Abstract
Although the presence of pathogenic Vibrio spp. in estuarine environments of northern New England has been known for some time (C. H. Bartley and L. W. Slanetz, Appl. Microbiol. 21: 965-966, 1971, and K. R. O'Neil, S. H. Jones, and D. J. Grimes, FEMS Microbiol. Lett. 60:163-167, 1990), their virulence and the relative threat they may pose to human health has yet to be evaluated. In this study, the virulence potential of 33 Vibrio parahaemolyticus isolates collected from the Great Bay Estuary of New Hampshire was assessed in comparison to that of clinical strains. The environmental isolates lack thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH), which are encoded by tdh and trh, respectively. Though not hemolytic, they do possess putative virulence factors, such type III secretion system 1, and are highly cytotoxic to human gastrointestinal cells. The expression of known and putative virulence-associated traits, including hemolysin, protease, motility, biofilm formation, and cytotoxicity, by clinical reference isolates correlated with increased temperature from 28°C to 37°C. In contrast, the environmental isolates did not induce their putative virulence-associated traits in response to a temperature of 37°C. We further identified a significant correlation between hemolytic activity and growth phase among clinical strains, whereby hemolysin production decreases with increasing cell density. The introduction of a tdh::gfp promoter fusion into the environmental strains revealed that they regulate this virulence-associated gene appropriately in response to temperature, indicating that their existing regulatory mechanisms are primed to manage newly acquired virulence genes.
Despite relatively cool water temperatures, estuarine environments in northern New England are a known niche for pathogenic species of the genus Vibrio (3, 39, 51). Shellfish beds within these habitats serve both as a natural reservoir where Vibrio spp. accumulate and as vectors for human infection. Recent outbreaks of Vibrio parahaemolyticus in New York, New Jersey, Connecticut, Washington, and Alaska highlight the importance of further research to understand the resident strains in these potentially significant habitats for this emergent pathogen (6, 32, 36).
Infections by V. parahaemolyticus are often acquired through the ingestion of raw or undercooked shellfish, including oysters. Upon colonization of the intestine, the products of the hemolysin genes (tdh and trh, encoding thermostable direct hemolysin [TDH] and TDH-related hemolysin) are believed to rapidly induce inflammatory gastroenteritis (20, 47, 49). Recently, a more infectious pandemic serotype (O3:K6) emerged that is often identified by the presence of the ORF8 gene (23). Not all strains of V. parahaemolyticus are human pathogens. Historically, acquisition by horizontal gene transfer (HGT) of the tdh gene within Vibrio pathogenicity island-7 (VPaI-7) (28) and/or the trh gene within the recently identified Vp-PAITH3996 (37) has been credited with the transition from noninfectious environmental to human pathogenic clinical strain. However, V. parahaemolyticus remains pathogenic even in the absence of these two thermostable hemolysins, indicating that other virulence traits exist (35, 43, 57). Furthermore, recent outbreaks have been caused by strains lacking tdh and/or trh (16). Although other virulence factors have been investigated, including type III secretion systems 1 and 2 (T3SS1 and T3SS2) and biofilm formation, the understanding of virulence remains incomplete (23, 52). Moreover, the contribution to pathogenicity of conserved virulence-associated factors present in a wide range of bacterial pathogens (e.g., protease, siderophore, and motility), the ability to regulate newly acquired virulence factors, and the capacity to adjust to a human host environment have yet to be revealed.
In order to assess the virulence potential of environmental V. parahaemolyticus isolates lacking the characteristic tdh/trh hemolysin genes, we examined their ability to express alternative, conserved virulence-associated factors at environmentally relevant and human body temperatures. The “environmental” strains in this study were isolated from environmental samples and have further been defined by the absence of either tdh or trh, whereas “clinical” strains were isolated either from patients or from the environment during an outbreak and harbor tdh and/or trh genes. Among the clinical strains examined, we found a strong correlation of the production of potential virulence-associated factors with human body temperature and an inverse correlation of hemolysin with cell density. In contrast, the environmental strains do not express their existing conserved virulence-associated traits in response to body temperature, but many of the environmental isolates were highly cytotoxic to human gastrointestinal cells. In addition, the environmental isolates lack the tdh gene but were able to express this gene's promoter appropriately in response to a temperature of 37°C. Although these environmental isolates are unlikely to cause a human infection, they do represent a population of bacteria whose existing regulatory networks can appropriately control newly acquired virulence genes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Thirty-three environmental strains of Vibrio parahaemolyticus isolated from New Hampshire's Great Bay Estuary from May through December 2007 (51) were used in this study, along with an additional two strains from other sources (Table 1). All New Hampshire isolates were negative for both tdh and trh based on multiplex PCR analysis (42, 51). The presence of various other proposed virulence genes, including T3SS2 apparatus gene vscC2 and effector vopP, a putative DNA methyltransferase, a homolog of the Escherichia coli cytotoxic necrotizing factor vopC, and a homolog of V. cholerae pathogenicity island gene VPA1376, was determined by PCR with published methods (4). A prepandemic tdh-positive strain, BB22, was used as a reference, and nine clinical strains from outbreaks worldwide were used for comparison (Table 1).
TABLE 1.
