Proposed use of a 50% limit of detection value in defining uncertainty limits in the validation of presence-absence microbial detection methods. http://www.fda.gov/ucm/groups/fdagov-public/@fdagov-foods-gen/documents/document/ucm088764.pdf.
- 2.Ayele, W. Y., P. Svastova, P. Roubal, M. Bartos, and I. Pavlik. 2005. Mycobacterium avium subspecies paratuberculosis cultured from locally and commercially pasteurized cow's milk in the Czech Republic. Appl. Environ. Microbiol. 71:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannantine, J. P., T. J. Radosevich, J. R. Stabel, S. Sreevatsan, V. Kapur, and M. L. Paustian. 2007. Development and characterization of monoclonal antibodies and aptamers against major antigens of Mycobacterium avium subsp. paratuberculosis. Clin. Vaccine Immunol. 14:518-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, S., D. Hinz, J. P. Bannantine, and F. T. Griffin. 2006. Isolation of high-affinity single-chain antibodies against Mycobacterium avium subsp. paratuberculosis surface proteins from sheep with Johne's disease. Clin. Vaccine Immunol. 13:1022-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favrin, S. J., S. A. Jassim, and M. W. Griffiths. 2001. Development and optimization of a novel immunomagnetic separation-bacteriophage assay for detection of Salmonella enterica serovar Enteritidis in broth. Appl. Environ. Microbiol. 67:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favrin, S. J., S. A. Jassim, and M. W. Griffiths. 2003. Application of a novel immunomagnetic separation-bacteriophage assay for the detection of Salmonella enteritidis and Escherichia coli O157:H7 in food. Int. J. Food Microbiol. 85:63-71. [DOI] [PubMed] [Google Scholar]
- 7.Foddai, A., C. T. Elliott, and I. R. Grant. 2009. Optimization of a phage amplification assay to permit accurate enumeration of viable Mycobacterium avium subsp. paratuberculosis cells. Appl. Environ. Microbiol. 75:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foddai, A., C. T. Elliott, and I. R. Grant. 2010. Rapid assessment of the viability of Mycobacterium avium subsp. paratuberculosis cells after heat treatment, using an optimized phage amplification assay. Appl. Environ. Microbiol. 76:1777-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant, I. R., H. J. Ball, and M. T. Rowe. 1998. Isolation of Mycobacterium paratuberculosis from milk by immunomagnetic separation. Appl. Environ. Microbiol. 64:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant, I. R., C. M. Pope, L. M. O'Riordan, H. J. Ball, and M. T. Rowe. 2000. Improved detection of Mycobacterium avium subsp. paratuberculosis in milk by immunomagnetic PCR. Vet. Microbiol. 77:369-378. [DOI] [PubMed] [Google Scholar]
- 11.Grant, I. R., H. J. Ball, and M. T. Rowe. 2002. Incidence of Mycobacterium paratuberculosis in bulk raw and commercially pasteurized cows' milk from approved dairy processing establishments in the United Kingdom. Appl. Environ. Microbiol. 68:2428-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagens, S., and M. J. Loessner. 2007. Application of bacteriophages for detection and control of foodborne pathogens. Appl. Microbiol. Biotechnol. 76:513-519. [DOI] [PubMed] [Google Scholar]
- 13.Hsih, H. Y., and H. Y. Tsen. 2001. Combination of immunomagnetic separation and polymerase chain reaction for the simultaneous detection of Listeria monocytogenes and Salmonella spp. in food samples. J. Food Prot. 64:1744-1750. [DOI] [PubMed] [Google Scholar]
- 14.Jeníková, G., J. Pazlarová, and K. Demnerová. 2000. Detection of Salmonella in food samples by the combination of immunomagnetic separation and PCR assay. Int. Microbiol. 3:225-229. [PubMed] [Google Scholar]
- 15.Karch, H., C. Janetzki-Mittmann, S. Aleksic, and M. Datz. 1996. Isolation of enterohemorrhagic Escherichia coli O157 strains from patients with hemolytic-uremic syndrome by using immunomagnetic separation, DNA-based methods, and direct culture. J. Clin. Microbiol. 34:516-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kretzer, J. W., R. Lehmann, M. Schmelcher, M. Banz, K.-P. Kim, C. Korn, and M. J. Loessner. 2007. Use of high-affinity cell wall-binding domains of bacteriophage endolysins for immobilization and separation of bacterial cells. Appl. Environ. Microbiol. 73:1992-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metzger-Boddien, C., D. Khaschabi, M. Schönbauer, S. Boddien, T. Schlederer, and J. Kehle. 2006. Automated high-throughput immunomagnetic separation-PCR for detection of Mycobacterium avium subsp. paratuberculosis in bovine milk. Int. J. Food Microbiol. 110:201-208. [DOI] [PubMed] [Google Scholar]
- 18.Moreno, Y., M. Hernández, M. A. Ferrús, J. L. Alonso, S. Botella, R. Montes, and J. Hernández. 2001. Direct detection of thermotolerant campylobacters in chicken products by PCR and in situ hybridization. Res. Microbiol. 152:577-582. [DOI] [PubMed] [Google Scholar]
- 19.NordVal. 2009. Protocol for the validation of alternative microbiological methods. National Veterinary Institute, Oslo, Norway. http://www.nmkl.org/NordVal/NordValprotocolmarch2009.pdf.
