Abstract
The catabolism of the disulfide 3,3′-dithiodipropionic acid (DTDP) is initiated by the reduction of its disulfide bond. Three independent Tn5::mob-induced mutants of Advenella mimigardefordensis strain DPN7T were isolated that had lost the ability to utilize DTDP as the sole source of carbon and energy and that harbored the transposon insertions in three different sites of the same dihydrolipoamide dehydrogenase gene encoding the E3 subunit of the pyruvate dehydrogenase multi-enzyme complex of this bacterium (LpdAAm). LpdAAm was analyzed in silico and compared to homologous proteins, thereby revealing high similarities to the orthologue in Ralstonia eutropha H16 (PdhLRe). Both bacteria are able to cleave DTDP into two molecules of 3-mercaptopropionic acid (3MP). A. mimigardefordensis DPN7T converted 3MP to 3-sulfinopropionic acid, whereas R. eutropha H16 showed no growth with DTDP as the sole carbon source but was instead capable of synthesizing heteropolythioesters using the resulting cleavage product 3MP. Subsequently, the genes lpdAAm and pdhLRe were cloned, heterologously expressed in Escherichia coli applying the pET23a expression system, purified, and assayed by monitoring the oxidation of NADH. The physiological substrate lipoamide was reduced to dihydrolipoamide with specific activities of 1,833 mkat/kg of protein (LpdAAm) or 1,667 mkat/kg of protein (PdhLRe). Reduction of DTDP was also unequivocally detected with the purified enzymes, although the specific enzyme activities were much lower: 0.7 and 0.5 mkat/kg protein, respectively.
In Advenella mimigardefordensis strain DPN7T (15, 42), three independent mutants with an insertion of Tn5::mob in the lpdA gene coding for the E3 component of the pyruvate dehydrogenase multi-enzyme complex revealed an interesting phenotype: these mutants were fully impaired in utilizing 3,3′-dithiodipropionic acid (DTDP) as the sole carbon and energy source, whereas the growth on no other tested carbon sources was affected (41). Our main interest in the catabolism of DTDP is to unravel the pathway and to identify the involved enzymes. Furthermore, the application of this disulfide as precursor substrate for biotechnological production of polythioesters (PTE) (22) is of interest. Since poly(3-mercaptopropionate) (PMP) biosynthesis depends hitherto on supplying the harmful thiol 3-mercaptopropionic acid (3MP) (35), an improvement of the recombinant Escherichia coli system by heterologous expression of enzymes capable of cleaving the less toxic DTDP symmetrically into two molecules of 3MP, which are then polymerized, could be an important achievement toward large-scale biotechnological production of PMP.
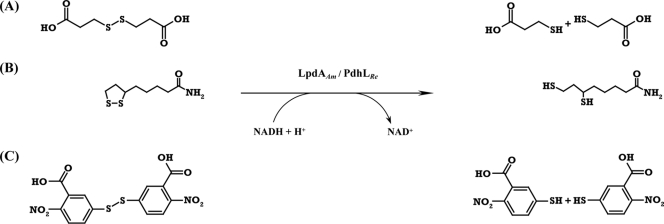
Two different enzyme systems catalyzing the conversion of disulfides into the corresponding thiols are already known and have been described in detail. (i) Enzymes belonging to the well-characterized family of pyridine-nucleotide disulfide oxidoreductases (25) contain a redox center formed by a disulfide bridge coupled to a flavin ring. They catalyze a simultaneous two-electron transfer via the enzymatic active disulfides associated with the pyridine nucleotides and flavin, toward the substrate (39, 40). (ii) An alternative disulfide reduction is catalyzed by enzymes using iron-sulfur clusters to cleave of disulfide substrates in two one-electron reduction steps (37). The disrupted gene in A. mimigardefordensis was designated lpdAAm (EC 1.8.1.4), and it encodes a homodimeric flavoprotein, the dihydrolipoamide dehydrogenase LpdAAm (i.e., the E3 component of the pyruvate dehydrogenase multi-enzyme complex of A. mimigardefordensis strain DPN7T) belonging to the above-mentioned family of pyridine nucleotide-disulfide oxidoreductases. Enzymes of this class share high sequence and structural similarities and catalyze reduction of compounds which are linked by disulfide bonds (38). Alkylhydroperoxide reductases, coenzyme A disulfide reductases, glutathione reductases, mycothione reductases, thioredoxin reductases, and trypanothione reductases also, in addition to dihydrolipoamide dehydrogenases, belong to this family (3, 38). The physiological function of LpdAAm is most probably the conversion of lipoamide to dihydrolipoamide, but the reduction of DTDP into two molecules of 3MP (Fig. 1) is also predicted, enabling the first step in DTDP catabolism in A. mimigardefordensis strain DPN7T (41).
FIG. 1.
Reactions catalyzed by LpdAAm and PdhLRe. Presented are the enzymatic conversions of DTDP into two molecules of 3MP (A), lipoamide into dihydrolipoamide (B), and DTNB into two molecules of NTB (C). Abbreviations: DTDP, 3,3′-dithiodipropionic acid; 3MP, 3-mercaptopropionic acid; DTNB, 5,5′-dithiobis-(2-nitrobenzoic acid); NTB, 2-nitro-5-thiobenzoic acid.