Bacterial strains with relevant pathogenicity markers
| Strain(s) | Origin | tdh | trh | ORF8a | vscC2 | vopP | MTase | vopC | VPA1376 | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Clinical | ||||||||||
| BB22 | Reference strain, waterb (India) | + | − | − | + | + | + | + | + | 31 |
| MDOH-03-17294-Vp | Patient (FL) | + | − | − | + | + | + | + | + | 11 |
| AQ4037 | Patient (Japan) | − | + | − | + | − | − | − | + | 12 |
| MDOH-03-17282-Vp | Patient (FL) | + | − | + | + | − | − | − | − | 11 |
| KE9967 | Patient (Japan) | + | − | − | + | + | − | + | + | 40c |
| TX2103 | Patient (TX) | + | − | − | + | + | + | + | − | 12 |
| VP81 | Clinical (India) | + | − | + | + | + | + | + | − | 29c |
| MDOH-04-5M732 | Patient (FL) | + | − | + | + | + | + | + | − | 11 |
| BAC98-03255 | Patient (NY) | + | + | − | + | + | + | + | + | 34 |
| F11-3A | Clamb (WA) | + | + | − | + | + | + | + | − | 12 |
| Environmental | ||||||||||
| EnvDOH-04-001 | Oyster (FL) | − | − | − | − | − | − | + | − | 11 |
| DAL1094 | Oysterb (TX) | − | − | − | + | − | − | − | − | 12 |
| G46, G151, G227 | Oyster (NH) | − | − | − | + | − | − | + | + | 51 |
| G246, G69 | Water (NH) | − | − | − | + | − | − | + | + | 51 |
| G10, G31, G43, G91, G251 | Oyster (NH) | − | − | − | + | − | − | − | + | 51 |
| G149 | Water (NH) | − | − | − | + | − | − | − | + | 51 |
| G26 | Clam (NH) | − | − | − | + | − | − | − | + | 51 |
| G74, G95, G237 | Oyster (NH) | − | − | − | − | − | − | − | − | 51 |
| G1, G7, G23, G242 | Oyster (NH) | − | − | − | − | − | − | − | + | 51 |
| G4, G6, G8, G235 | Water (NH) | − | − | − | − | − | − | − | + | 51 |
| G25 | Mussel (NH) | − | − | − | − | − | − | − | + | 51 |
| G12, G13, G79, G145, G259, G277 | Oyster (NH) | − | − | − | − | − | − | − | − | 51 |
| G61, G62, G255 | Water (NH) | − | − | − | − | − | − | − | − | 51 |
Associated with pandemic isolates.
Associated with outbreak.
Provided by E. F. Boyd.
Bacteria were grown in heart infusion (HI) medium at a pH of 7.3 (Fluka, Buchs, Switzerland) at 28°C, a temperature often used to cultivate many environmental Vibrio spp., and 37°C, human body temperature, for all phenotypic assays. Uniform growth patterns were observed for environmental and clinical strains grown in rich medium, as determined from growth curves in HI medium at 28°C and 37°C using a Tecan infinite M200 plate reader (Männedorf, Switzerland) for 24 h. For experiments measuring tdh promoter activity, plasmid pVCW10C1 was maintained by the addition of chloramphenicol (CHL) at a final concentration of 25 μg/ml. Triparental matings were performed on super optimal broth (SOB) plates containing 2% Bactotryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgS04, and 1.5% agar at 37°C for 12 h, and outgrowth was performed on HI-CHL at 28°C. For quantitative hemolysin assays conducted in conditioned broth, strain BB22 was grown to an optical density at 600 nm (OD600) of 1.0, the cells pelleted by centrifugation and discarded, and the cleared broth filter sterilized. Strains were inoculated into a 1:1 mix of rich HI medium and BB22 cell-free supernatant.
Characterization of virulence-associated phenotypes.
All phenotypic characterization assays were conducted a minimum of three times. Plate assays were conducted for motility, protease, and siderophore in cultures grown in HI with shaking and then standardized to an OD600 of 0.5 prior to inoculation. Throughout our phenotypic characterization experiments, the OD600 was often determined from 150 μl of culture in a 96-well plate, and the value converted to a standard OD600 per ml based on an empirically determined conversion factor so that our data would be comparable to those of other studies. Motility was measured using 25-ml soft agar plates containing 10 g tryptone, 20 g NaCl, and 3.35 g Bacto agar per liter as previously described (24). The plates were inoculated with 2 μl of each isolate in triplicate. After 24 h, the diameter of the zone of bacterial migration was measured in millimeters. Protease production was assessed with HI plates to which 4 g/liter sterilized skim milk was added following autoclaving and cooling to 50°C. The plates were inoculated in triplicate with 5 μl of each isolate. A positive result was indicated by a clearing around the inoculum after 24 h. Siderophore production was determined from cultures spotted onto artificial-seawater-based chrome azurol S (CAS) agar buffered with 100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.8 (46). A positive result was indicated by an orange halo around siderophore-producing cells. Hemolytic activity was determined qualitatively using Wagatsuma agar (33). Cultures were grown either to mid-log or overnight, and 5-μl amounts were spotted on the plates in triplicate. Plates were examined at 24 h, and a beta-hemolytic clearing around the colony indicated a positive result.
Biofilm assays were performed according to a standard protocol (41), with slight modifications. Strains were inoculated into 200 μl of HI medium per well of a polystyrene microplate (Costar, St. Louis, MO) and grown for 4 h at either 28°C or 37°C with shaking at 200 rpm. The OD595 was determined for baseline optical density. The cultures were then expelled from the plate, which was washed twice with 200 μl of distilled water per well. The plates were allowed to dry, and then the cells were fixed at 80°C for 30 min. Following staining with 0.1% crystal violet for 20 min, the wells were destained with 200 μl 95% ethanol. The OD595 was measured, and the biofilm quantified as absorbance normalized to the cell density of the initial culture (OD600). When cultures were grown for more than 4 h, the biofilm production was so great that clumped cells were sometimes expelled with the medium, making biofilm measurements highly variable.