- 20.Possé, B., L. De Zutter, M. Heyndrickx, and L. Herman. 2008. Quantitative isolation efficiency of O26, O103, O111, O145 and O157 STEC serotypes from artificially contaminated food and cattle faeces samples using a new isolation protocol. J. Appl. Microbiol. 105:227-235. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen, H. N., O. F. Rasmussen, H. Christensen, and J. E. Olsen. 1995. Detection of Yersinia enterocolitica O:3 in faecal samples and tonsil swabs from pigs using IMS and PCR. J. Appl. Microbiol. 78:563-568. [DOI] [PubMed] [Google Scholar]
- 22.Rijpens, N., L. Herman, F. Vereecken, G. Jannes, J. De Smedt, and L. De Zutter. 1999. Rapid detection of stressed Salmonella spp. in dairy and egg products using immunomagnetic separation and PCR. Int. J. Food Microbiol. 46:37-44. [DOI] [PubMed] [Google Scholar]
- 23.Rybniker, J., S. Kramme, and P. L. Small. 2006. Host range of 14 mycobacteriophages in Mycobacterium ulcerans and seven other mycobacteria including Mycobacterium tuberculosis—application for identification and susceptibility testing. J. Med. Microbiol. 55:37-42. [DOI] [PubMed] [Google Scholar]
- 24.Slana, I., P. Kralik, A. Kralova, and I. Pavlik. 2008. On-farm spread of Mycobacterium avium subsp. paratuberculosis in raw milk studied by IS900 and F57 competitive real time quantitative PCR and culture examination. Int. J. Food Microbiol. 128:250-257. [DOI] [PubMed] [Google Scholar]
- 25.Spanová, A., B. Rittich, R. Karpiskova, L. Cechova, and D. Skapova. 2000. PCR identification of Salmonella cells in food and stool samples after immunomagnetic separation. Bioseparation 9:379-384. [DOI] [PubMed] [Google Scholar]
- 26.Stanley, E. C., R. J. Mole, R. J. Smith, S. M. Glenn, M. R. Barer, M. McGowan, and C. E. D. Rees. 2007. Development of a new, combined rapid method using phage and PCR for detection and identification of viable Mycobacterium paratuberculosis bacteria within 48 hours. Appl. Environ. Microbiol. 73:1851-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephan, R., S. Schumacher, T. Tasara, and I. R. Grant. 2007. Prevalence of Mycobacterium avium subsp. paratuberculosis in Swiss raw milk cheeses collected at retail level. J. Dairy Sci. 90:3590-3595. [DOI] [PubMed] [Google Scholar]
- 28.Stratmann, J., K. Dohmann, J. Heinzmann, and G. F. Gerlach. 2006. Peptide aMptD-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in bulk milk samples. Appl. Environ. Microbiol. 72:5150-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stratmann, J., B. Strommenger, R. Goethe, K. Dohmann, G. F. Gerlach, K. Stevenson, L. L. Li, Q. Zhang, V. Kapur, and T. J. Bull. 2004. A 38-kilobase pathogenicity island specific for Mycobacterium avium subsp. paratuberculosis encodes cell surface proteins expressed in the host. Infect. Immun. 72:1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stratmann, J., B. Strommenger, K. Stevenson, and G. F. Gerlach. 2002. Development of a peptide-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in milk. J. Clin. Microbiol. 40:4244-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uyttendaele, M., I. Van Hoorde, and J. Debevere. 2000. The use of immuno-magnetic separation (IMS) as a tool in a sample preparation method for direct detection of L. monocytogenes in cheese. Int. J. Food Microbiol. 54:205-212. [DOI] [PubMed] [Google Scholar]
- 32.Vytřasová, J., I. Zachová, L. Červenka, J. Štĕpánková, and M. Pejchalová. 2005. Non-specific reactions during immunomagnetic separation of Listeria. Food Technol. Biotechnol. 43:397-401. [Google Scholar]