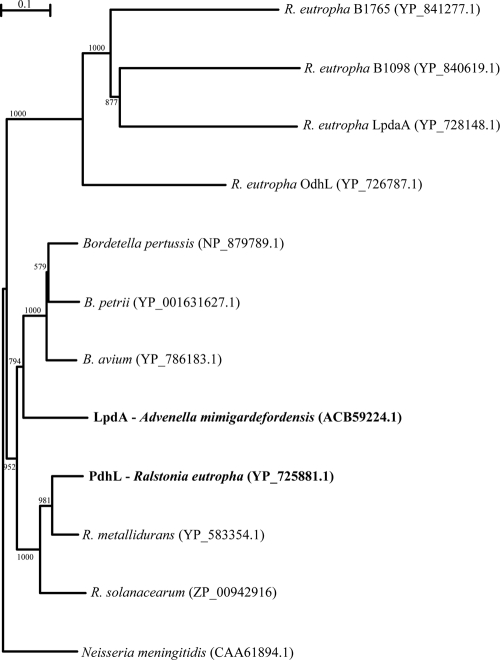
Ralstonia eutropha H16 synthesizes copolymers of 3-hydroxybutyrate and 3MP, if 3MP (23) or DTDP (22) is supplied as a precursor in addition to a second utilizable carbon source. Although R. eutropha is not able to grow with DTDP as the sole carbon source, it must be capable of cleaving this organic disulfide symmetrically, because it synthesizes from it heteropolymers containing the resulting 3MP. Thus, R. eutropha must possess at least one gene encoding a DTDP-cleaving enzyme. Five genes coding for homologues of a dihydrolipoamide dehydrogenase (DHLDH), which in A. mimigardefordensis DPN7T is obviously involved in DTDP degradation, are known to exist in the genome of R. eutropha H16 (27; M. Raberg, J. Bechmann, U. Brandt, J. Schlüter, B. Uischner, and A. Steinbüchel, unpublished data). Therefore, LpdAAm and the five DHLDH paralogues of R. eutropha H16 were aligned and compared (Fig. 2). Subsequently, lpdAAm and the gene encoding the DHLDH belonging to the pyruvate dehydrogenase complex of R. eutropha H16 (pdhLRe) were cloned, heterologously expressed in Escherichia coli, purified, and assayed.
FIG. 2.
Phylogenetic relationships of the A. mimigardefordensis strain DPN7T LpdA (boldface), R. eutropha H16 PdhL (boldface), and homologues. The neighbor-joining plot was derived from a CLUSTAL X alignment of amino acid sequences closely related to LpdAAm. The amino acid sequence of the outer membrane protein P64K from Neisseria meningitidis was used as the outgroup. GenBank accession numbers are given in parentheses. Scale bar, 10% sequence divergence.
MATERIALS AND METHODS
Sulfur-containing chemicals.
DTDP was purchased from Acros Organics (Geel, Belgium). 3MP, DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)], 4,4′-dithiodibutyric acid (DTDB), glutathione-disulfide (GSSG) lipoic acid, and lipoamide were purchased from Sigma-Aldrich Chemie (Steinheim, Germany).
Strains and cultivation conditions.
The bacterial strains and plasmids used in the present study are listed in Table 1 . Cells of R. eutropha H16, A. mimigardefordensis strain DPN7T and relevant Tn5::mob-induced mutants were grown aerobically in 0.8% (wt/vol) nutrient broth (NB) or in mineral salts medium (MSM) (34) at 30°C containing the indicated carbon source. The latter were prepared as filter sterilized 20% (wt/vol) stock solutions and adjusted to pH 7.0. Solid medium contained 1.8% (wt/vol) purified agar-agar. Escherichia coli strains were cultivated under oxic conditions in Luria-Bertani (LB) medium (31) at 37°C in the presence of antibiotics at the following concentrations to maintain plasmids: ampicillin, 75 μg/ml; carbenicillin, 75 μg/ml; and chloramphenicol, 34 μg/ml.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristica | Source or referenceb |
|---|---|---|
| Strains | ||
| A. mimigardefordensis strain DPN7T | Wild type, DTDP-degrading | 42; DSM 17166; LMG 22922 |
| Tn5::mob-induced mutants of A. mimigardefordensis (JHW51c, JHW90a, and JHW101b) | No growth with DTDP as the sole carbon source; insertion of Tn5::mob in lpdAAm | 41 |
| R. eutropha H16 | Wild type, produces polythioester | DSM 428 |
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) rpsL nupG φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK endA1 | Invitrogen, Carlsbad, CA |
| E. coli One-Shot Mach 1-T1 | lacZΔM15 hsdR lacX74 recA endA tonA | Invitrogen, Carlsbad, CA |
| E. coli (DE3)BL21/pLysS | F−ompT hsdSB (rB− mB−) gal dcm (DE3)/pLysS (Cmr) | Novagen, Madison, WI |
| Plasmids | ||
| pGEM-T Easy | Apr, lacPOZ′ | Promega, Madison, WI |
| pCR2.1TOPO | pUCori lacZ, f1 ori; Kmr Apr | Invitrogen, Carlsbad, CA |
| pET23a(+) | pBR322 ori, f1 ori His6; Apr T7lac | Novagen, Madison, WI |
| Primers | ||
| Forward-lpdANdeI | CATATGAGTCAAGTAGAAATCAAGGTG | MWG-Biotech AG |
| Reverse-lpdAXhoI | CTCGAGCTTGCGTTTGACCGGAGGC | MWG-Biotech AG |
| Forward-pdhLNdeI | AAAAACATATGAGTGTGATCGAAGTCAAGGTGCC | MWG-Biotech AG |
| Reverse-pdhLXhoI | AAAAACTCGAGGCGCTTGCGCGGCGGC | MWG-Biotech AG |
Cmr, chloramphenicol resistance; Apr, ampicillin resistance. For E. coli genotypes, see reference 4; LMG, Laboratorium Microbiologie Universiteit Gent.