A quantitative hemolysin assay was adapted from a previously published assay (15). In brief, 150-μl amounts of cultures taken throughout the growth cycle were combined with 800 μl of a blood mix of 40 ml 1× phosphate-buffered saline (PBS) plus 500 μl defibrinated sheep blood (Northeast Laboratories Services, Winslow, ME), and cells were pelleted by centrifugation at 9,000 rpm (6,605 × g) for 4 min. The tubes were incubated for 18 h, and the cell wall debris removed by centrifugation at 13,000 rpm. The amount of heme present in cleared cell lysate as determined by the OD415 was used as a measure of red blood cell lysis and normalized to the bacterial cell density (OD600).
In order to determine the potential role of late-growth-phase extracellular or cell-associated proteases in the degradation of hemolysins, we performed a time course hemolysin assay with exposure to either late-phase cell-free supernatant or sonicated whole cells. Strain MDOH-03-17282-Vp was grown to an OD600 value of 1.0, when hemolysin is optimally produced, and an OD600 value of 2.5, when hemolysin activity was substantially decreased. The cells were pelleted, and the supernatants were purified with a 0.2-μm polyethersulfone filter (VWR, West Chester, Pennsylvania). The cell pellet was then sonicated in 1 ml of sterile 1× PBS to release cell-associated proteases. The supernatant or sonicated cells were then combined with 1 ml of extracted hemolysin, and hemolytic activity against blood mix was assayed as described above every 30 min for 3 h. As a control, cell-free hemolysin was also incubated in HI.
Cytotoxicity assays were developed using a human gastrointestinal epithelial cell line (CaCo-2) grown in Eagle's minimal essential medium (Mediatech, Inc., Herndon, VA) modified with HEPES (J.T.Baker, Mumbai, India) and supplemented with 0.7% Leibovitz's L-15 (Mediatech, Inc., Herndon, VA), 0.03% l-glutamine (Lonza, Basel, Switzerland), 0.08% sodium bicarbonate (J.T.Baker, Phillipsburg, NJ), 1% nonessential amino acids (Thermo Scientific HyClone, Logan, UT), 10% fetal bovine serum (Lonza, Basel, Switzerland), and 1% penicillin-streptomycin-amphotericin B (100×) (Lonza, Basel, Switzerland) at a pH of 7.0 with no CO2. Preliminary experiments were conducted to determine the optimal multiplicity of infection (MOI), using strain BB22 as a positive control, because it was previously determined to be highly cytotoxic (L. L. McCarter, personal communication), and HI broth as a negative control. Exposure to each V. parahaemolyticus strain, grown overnight with shaking at an MOI of 100, was carried out in triplicate in a 384-well plate. Following incubation for at least 5 h at 37°C with 0% CO2, the relative cytotoxicity was measured by lactate dehydrogenase release, quantified by using a Cytotox-Fluor cytotoxicity assay, following the manufacturer's protocols (Promega, Madison, WI), and read on a Tecan Infinite M200 plate reader. The experiment was conducted four times. Each experiment had low within-experiment variability, and the data from one representative experiment are presented.
Analysis of significance of differences in cytotoxicity and correlations of phenotypes with temperature using replicate experiments were conducted by one-way analysis of variance (ANOVA) using the statistical software SPSS (SPSS, Inc., Chicago, IL).
Expression of tdh.
To measure tdh gene expression, a tdh::gfp promoter fusion was generated by splicing-by-overlap extension (SOE)-PCR following published protocols (22), using the Expand long template PCR system (Roche Applied Science, Indianapolis, IN). The outer forward primer (tdhF1, TATGTCGACCAGATTTCTCGCTTGTGC) was designed to the tdh gene (VPA1314) from the RIMD2210633 published sequence. The outer reverse primer (gfpR1, TATCTGCAGGTTGTACAGTTCATCCATGC) was designed to the gfp gene from pQBI63 (Quantum Biotechnology, Montreal, Canada). Internal forward and reverse SOE primers were designed to fuse the gfp gene to the tdh promoter at its ribosomal binding site, ensuring correct spacing of the gene within the transcript (tdhSOEF6, AAGTTATTAATCAATTCAGAGGAGGAGAATACTAATGGC, and tdhSOER6, GCCATTAGTATTCTCCTCCTCTGAATTGATTAATAACTT). The first PCR amplicons were generated with primers tdhF1 and tdhSOER6 using strain BB22 as a template and with tdhSOEF6 and gfpR1 using pVSV102 as a template for gfp (14), and the second round of SOE was performed with the outer primers (tdhF1 and fgpR1) only. The amplicon was directionally cloned into the low-copy-number plasmid pVSV105 (14) so that expression was driven by the native tdh promoter only. The newly generated construct (pVCW10C1) was then introduced into strains BB22, DAL1094, MDOH-03-17282-Vp, MDOH-04-5M732, KE9967, G1, G91, and G145 by conjugation.
The expression of the tdh gene was determined throughout the growth cycle of each strain inoculated in quadruplicate into 150 μl HI-CHL in the wells of a clear-bottom black polystyrene microplate (Costar, St. Louis, MO) and grown at either 28°C or 37°C for 24 h. At each time point, the OD600 was used to determine cell growth, and tdh expression was determined by emission at a wavelength of 515 nm following excitation at a wavelength of 480 nm using a Tecan Infinite M200 plate reader with a manual gain of 175. The background fluorescence produced by reference strain BB22 was subtracted from each value, and the fluorescence normalized to the OD600. Upon determination of the induction profile of gfp in each strain, a single postinduction time point (OD600 = 1) was chosen for direct comparison between strains at each temperature.
RESULTS AND DISCUSSION
Although the environmental strains examined in this study were confirmed to lack the tdh/trh virulence-associated genes, we further genotypically characterized these strains for five genes putatively involved in virulence, as previously described, most notably in other environmental isolates without the classical virulence markers tdh and trh (4). Although none harbored the putative T3SS2 effector protein vopP, some strains were positive for a gene involved in its secretory apparatus, as well as a homolog of a gene found within a Vibrio cholerae pathogenicity island (Table 1). Despite the presence of one or more of these putative virulence genes in a small subset of environmental strains, which could prove to be relevant to their virulence potential, this cohort did not display enhanced or altered virulence patterns in the phenotypic characterization assays. This may be the result of genes that are present in a genome but not functionally expressed or, possibly, a lack of connection between these putative virulence genes and the phenotypes for which we screened.
The environmental strains did show evidence of more conserved virulence-associated mechanisms, such as motility and biofilm, siderophore, and protease production (27, 52, 53, 54). Specifically, the environmental and clinical strains were motile and produced extracellular proteases, biofilm, and siderophore at both 28°C and 37°C (see Table S1 in the supplemental material). Unlike the environmental isolates, the clinical group showed a notable difference in response to the human host temperature of 37°C (Table 2). In particular, we observed a significant increase in motility and in biofilm and protease production at 37°C compared to the results at 28°C in the clinical strains, whereas the environmental isolates showed no significant difference in motility or biofilm production and showed a significant decrease in protease production at 37°C. Further underscoring the difference between the environmental and clinical strains, a direct comparison of each population at 37°C indicates that the environmental group produces significantly less protease (P < 0.001) and less biofilm (P = 0.02) than the clinical group, although due to high variability among strains, they were not significantly different in motility. Siderophore was detected in all strains at both 28°C and 37°C, with the exception of the reference strain BB22 (see Table S1 in the supplemental material). Although siderophore production in other Vibrio spp., specifically Vibrio (Aliivibrio) salmonicida, has been observed only in response to disease-permissive temperatures (9), the production of siderophore by V. parahaemolyticus at both temperatures indicates that it may play a role both in infection and in environmental fitness.
TABLE 2.
Phenotypic characterization of both the environmental and the clinical population
| Population, phenotypic characteristic | Effect of temp shift (28°C to 37°C) | Avg fold Δ | P value |
|---|---|---|---|
| Clinical isolates from outbreaks | |||
| Motility | Significant increase at 37°C | 1.2 | <0.0001 |
| Biofilm production | Significant increase at 37°C | 9 | 0.01 |
| Siderophore production | No trend observed | 0 | |
| Protease production | Significant increase at 37°C | 4 | 0.001 |
| Environmental isolates from New Hampshire | |||
| Motility | No trend observed | 0 | 0.336 |
| Biofilm production | No trend observed | 0 | 0.092 |
| Siderophore production | No trend observed | 0 | |
| Protease production | Significant decrease at 37°C | 2 | <0.0001 |
Bacterial pathogens that oscillate between the natural habitat and a mammalian host commonly utilize environmental cues to coordinate virulence-associated gene expression (48). Though a variety of signals exist within the human host, temperature has been implicated as an important activator of virulence gene expression in several gastrointestinal pathogens, including Shigella spp., Yersinia spp., and pathogenic Escherichia coli (18, 30, 38). Pathogenic Shigella spp., for example, are unable to successfully penetrate human epithelial cells and have attenuated virulence at 30°C that is fully restored upon a temperature increase to 37°C (30). Furthermore, Vibrio parahaemolyticus strains that are positive for the virulence-associated gene urease exhibit an increase in urease expression at 37°C compared to its expression at 30°C (45). The specific response of virulence phenotypes, including hemolysin (see Table S1 in the supplemental material), to host temperature seen in this study may be reflective of the fundamental regulatory differences between environmental and clinical strains. This unique regulatory switch could provide a selective advantage in the human host environment, promoting infection and disease. Despite the presence of some functional virulence-associated factors in the environmental population, the strains are apparently unable to coordinate the production of these factors in response to this host cue. The data presented herein support previous work implicating temperature-dependent regulation in the appropriate expression of putative virulence genes in other bacterial pathogens and further clarify the differences between environmental and clinical strains of V. parahaemolyticus.