DSM, Deutsche Sammlung von Mikroorganismen. Start codons are in boldface type and introduced restriction sites are underlined.
Isolation, transfer, and amplification of DNA.
Chromosomal DNA was isolated as described by Marmur (24). Plasmid DNA was isolated by the method of Birnboim and Doly (5). Restriction enzymes and ligases were used according to the instructions of the manufacturers. T4-DNA-ligase and Taq polymerase were purchased from Invitrogen (Karlsruhe, Germany). E. coli competent cells were prepared and transformed by the CaCl2 procedure (18).
Amplification of DNA was accomplished as described by Innis et al. (20). PCR was conducted in an Omnigene HBTR3CM DNA thermal cycler (Hybaid, Heidelberg, Germany). PCR products were isolated from an agarose gel and purified by using a NucleoTrap kit (Machery and Nagel, Düren, Germany) according to the manufacturer's instructions. Primers (Table 1) were synthesized by MWG-Biotech AG (Ebersberg, Germany).
DNA sequencing and sequence data analysis.
For DNA sequencing (32) the BigDye Terminator v3.1 cycle sequencing kit was used according to the manufacturer's instructions (Applied Biosystems, Darmstadt, Germany). Samples were submitted to the Universitätsklinikum Münster for purification of the extension products and sequencing in an ABI Prism 3700 DNA Analyzer (Applied Biosystems). Sequences were analyzed by using the program BLAST (National Centre for Biotechnology Information; http://www.ncbi.nlm.nih.gov/BLAST/) (2). The program BioEdit (17) was used for multiple alignments and contig assembly. Phylogenetic trees of deduced amino acid sequences were generated with CLUSTAL X (36), displayed with TreeView (26), and calculated by using the neighbor-joining method (30). The DNA sequence and deduced amino acid sequence of lpdAAm have been deposited in the GenBank database under accession number EU423868.
Cloning of lpdA and pdhL from A. mimigardefordensis strain DPN7 and R. eutropha H16, respectively.
lpdA was amplified from genomic DNA of A. mimigardefordensis strain DPN7 (lpdAAm) by PCR using Taq DNA polymerase (Invitrogen, Karlsruhe, Germany) and the oligonucleotides Forward-lpdANdeI and Reverse-lpdAXhoI (Table 1). The PCR product was isolated from agarose gels by using the NucleoTrap kit (Machery and Nagel) and ligated with pGEM-T Easy DNA (Promega, Madison, WI). E. coli TOP10 cells were transformed with the ligation products, and transformants were selected on LB agar plates containing IPTG (isopropyl-β-d-thiogalactopyranoside), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and ampicillin. For heterologous expression in the T7 promoter/polymerase-based expression vector pET23a, lpdAAm was obtained by restriction of plasmid pGEM-T Easy::lpdAAm with NdeI and XhoI and purified from an agarose gel by using a NucleoTrap kit. After ligation into expression vector pET23a, which was linearized with the same restriction endonucleases, the ligation product was used for transformation of cells of E. coli TOP10. Transformants were selected on LB medium containing ampicillin, and hybrid plasmids were isolated, analyzed by sequencing, and used for the transformation of cells of E. coli (DE3) strain BL21/pLysS.
The gene encoding PdhLRe (H16_A1377; that is, the lipoamide dehydrogenase E3 component of the pyruvate dehydrogenase multi-enzyme complex of R. eutropha H16) was amplified from genomic DNA of R. eutropha H16 using the primers described in Table 1. The resulting PCR product was processed as described above for lpdAAm except that subcloning for verification of the error-free amplification product was done with pCR2.1TOPO (Invitrogen, Carlsbad, CA). Cells of E. coli One-Shot Mach 1-T1 were transformed with the ligation constructs of pET23a::pdhLRe to isolate, validate, and select the correct expression vector.
Preparation of crude extracts.