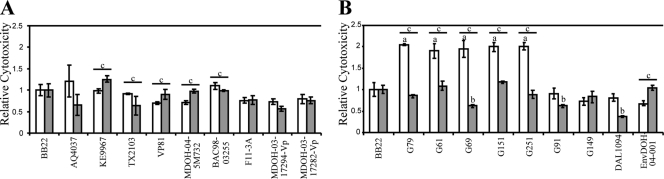
Cytotoxicity toward human gastrointestinal cells is widely used to assess bacterial virulence and was used here to assess the virulence of both clinical and environmental strains (19). Clinical isolates of V. parahaemolyticus contain a conserved T3SS1 (28) and horizontally acquired genes, including tdh and T3SS2 (25, 44), that are known to contribute to the lysis of epithelial cells through disruption of cytoskeletal structures and a loss of membrane integrity, possibly leading to secretory diarrhea (5, 35). The clinical strains all displayed significant cell lysis ability at both 28°C and 37°C, which perhaps implies that this mechanism is important both in the natural environment and during human infection (Fig. 1A). In comparison, the majority of environmental strains were as cytotoxic as a reference strain at 37°C but had overall increased cytotoxicity at 28°C (Fig. 1B). The upregulation of a virulence mechanism at a more environmentally relevant temperature further conflicts with the behavior of the virulent clinical strains. Strains G91 and G149 represent a small subgroup of environmental isolates that showed a different trend, with low cytotoxicity at both temperatures (Fig. 1B). The elevated cytotoxicity seen in the environmental group in response to a cooler temperature (28°C) may imply the utility of cytotoxicity in the natural environment, perhaps during interactions with a eukaryotic marine host. However, the high level of cytotoxicity in environmental strains may also indicate that cytotoxicity is not an accurate measure of the virulence capacity of a given strain of V. parahaemolyticus and that the development of other in vitro or in situ models is critical for understanding the pathogenic capability of this emergent pathogen.
FIG. 1.
Cytotoxicities of clinical strains and select environmental isolates relative to that of reference strain BB22. (A and B) Average cytotoxicity of clinical (A) and environmental (B) strains toward human Caco-2 cells at 28°C (white bars) and 37°C (gray bars) relative to the cytotoxicity of BB22. Cytotoxicity was determined in four separate experiments; however, because of a high level of variability, the values for one representative experiment are shown (the trends were consistent for the strains shown here throughout all experiments). Values represent the averages of three replicates; error bars show standard errors. One-way ANOVAs were performed for all strains, and a significant difference relative to the result for BB22 is denoted by “a” for a significant increase in relative cytotoxicity at 28°C and “b” for a significant decrease at 37°C; “c” denotes a statistically significant change in a single strain at 37°C compared to its cytotoxicity at 28°C.
Unlike the conserved, virulence-associated traits we investigated in both environmental and clinical strains, the putative virulence factor hemolysin is likely the result of horizontal acquisition of the tdh gene on Vibrio pathogenicity island-7 (8, 34) and/or the trh gene on Vp-PAITH3996,. These pore-forming toxins are solely responsible for the Kanagawa phenomenon that is the defining characteristic of many clinical strains (8). The environmental isolates used in this work lacked tdh and trh (Table 1), and as expected, they showed no hemolytic activity at either 28°C or 37°C on Wagatsuma agar (see Table S1 in the supplemental material) (33). The clinical strains were not hemolytic at 28°C but were strongly hemolytic at 37°C (see Table S1 in the supplemental material). Although no comprehensive study has been completed on temperature regulation of hemolysin in V. parahaemolyticus, some observations have been made as part of other studies. For instance, V. parahaemolyticus induced to the viable but nonculturable state by cold and then exposed to temperature upshifts to 37°C did not show tdh expression (10), but heat shock at 42°C did induce TDH production in a separate study (7). The dramatic response of this putative virulence phenotype to human host temperature across a group of diverse clinical strains agrees with its role in pathogenesis and provides us with new insights about host cues, specifically temperature, involved in hemolysin activation.
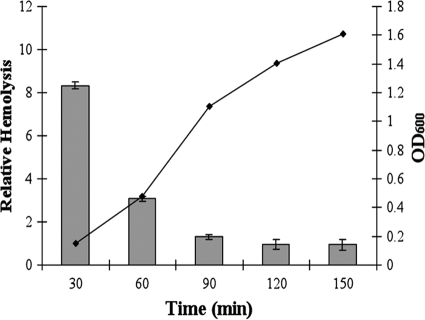
In addition to activation in response to increased temperature, hemolysin production correlated inversely with culture density (Fig. 2). Specifically, hemolysins are present at high levels during early log phase and decrease as the OD600 increases. Virulence gene repression at high densities in both Vibrio harveyi and Vibrio parahaemolyticus is known to result from quorum-sensing regulation (17). In order to assess the possible involvement of quorum sensing in the decrease of hemolysin during late growth phase, hemolysin assays were completed with cultures grown in medium conditioned with cell-free supernatants from late-log-phase cultures of the reference strain BB22. We anticipated that if the repression of hemolysin was in response to quorum-sensing signals that accumulate in the medium, then hemolytic activity would decrease more rapidly in conditioned broth. We saw no effect on hemolysin production (data not shown), indicating that hemolysin regulation is likely the result of a cue(s) separate from quorum sensing. Next, we examined the transcription of tdh using a tdh::gfp promoter fusion normalized to cell density, which showed no decline in expression as cell density increased but, rather, showed a plateau in expression, supporting the idea that the decrease in hemolysis was not transcriptional (data not shown). Finally, we tested the role of extracellular and/or cell-associated proteases in hemolysin degradation. Complete loss of hemolysis was achieved after incubation with late-log-phase cell-free supernatants (data not shown), indicating that proteolysis is the likely mechanism of hemolysin turnover.
FIG. 2.
Quantitative hemolysis by a select clinical strain at 37°C compared to cell density. Red blood cell lysis was determined over time, and relative hemolytic activity was determined (OD415/OD600) for one representative strain, MDOH-03-17282-Vp (bars), throughout the growth cycle (black line; OD600).