Cells from 50- to 500-ml cultures were harvested by centrifugation (20 min, 4°C, and 3,400 × g), washed twice, and resuspended in an appropriate buffer. For the histidine-tagged fusion proteins, the binding buffers were prepared as recommended by the manufacturer of the His Spin-Trap affinity columns (GE Healthcare, Uppsala, Sweden). All cells were disrupted by a 3-fold passage through a French press (100 × 106 Pa). Lysates with the soluble protein fractions were obtained after centrifugation (30 min, 4°C, 16,000 × g) of the crude extracts and used for enzyme purifications. Protein concentrations were determined as described by Bradford (6).
Immobilized metal-chelate affinity chromatography (IMAC).
To obtain purified histidine-tagged fusion proteins, His Spin-Trap affinity columns (GE Healthcare, Uppsala, Sweden) were used according to the instructions of the manufacturer.
Enzyme assays.
Standard in vitro activity of DHLDH was assayed by incubating and monitoring 1.0 to 300 μg of purified enzyme or protein of the soluble fraction from cell extracts for 3 to 10 min at 30°C in the presence of the following components: 2 mM lipoamide or lipoic acid or 0.1 to 10 mM DTDP or DTDB (the C8-analogon of DTDP), 200 μM NADH or NADPH, 2 mM EDTA, and 100 mM BisTris/Tris buffer (pH 6.5). Additional buffers with pH ranges between 5 and 9 were first applied to find optimal conditions: BisTris/Tris buffer, potassium phosphate buffer, (2-[N-morpholino]ethanesulfonic acid) buffer (MES), and 3-(N-morpholino)propanesulfonic acid (MOPS) buffer. The assays were monitored spectrophotometrically at 340 nm in an Evolution 100 photometer (Thermo Fisher Scientific, Cambridge, United Kingdom) and evaluated with the software VISIONlite (Thermo Fisher Scientific, Cambridge, United Kingdom), using the extinction coefficient for NADH of 6.22 mM−1 cm−1. In addition, cleavage of lipoamide and DTDP was measured in the presence of 1.125 mM DTNB by recording the increase of absorbancy at 412 nm. Controls were done with inactivated enzyme, without substrate, without enzyme or with inactivated soluble fraction of cell extracts from A. mimigardefordensis; the slopes of the time course of decrease or increase of absorption in controls were subtracted from the slopes of the samples containing active enzymes.
RESULTS
Characterization of the lpdAAm translational product and comparison with orthologues.
LpdAAm consists of 613 amino acids and has a calculated molecular mass of 63.849 kDa (isotopically averaged) and a calculated pI of 5.54. Sequence alignments of LpdAAm orthologues from various related species revealed high similarities of the primary structures. Only the flexible hinge region showed a significant divergence of the primary sequence over an extended range. In contrast, the disulfide-active site, the NAD-binding motif, and the active-site amino acid pair “His-Glu” were highly conserved, exhibiting 100% identical amino acids within the compared sequences (see Fig. S1 in the supplemental material). As a further consolidated characteristic, all orthologous proteins compared in Fig. 2, excluding four of the five paralogues of R. eutropha H16, have lipoyl domains. Only PdhLRe, which has been previously identified as a new type of DHLDH (19), also possesses an own lipoyl domain at the amino-terminal region separated from the other part of the enzyme by a flexible hinge region. Lipoyl attachment sites are characterized by a conserved lysine-containing motif to which lipoamide is covalently bound through an amide linkage (28).
R. eutropha possesses all five known types of 2-oxoacid multi-enzyme complexes and five paralogues of DHLDH encoding genes, as revealed by in silico genome analysis (Raberg et al., unpublished). The presence of all genes encoding the respective E3 subunits of each corresponding oxoacid multi-enzyme complex is an uncommon observation, considering the data of completely sequenced organisms. In the R. eutropha genome the genes encoding the E3 subunits of the pyruvate dehydrogenase complex (pdhLRe) and the 2-oxoglutarate dehydrogenase complex (odhL) are clustered with the corresponding E1 and E2 genes. The other three paralogues (lpdaA, H16_B1098, and H16_B1765) are singularly spread over the genome. Comparison of phylogenetic relationship (Fig. 2) and analyses of the amino acid sequences by multiple sequence alignment (see Fig. S1 in the supplemental material) confirmed significant homologies between LpdAAm and PdhLRe. Both proteins are orthologues with high structural similarities; consequently, pdhLRe was further processed and investigated in the present study.
Enzyme activity assays with the soluble protein fractions of the mutants JHW51c, JHW90a, JHW101b, and the wild-type A. mimigardefordensis strain DPN7.