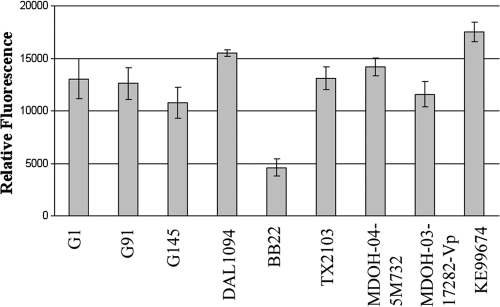
Not only do the environmental strains lack the tdh/trh putative virulence genes, but they also lack the ability to regulate other conserved virulence-associated traits in response to human body temperature. The underlying questions of how existing and acquired virulence genes are appropriately regulated in response to temperature and, more specifically, whether this regulation is ancient or obtained horizontally with acquired virulence genes remain unanswered. We addressed this question by introducing a tdh::gfp promoter fusion into various environmental and clinical strains and assessing the activity of the promoter at the two experimental temperatures. Green fluorescent protein (GFP) was only detected above background levels at 37°C and not at 28°C degrees in all environmental and clinical strains tested (Fig. 3). The early induction of GFP in all strains by the time the OD600 reached 0.5 and the rapid plateau of GFP by the time the OD600 reached 1.0 are in agreement with the hemolysin phenotype seen in the quantitative assays (Fig. 2) and could mirror early infection. Virulence traits that are acquired horizontally must be regulated in coordination with the rest of the genome in order to confer enhanced pathogenicity. During the transition from an environmental setting to a host environment, the bacterial cell must integrate an assortment of signals and respond with appropriate activation or repression of both conserved and newly acquired genes. Although a variety of unique regulatory mechanisms exist that direct the expression of ancient genes, the mechanisms that regulate newly acquired HGT components are less clear (26). In recent years, a body of evidence has grown showing that a predominant regulatory mechanism for both existing and horizontal genes is the temperature-sensitive histone-like nucleoid-structuring protein (H-NS) (2, 13, 56). H-NS is a globally acting repressor that appears to target and “police” the expression of new DNA (1). Upon exposure to a signal, such as increased temperature, H-NS silencing is relieved and the transcription of virulence genes is activated (50, 55). Given that the tdh gene is located within a horizontally acquired pathogenicity island (23) and is induced at higher temperatures, H-NS silencing is a probable mechanism for the temperature-dependent expression we observed. If all virulence phenotypes are under the regulatory control of H-NS, then the disparity between environmental strain regulation of tdh but not conserved virulence-associated traits in response to temperature indicates that the regulation of virulence is more complex, perhaps suggestive of different niche adaptations.
FIG. 3.
Induction of the tdh promoter in clinical and environmental strains. Promoter activity of a tdh::gfp promoter fusion harbored in trans in a low-copy-number plasmid is presented as relative fluorescence (fluorescence/OD600) for various strains during maximal production (OD600 ∼ 1.0) as determined from time course assays for cultures grown at 37°C.
Since the majority of the environmental strains in this study were isolated from the same cooler Northern population, it is possible that the temperature dysregulation we observed in environmental strains is unique to this population. It is, however, notable that the one clinical strain isolated from Washington, also a cooler climate, did trend more closely with the clinical strains and not with the cool-water environmental strains (see Table S1 in the supplemental material), supporting the hypothesis that temperature regulation is an attribute of clinical strains. Still, the two environmental strains isolated from warmer climates (both of which were environmentally isolated but associated with an outbreak) did show some temperature regulation similar to that in the clinical group (see Table S1 in the supplemental material). A broader sampling of environmental strains will be necessary to fully address the question of whether northern strains are unique in their regulation. Ultimately, temperature regulation could still be an important consideration in determining the virulence potential of strains from such climates.
Although the environmental strains do not contain tdh or the VPaI-7 island, they do have the capacity to regulate tdh in response to an increase in temperature and growth phase. Thus, the regulatory mechanisms involved in managing newly acquired virulence elements are present even in nonclinical strains of V. parahaemolyticus. This provides evidence that the control of new virulence genes is ancient and that horizontally acquired components are incorporated into existing regulatory networks for proper expression.
Supplementary Material
Acknowledgments
We thank Brian Schuster, Megan Striplin, and Colin Edwards for assistance with collection of environmental V. parahaemolyticus and Vaughn S. Cooper for helpful suggestions.
This work was funded by Sea Grant number R/CE-137, National Institutes of Health grants R03AI081102 and R03AI076831, and a fellowship from the Great Bay Stewards.
Footnotes
Published ahead of print on 1 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Banos, R. C., A. Vivero, S. Aznar, J. Garcia, M. Pons, C. Madrid, and A. Juarez. 2009. Differential regulation of horizontally acquired and core genome genes by the bacterial modulator H-NS. PLoS Genet. 5:e1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartley, C. H., and L. W. Slanetz. 1971. Occurrence of Vibrio parahaemolyticus in estuarine waters and oysters of New Hampshire. Appl. Microbiol. 21:965-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caburlotto, G., M. Gennari, V. Ghidini, M. Tafi, and M. Lleo. 2009. Presence of T3SS2 and other virulence-related genes in tdh-negative Vibrio parahaemolyticus environmental strains isolated from marine samples in the area of the Venetian Lagoon, Italy. FEMS Microb. Ecol. 70:506-514. [DOI] [PubMed] [Google Scholar]
- 5.Caburlotto, G., M. M. Lleò, T. Hilton, A. Huq, R. R. Colwell, and J. B. Kaper. 2010. Effect on human cells of environmental Vibrio parahaemolyticus strains carrying type III secretion system 2. Infect. Immun. 78:3280-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2006. Vibrio parahaemolyticus infections associated with consumption of raw shellfish—three states, 2006. MMWR Morb. Mortal. Wkly. Rep. 55:854-856. [PubMed] [Google Scholar]
- 7.Chiang, M. L., and C. C. Chou. 2008. Expression of superoxide dismutase, catalase and thermostable direct hemolysin by, and growth in the presence of various nitrogen and carbon sources of heat-shocked and ethanol-shocked Vibrio parahaemolyticus. Int. J. Food Microbiol. 121:268-274. [DOI] [PubMed] [Google Scholar]
- 8.Chun, D., J. K. Chung, R. Tak, and S. Y. Seol. 1975. Nature of the Kanagawa phenomenon of Vibrio parahaemolyticus. Infect. Immun. 12:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colquhoun, D. J., and H. Sørum. 2001. Temperature dependent siderophore production in Vibrio salmonicida. Microb. Pathog. 31:213-219. [DOI] [PubMed] [Google Scholar]
- 10.Coutard, F., S. Lozach, M. Pommepuy, and D. Hervio-Heath. 2007. Real-time reverse transcription-PCR for transcriptional expression analysis of virulence and housekeeping genes in viable but nonculturable Vibrio parahaemolyticus after recovery of culturability. Appl. Env. Microbiol. 73:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, C. R., D. L. Wingfield, K. K. Peak, W. Veguilla, P. T. Amuso, A. C. Cannons, and J. Cattani. 2007. Molecular characterization of Vibrio parahaemolyticus strains associated with foodborne illness in Florida. J. Food Prot. 70:2396-2401. [DOI] [PubMed] [Google Scholar]
- 12.DePaola, A., J. Ulazek, C. A. Kaysner, B. J. Tenge, J. L. Nordstrom, J. Wells, N. Purh, and S. M. Gendel. 2003. Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Environ. Microbiol. 69:3999-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, A. K., D. S. Millikan, D. M. Adin, J. L. Bose, and E. V. Stabb. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan, J.-J., C.-P. Shao, Y.-C. Ho, C.-K. Yu, and L.-I. Hor. 2001. Isolation and characterization of a Vibrio vulnificus mutant deficient in both extracellular metalloprotease and cytolysin. Infect. Immun. 69:5943-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia, K., R. Torres, P. Uribe, C. Hernandez, M. L. Rioseco, J. Romero, and R. T. Espejo. 2009. Dynamics of clinical and environmental Vibrio parahaemolyticus strains during seafood-related summer diarrhea outbreaks in southern Chile. Appl. Environ. Microbiol. 75:7482-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henke, J. M., and B. L. Bassler. 2004. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 186:3794-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herbst, K., M. Bujara, A. K. Heroven, W. Opitz, M. Weichert, A. Zimmermann, and P. Dersch. 2009. Intrinsic thermal sensing controls proteolysis of Yersinia virulence regulator RovA. PLoS Pathog. 5:e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiyoshi, H., T. Kodama, T. Iida, and T. Honda. 2010. Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect. Immun. 78:1772-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda, T., and T. Iida. 1993. The pathogenicity of Vibrio parahaemolyticus and the role of thermostable direct hemolysin and related hemolysins. Rev. Med. Microbiol. 4:106-113. [Google Scholar]
- 21.Reference deleted.
- 22.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528-535. [PubMed] [Google Scholar]
- 23.Hurley, C. C., A. Quirke, F. J. Reen, and E. F. Boyd. 2006. Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates. BMC Genomics 7:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaques, S., and L. L. McCarter. 2006. Three new regulators of swarming in Vibrio parahaemolyticus. J. Bacteriol. 188:2625-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kodama, T., M. Rokuda, K. S. Park, V. V. Cantarelli, S. Matsuda, T. Iida, and T. Honda. 2007. Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system. Cell. Microbiol. 9:2598-2609. [DOI] [PubMed] [Google Scholar]
- 26.Lercher, M. J., and C. Pal. 2008. Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol. Biol. Evol. 25:559-567. [DOI] [PubMed] [Google Scholar]
- 27.Luan, X., J. Chen, X. H. Zhang, Y. Li, and G. Hu. 2007. Expression and characterization of a metalloprotease from a Vibrio parahaemolyticus isolate. Can. J. Microbiol. 53:1168-1173. [DOI] [PubMed] [Google Scholar]
- 28.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of Vibrio cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Urtaza, J., A. Lozano-Leon, A. DePaola, M. Ishibashi, K. Shimada, M. Nishibuchi, and E. Liebana. 2004. Characterization of pathogenic Vibrio parahaemolyticus isolates from clinical sources in Spain and comparison with Asian and North American pandemic isolates. J. Clin. Microbiol. 42:4672-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurelli, A. T., B. Blackmon, and R. Curtiss III. 1984. Temperature-dependent expression of virulence genes in Shigella species. Infect. Immun. 43:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarter, L. 1998. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 180:3166-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLaughlin, J. B., A. DePaola, C. A. Bopp, K. A. Martinek, N. P. Napolilli, C. G. Allison, S. L. Murray, E. C. Thompson, M. M. Bird, and J. P. Middaugh. 2005. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N. Engl. J. Med. 353:1463-1470. [DOI] [PubMed] [Google Scholar]