Cells were first cultivated for 48 h in liquid MSM containing 0.5% (wt/vol) sodium gluconate and then washed, transferred into fresh MSM with 0.5% (wt/vol) sodium gluconate in addition to 0.6% (wt/vol) DTDP, and cultivated for another 48 h. The obtained soluble protein fractions were used for enzyme activity assays with BisTris/Tris buffer (pH 6.5). Specific enzyme activity of each soluble protein fraction was determined using lipoamide or DTDP as substrates. Activities were determined by after the oxidation of NADH. Applying lipoamide, different specific activities of the soluble whole-cell protein fractions between the wild type and mutants were observed. The soluble protein extracts of the two mutants JHW51c and JHW101b, whose LpdAAm is disrupted in the interface domain (see Fig. S1 in the supplemental material), exhibited only ca. 70% of the specific activity in comparison to the activity in the soluble protein fraction of wild-type cells. Tn5::mob-insertion disrupted the LpdAAm of mutant JHW90a at the very beginning of the central domain (see Fig. S1 in the supplemental material), resulting in a loss of specific activity of slightly more than 50% of the activity measured in samples obtained from cells of the wild type (data not shown). In all fractions, no detectable decrease of the absorbance was apparent with DTDP as a substrate.
Enzyme activity assays of purified LpdAAm and PdhLRe.
The DHLDH genes lpdAAm and pdhLRe were heterologously expressed by using the T7-promoter/polymerase-based expression vector pET23a and E. coli (DE3)BL21/pLysS as a host strain. Both overexpressed enzymes were purified as hexahistidine-tagged proteins by IMAC and afterward subjected to in vitro enzyme assays.
Specific activities were determined by using lipoamide, lipoic acid, DTNB, DTDP, DTDB, and GSSG as substrates (Table 2). LpdAAm and PdhLRe were highly specific for NADH, and no reaction was observed if NADPH was used as a cofactor. Optimal buffers for enzyme activity were identified by using different buffers possessing pH values between 5 and 9. The highest enzyme activity was measured at pH 6.5 with BisTris/Tris buffer.
TABLE 2.
Disulfide reductase activities of purified LpdAAm and PdhLRe with different substrates
| Disulfide substratea | Mean specific enzyme activity (mkat/kg of protein) ± SDb |
|
|---|---|---|
| LpdAAm | PdhLRe | |
| (±)-α-Lipoamide | 1,833.3 ± 19.2 | 1,666.7 ± 57.8 |
| (±)-α-Lipoic acid | 130.0 ± 1.5 | 78.3 ± 2.8 |
| DTNB | 3.2 ± 0.5 | 2.8 ± 0.3 |
| DTDP | 0.7 ± 0.2 | 0.5 ± 0.1 |
| DTDB | - | - |
| GSSG | - | - |
DTNB, 5,5′-dithiobis-(2-nitrobenzoic acid); DTDP, 3,3′-dithiodipropionic acid; DTDB, 4,4′-dithiodibutyric acid; GSSG, glutathione-disulfide.
Each assay was performed in triplicate. -, below detection limit.
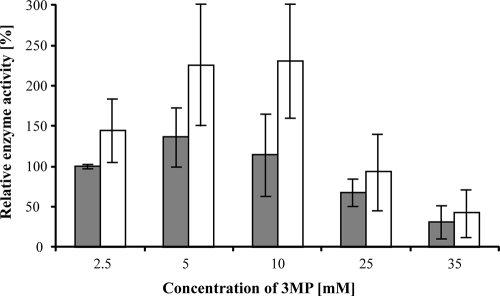
Initially, DTNB should be used for the detection of free thiol groups occurring during the enzyme reaction. However, DTNB was obviously recognized as a substrate by LpdAAm and also by PdhLRe (Table 2; Fig. 1). Not surprisingly, both enzymes revealed the highest activities with their physiological substrate lipoamide. Lipoic acid was also reduced by LpdAAm and PdhLRe, but in comparison to lipoamide with a 14- or a 20-fold-lower specific enzyme activity, respectively. No reaction was observed when applying DTDB or GSSG as substrates (Table 2). As predicted, DTDP was recognized as a substrate by both purified DHLDH, showing low, but conspicuous activities (Table 2). The presence of the highly reactive thiol 3MP, which is released during the cleavage of DTDP, might cause a strong inhibitory effect as described previously for other enzymes putatively involved in conversion of 3MP (9, 10, 12, 13, 29). 3MP was verified as a potential inhibitor by adding it at increasing concentrations (25 μM to 250 mM) to the enzyme activity assay using lipoamide as an optimal substrate. Figure 3 shows the relative enzyme activities of LpdAAm and PdhLRe in the presence of 2.5, 5, 10, 25, and 35 mM 3MP. An inhibiting effect caused by 3MP was observed at concentrations higher than 10 mM (LpdAAm) or 25 mM (PdhLRe); increasing amounts of 3MP in the assays gave a concomitant decrease of enzyme activity. However, due to lower applied concentrations of DTDP in the assays or cultivations, the corresponding concentration of the cleavage product 3MP could not reach the amount required for complete inhibition.
FIG. 3.
Effect of increasing concentrations of 3MP on enzyme activities of LpdAAm (░⃞) and PdhLRe (□). The increases and decreases in specific enzyme activities were monitored in subsequent assays (three repetitions each) using lipoamide as a substrate and various concentrations of 3MP (2.5 to 35 mM).