- 33.Miyamoto, Y., K. Teiji, O. Yasushi, A. Shoichi, K. Akiyama, T. Kinjiro, and Y. Shiro. 1969. In vitro hemolytic characteristic of Vibrio parahaemolyticus: its close correlation with human pathogenicity. J. Bacteriol. 100:1147-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers, M. L., G. Panicker, and A. K. Bej. 2003. PCR detection of a newly emerged pandemic Vibrio parahaemolyticus O3:K6 pathogen in pure cultures and seeded waters from the Gulf of Mexico. Appl. Environ. Microbiol. 69:2194-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishibuchi, M., A. Fasano, R. G. Russell, and J. B. Kaper. 1992. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect. Immun. 60:3539-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nolan, C. M., J. Ballard, C. A. Kaysner, J. L. Lilja, L. P. Williams, Jr., and F. C. Tenover. 1984. Vibrio parahaemolyticus gastroenteritis. An outbreak associated with raw oysters in the Pacific Northwest. Diagn. Microbiol. Infect. Dis. 2:119-128. [DOI] [PubMed] [Google Scholar]
- 37.Okada, N., T. Iida, K. S. Park, N. Goto, T. Yasunaga, H. Hiyoshi, S. Matsuda, T. Kodama, and T. Honda. 2009. Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect. Immun. 77:904-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsen, P. B., M. A. Schembri, D. L. Gally, and P. Klemm. 1998. Differential temperature modulation by H-NS of the fimB and fimE recombinase genes which control the orientation of the type 1 fimbrial phase switch. FEMS Microbiol. Lett. 162:17-23. [DOI] [PubMed] [Google Scholar]
- 39.O'Neil, K. R., S. H. Jones, and D. J. Grimes. 1990. Incidence of Vibrio vulnificus in northern New England water and shellfish. FEMS Microbiol. Lett. 60:163-167. [DOI] [PubMed] [Google Scholar]
- 40.Osawa, R., A. Iguchi, E. Arakawa, and H. Watanabe. 2002. Genotyping of pandemic Vibrio parahaemolyticus O3:K6 still open to question. J. Clin. Microbiol. 40:2708-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to the study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 42.Panicker, G., R. Douglas, M. Call, M. J. Krug, and A. K. Bej. 2004. Detection of pathogenic Vibrio spp. in shellfish by using multiplex PCR and DNA microarrays. Appl. Environ. Microbiol. 70:7436-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park, K. S., T. Iida, Y. Yaaichi, T. Oyagi, K. Yamamoto, and T. Honda. 2000. Genetic characterization of DNA region containing the trh and ure genes of Vibrio parahaemolyticus. Infect. Immun. 68:5742-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park, K. S., T. Ono, M. Rokuda, M. Jang, K. Okada, T. Iida, and T. Honda. 2004. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72:6659-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, K. S., S. J. Lee, Y. H. Chung, T. Iida, and T. Honda. 2009. Temperature-dependency urease activity in Vibrio parahaemolyticus is related to transcriptional activator UreR. J. Microbiol. Biotechnol. 19:1456-1463. [DOI] [PubMed] [Google Scholar]
- 46.Payne, S. 1994. Detection, isolation and characterization of siderophores. Methods Enzymol. 235:329-344. [DOI] [PubMed] [Google Scholar]
- 47.Raimondi, F., J. P. Kao, J. B. Kaper, S. Guandalini, and A. Fasano. 1995. Calcium-dependent intestinal chloride secretion by Vibrio parahaemolyticus thermostable direct hemolysin in a rabbit model. Gastroenterology 109:381-386. [DOI] [PubMed] [Google Scholar]
- 48.Roux, A., S. M. Payne, and M. S. Gilmore. 2009. Microbial telesensing: probing the environment for friends, foes, and food. Cell Host Microbe 6:115-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiraih, H., H. Ito, T. Hirayama, Y. Nakamoto, N. Nakabayashi, K. Kumagai, Y. Takeda, and M. Nishibuchi. 1990. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58:3568-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoebel, D. M., A. Free, and C. J. Dormam. 2008. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 154:2533-2545. [DOI] [PubMed] [Google Scholar]
- 51.Striplin, M., J. C. Mahoney, V. S. Cooper, C. A. Whistler, and S. H. Jones. 2008. Identification of Vibrio spp. found in oysters and water from the Great Bay Estuary, abstr. N-304, p. 283. Abstr. 109th Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 52.Tamayo, R., B. Patimalla, and A. Camilli. 2010. Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect. Immun. 78:3560-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsou, A. M., E. M. Frey, A. Hsiao, Z. Liu, and J. Zhu. 2008. Coordinated regulation of virulence by quorum sensing and motility pathways during the initial stages of Vibrio cholerae infection. Commun. Integr. Biol. 1:42-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, Q., Q. Liu, X. Cao, M. Yang, and Y. Zhang. 2010. Characterization of two TonB systems in marine fish pathogen Vibrio alginolyticus: their roles in iron utilization and virulence. Arch. Microbiol. 190:595-603. [DOI] [PubMed] [Google Scholar]
- 55.White-Ziegler, C. A., M. L. Angus Hill, B. A. Braaten, M. W. van der Woude, and D. A. Low. 1998. Thermoregulation of Escherichia coli pap transcription: H-NS is a temperature-dependent DNA methylation blocking factor. Mol. Microbiol. 28:1121-1137. [DOI] [PubMed] [Google Scholar]
- 56.White-Ziegler, C. A., and T. R. Davis. 2009. Genome-wide identification of H-NS-controlled, temperature-regulated genes in Escherichia coli K-12. J. Bacteriol. 191:1106-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, M., K. Yamamoto, T. Honda, and X. Ming. 1994. Construction and characterization of an isogenic mutant of Vibrio parahaemolyticus having a deletion in the thermostable direct hemolysin-related hemolysin gene (trh). J. Bacteriol. 176:4757-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.