Interestingly, concentrations of 3MP between 2.5 and 25 mM (lower than necessary for an inhibiting effect) enhanced PdhLRe enzyme activity (Fig. 3). For LpdAAm the activity increase was weaker in comparison to PdhLRe, with a maximum at 5 mM 3MP (Fig. 3). To verify whether the enhancing and inhibiting effect is primarily due to the sulfhydryl group of 3MP, the close structural analogues 2-mercaptoethanol (2ME) and 2-(methylthio)ethanol (Fig. 4) were also used in in vitro assays of both enzymes. Inhibition by 2ME occurred only after adding unphysiologically high concentrations: more than 50 or 150 mM 2ME was necessary for a slight decrease of LpdAAm or PdhLRe, respectively. Lower concentrations of 2ME even enhanced the respective enzyme activities (data not shown). In contrast, the addition of increasing concentrations of 2-(methylthio)ethanol (up to 100 mM) had no effect on the enzyme activity of both disulfide reductases (data not shown); therefore, the presence of a thiol group has an effect on DHLDH activity, with 3MP exerting a stronger effect than 2ME.
FIG. 4.
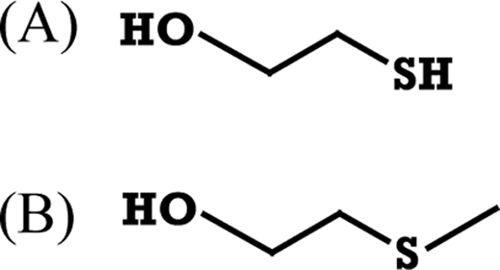
Structural formulas of 2-mercaptoethanol (A) and 2-(methylthio)ethanol (B).
DISCUSSION
LpdAAm and PdhLRe were purified, and the activities with different disulfide substrates were verified. The reduction of DTDP was obvious, although the detected specific enzyme activities were much lower than with the physiological substrate lipoamide. LpdAAm is therefore evidently involved in the catabolism of DTDP in A. mimigardefordensis strain DPN7T. Otherwise, it cannot be explained why the respective Tn5::mob insertions are located in different sites of lpdAAm in three independent mutants impaired in utilizing DTDP (41). The translated lpdAAm gene products of the mutants probably lost their ability to dimerize and became inactive, as also described earlier (21). Genes encoding respective paralogues have been identified in many bacterial genomes, and sharing of E3 subunits by different oxo-acid multi-enzyme complexes occurs frequently in prokaryotes (11). Putatively, the indispensable pyruvate dehydrogenase multi-enzyme complex is completed by an LpdA-paralogue, but activities or specificities would be insufficient for growth of the mutants on solely DTDP.
Several enzymes affiliated to the family of pyridine nucleotide-disulfide oxidoreductases exhibit broad substrate specificities (7, 16), but there are also representatives with very narrow substrate specificities such as the glutathione reductases (33). Enzymes involved in systems to ensure the correct homeostasis of the thiol redox environment of cells were suspected to cleave DTDP. In fact, the measured enzyme activity of purified LpdAAm turned out to be quite low for the observed cell growth and the corresponding turnover of DTDP during degradation. Specific enzyme activities of at least 2.5 mkat/kg protein were theoretically required for typical cell growth of A. mimigardefordensis strain DPN7T in MSM containing DTDP as the sole carbon source (41). The activity obtained with purified LpdAAm (0.7 mkat/kg of protein) was therefore not sufficient, although the conditions during the in vitro assay might have been suboptimal for reduction of DTDP and might be higher in vivo.
The nonspecific LpdAAm must cleave in vivo DTDP at a low rate. The resulting 3MP is apparently partially exported and detectable in millimolar concentrations (up to 25 mM) in the cultures (41). Under ordinary conditions the extracellular environment is rich in stabilizing disulfides, and also the periplasm of Gram-negative bacteria is maintained in an oxidized state (14). The inevitably occurring 3MP causes most likely problems in the redox state outside the cytoplasm of the cells, such as higher concentrations of DTDP inside the cells. Within the cytoplasm, 3MP could, for example, interact with free sulfhydryl groups of proteins, thereby evoking a heat shock response, or activated forms of 3MP could cause inhibitory effects (9, 10, 12, 13, 29, 41). In contrast, moderate concentrations of 3MP (between 5 and 10 mM) would enhance lipoamide dehydrogenase activity, as shown in the in vitro assays (Fig. 3).
3MP is one of the most abundant thiols in natural aquatic environments (1, 43). In contrast, there are no reports about the occurrence of DTDP in nature. DTDP is easily formed only under laboratory conditions by an oxidative dimerization of two 3MP molecules. Therefore, specific enzymes for cleavage of DTDP might have not evolved during evolution, and reduction of DTDP is presumably done by enzymes with other physiological functions and with low specificities for DTDP. Once DTDP has been transported into the reductive environment of the cytoplasm, initial cleavage is done by LpdAAm or PdhLRe. Subsequently, 3MP is converted and either incorporated into PTE (R. eutropha) or used for growth (A. mimigardefordensis) (8). Excessive 3MP is also excreted via the periplasm, probably causing the redox control systems to become profoundly involved and supporting the reduction of DTDP. Enzymes, which cleave DTDP into 3MP, could be applied for optimized production of PMP. Successive reduction of the less-toxic DTDP would facilitate fermentation processes, since the disulfide can be used at much higher concentrations than 3MP.
Supplementary Material
Acknowledgments
We thank Fred-Bernd Oppermann-Sanio for critical and constructive discussions.
Footnotes
Published ahead of print on 10 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Al-Farawati, R., and C. M. G. van den Berg. 2001. Thiols in coastal waters of the North Sea and English Channel. Environ. Sci. Tech. 35:1902-1911. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped blast and psi blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argyrou, A., and J. S. Blanchard. 2004. Flavoprotein disulfide reductases: advances in chemistry and function. Prog. Nucleic Acids Res. Mol. Biol. 78:89-142. [DOI] [PubMed] [Google Scholar]
- 4.Bachmann, B. J. 1987. Linkage map of Escherichia coli K-12, p. 807-876. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 7th ed., vol. 2. American Society for Microbiology, Washington, DC. [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Bruchhaus, I., S. Richter, and E. Tannich. 1998. Recombinant expression and biochemical characterization of an NADPH:Flavin oxidoreductase from Entamoeba histolytica. Biochem. J. 330:1217-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruland, N., J. H. Wübbeler, and A. Steinbüchel. 2009. 3-Mercaptopropionate dioxygenase, a cysteine dioxygenase homologue, catalyzes the initial step of 3-mercaptopropionate catabolism in the 3,3′-thiodipropionic acid degrading bacterium Variovorax paradoxus. J. Biol. Chem. 284:660-672. [DOI] [PubMed] [Google Scholar]
- 9.Chai, S. C., A. A. Jerkins, J. J. Banik, I. Shalev, J. L. Pinkham, P. C. Uden, and M. J. Maroney. 2004. Heterologous expression, purification, and characterization of recombinant rat cysteine dioxygenase. J. Biol. Chem. 280:9865-9869. [DOI] [PubMed] [Google Scholar]
- 10.Cuebas, D., J. D. Beckmann, F. E. Frerman, and H. Schulz. 1985. Mitochondrial metabolism of 3-mercaptopropionic acid. J. Biol. Chem. 260:7330-7336. [PubMed] [Google Scholar]
- 11.De Kok, A., A. F. Hengeveld, A. Martin, and A. H. Westphal. 1998. The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria. Biochim. Biophys. Acta 1385:353-366. [DOI] [PubMed] [Google Scholar]
- 12.Ding, R., Tsunekawa, F., and K. Obata. 2004. Cleft palate by picrotoxin or 3-MP and palatal shelf elevation in GABA-deficient mice. Neurotoxicol. Teratol. 26:587-592. [DOI] [PubMed] [Google Scholar]
- 13.Dominy, J. E., Jr., C. R. Simmons, P. A. Karplus, A. M. Gehring, and M. H. Stipanuk. 2006. Identification and characterization of a bacterial cysteine dioxygenase: a new route of cysteine degradation for eubacteria. J. Bacteriol. 188:5561-5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eser, M., L. Masip, H. Kadokura, G. Georgiou, and J. Beckwith. 2009. Disulfide bond formation by exported glutaredoxin indicates glutathione's presence in the Escherichia coli periplasm. Proc. Natl. Acad. Sci. U. S. A. 106:1572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibello, A., A. I. Vela, M. Martin, A. Barra-Caracciolo, P. Grenni, and J. F. Fernández-Garayzábal. 2009. Reclassification of the members of the genus Tetrathiobacter Ghosh et al. 2005 to the genus Advenella Coenye et al. Int. J. Syst. Evol. Microbiol. 59:1914-1918. [DOI] [PubMed] [Google Scholar]
- 16.Gromer, S., J. Wissing, D. Behnke, K. Ashman, R. H. Schirmer, and K. Becker. 1998. A hypothesis on the catalytic mechanism of the selenoenzyme thioredoxin reductase. Biochem. J. 332:591-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, T. A. 1999. BioEdit: a user friendly biological Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 18.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 136:557-580. [DOI] [PubMed] [Google Scholar]
- 19.Hein, S., and A. Steinbüchel. 1994. Biochemical and molecular characterization of the Alcaligenes eutrophus pyruvate dehydrogenase complex and identification of a new type of dihydrolipoamide dehydrogenase. J. Bacteriol. 176:4394-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Innis, M. A., D. H. Gelfaud, J. J. Suinsky, and T. J. White. 1990. PCR-Protocol: a guide to methods and applications. Academic Press, Inc., San Diego, CA.
- 21.Kanagalaghatta, R. R., R. Bryk, R. Kniewel, J. A. Buglino, C. F. Nathan, and C. D. Lima. 2005. Crystal structure and functional analysis of lipoamide dehydrogenase from Mycobacterium tuberculosis. J. Biol. Chem. 280:33977-33983. [DOI] [PubMed] [Google Scholar]
- 22.Lütke-Eversloh, T., and A. Steinbüchel. 2003. Novel precursor substrates for polythioesters (PTE) and limits of PTE biosynthesis in Ralstonia eutropha. FEMS Microbiol. Lett. 221:191-196. [DOI] [PubMed] [Google Scholar]
- 23.Lütke-Eversloh, T., K. Bergander, H. Luftmann, and A. Steinbüchel. 2001. Identification of a new class of biopolymer: bacterial synthesis of sulfur-containing polymer with thioester linkages. Microbiology 147:11-19. [DOI] [PubMed] [Google Scholar]
- 24.Marmur, J. 1961. A procedure for the isolation of desoxyribonucleic acid from microorganisms. J. Mol. Biol. 1:208-218. [Google Scholar]
- 25.Massey, V. 1963. Lipoyldehydrogenase, p. 275-306. In P. D. Boyer, H. Lardy, and K. Myrback (ed.), The enzymes, 2nd ed., vol. 7. Academic Press, Inc., New York, NY. [Google Scholar]
- 26.Page, R. D. M. 1996. Treeview: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 27.Pohlmann, A., W. F. Fricke, F. Reinecke, B. Kusian, H. Liesegang, R. Cramm, T. Eitinger, C. Ewering, M. Pötter, E. Schwartz, A. Strittmatter, I. Voß, G. Gottschalk, A. Steinbüchel, B. Friedrich, and B. Bowien. 2007. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 24:1257-1262. [DOI] [PubMed] [Google Scholar]
- 28.Russel, G. C., and J. R. Guest. 1991. Sequence similarities within the family of dihydrolipoamide acyltransferases and discovery of a previously unidentified fungal enzyme. Biochim. Biophys. Acta 1076:225-232. [DOI] [PubMed] [Google Scholar]
- 29.Sabbagh, E., D. Cuebas, and H. Schulz. 1985. 3-mercaptopropionic acid, a potent inhibitor of fatty acid oxidation in rat heart mitochondria. J. Biol. Chem. 260:7337-7342. [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 32.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schirmer, R. H., R. L. Krauth-Siegel, and G. E. Schulz. 1988. Glutathione reductase, p. 553-596. In D. Dolphin, R. Poulson, and O. Avramovic (ed.), Glutathione: chemical, biochemical and medical aspects, part A. John Wiley & Sons, Inc., New York, NY.
- 34.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch. Mikrobiol. 38:209-222. [PubMed] [Google Scholar]
- 35.Thakor, N., T. Lütke-Eversloh, and A. Steinbüchel. 2005. Application of the BPEC pathway for large-scale biotechnological production of poly(3-mercptopropionate) by recombinant Escherichia coli including a novel in situ isolation method. Appl. Environ. Microbiol. 71:835-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walters, E. M., and M. K. Johnson. 2004. Ferredoxin:thioredoxin reductase: disulfide reduction catalyzed via novel site-specific [4Fe-4S] cluster chemistry. Photosynth. Res. 79:249-264. [DOI] [PubMed] [Google Scholar]
- 38.Williams, C. H., Jr. 1992. Lipoamine dehydrogenase, glutathione reductase, thioredoxin reductase, and mercuric ion reductase—a family of flavoenzyme transhydrogenases, p. 121-211. In F. Muller (ed.), Chemistry and biochemistry of flavoenzymes, vol. III. CRC Press, Boca Raton, FL. [Google Scholar]
- 39.Williams, C. H. 1995. Flavoprotein structure and mechanism. 6. Mechanism and structure of thioredoxin reductase from Escherichia coli. FASEB J. 9:1267-1276. [DOI] [PubMed] [Google Scholar]
- 40.Williams, C. H., L. D. Arscott, S. Muller, B. W. Lennon, M. L. Ludwig, P. F. Wang, D. M. Veine, K. Becker, and R. H. Schirmer. 2000. Thioredoxin reductase two modes of catalysis have evolved. Eur. J. Biochem. 267:6110-6117. [DOI] [PubMed] [Google Scholar]
- 41.Wübbeler, J. H., N. Bruland, K. Kretschmer, and A. Steinbüchel. 2008. A novel pathway for the catabolism of the organic sulfur compound 3,3′-dithiodipropionic acid via 3-mercaptopropionic acid and 3-sulfinopropionic acid to propionyl-CoA by the aerobic bacterium Tetrathiobacter mimigardefordensis strain DPN7. Appl. Environ. Microbiol. 74:4028-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wübbeler, J. H., T. Lütke-Eversloh, P. Vandamme, S. Van Trappen, and A. Steinbüchel. 2006. Tetrathiobacter mimigardefordensis sp. nov., isolated from compost, a betaproteobacterium capable of utilizing the organic disulfide 3,3′-dithiodipropionic acid. Int. J. Syst. Evol. Microbiol. 56:1305-1310. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, J., F. Wang, J. D. House, and B. Page. 2004. Thiols in wetland interstitial waters and their role in mercury and methylmercury speciation. Limnol. Oceanogr. 49:2276-2286. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.