Abstract
To understand how microbial communities and functional genes respond to arsenic contamination in the rhizosphere of Pteris vittata, five soil samples with different arsenic contamination levels were collected from the rhizosphere of P. vittata and nonrhizosphere areas and investigated by Biolog, geochemical, and functional gene microarray (GeoChip 3.0) analyses. Biolog analysis revealed that the uncontaminated soil harbored the greatest diversity of sole-carbon utilization abilities and that arsenic contamination decreased the metabolic diversity, while rhizosphere soils had higher metabolic diversities than did the nonrhizosphere soils. GeoChip 3.0 analysis showed low proportions of overlapping genes across the five soil samples (16.52% to 45.75%). The uncontaminated soil had a higher heterogeneity and more unique genes (48.09%) than did the arsenic-contaminated soils. Arsenic resistance, sulfur reduction, phosphorus utilization, and denitrification genes were remarkably distinct between P. vittata rhizosphere and nonrhizosphere soils, which provides evidence for a strong linkage among the level of arsenic contamination, the rhizosphere, and the functional gene distribution. Canonical correspondence analysis (CCA) revealed that arsenic is the main driver in reducing the soil functional gene diversity; however, organic matter and phosphorus also have significant effects on the soil microbial community structure. The results implied that rhizobacteria play an important role during soil arsenic uptake and hyperaccumulation processes of P. vittata.
Arsenic (As) is an abundant and widespread trace metalloid element present in virtually all environmental media and is well known to be carcinogenic even at low levels (24). Arsenic contaminations in soil and groundwater have been reported in many parts of the world (2, 29, 34). Recently, in parts of Asia, including China, chronic drinking of arsenic-contaminated groundwater has caused endemic arsenicosis, which has become a major threat to public health (36). Soil arsenic contamination also affects the physiology, growth, and grain quality of crops. For example, high arsenic concentrations were found in rice seeds from Chenzhou, Hunan province, which exceeded the maximal permissible limit of 0.5 mg/kg (dry weight) (21). Hence, remediation of arsenic-contaminated soil and water is one of the major challenges in environmental science and public health. Low-cost, efficient, and environmentally friendly remediation technologies to remove arsenic from contaminated soil and water are urgently needed.
Phytoremediation, the use of plants to restore contaminated soil, has attracted great attention recently. A pivotal step toward the phytoremediation of arsenic-contaminated soils is the discovery of the arsenic hyperaccumulator Pteris vittata L. (Chinese brake fern), which possesses high arsenic tolerance and produces a large biomass. This plant species holds great promise for the phytoremediation of arsenic-contaminated soils. It was shown previously that the leaflets of P. vittata were able to accumulate about 100-fold of arsenic from soils (22). Plant arsenic uptake depends mainly on the arsenic source and bioavailability (25). P. vittata remediates arsenic contamination mainly by taking up arsenate [As(V)] via phosphate transport systems, whereas arsenite [As(III)] is very slowly taken up by P. vittata, at 1/10 of the rate of that for arsenate in the absence of phosphate (41). However, the uptake mechanisms still remain largely unknown.
Microorganisms play a crucial role in arsenic geochemical cycling through microbial transformation processes, including reduction, oxidation, and methylation (2, 11, 31, 33, 40). Although the impacts of microbial metabolisms were previously reported to be associated with arsenic cycling of soil and water (7, 29), little is known about how rhizobacterial communities of P. vittata respond to arsenic. Recently, we found that inoculating arsenic resistance bacteria increased the arsenic accumulation efficiency of P. vittata by 13 to 110% (46). Therefore, rhizobacteria may play an important role during arsenic uptake and accumulation processes by P. vittata. Thus, it is important to elucidate the microbially diverse populations and functional genes associated with arsenic mobility and transport in the P. vittata rhizosphere. However, to fully understand the ecology of such complex rhizosphere-contaminated soils, it is necessary to analyze different microbial populations simultaneously.
Our hypothesis is that the arsenic-hyperaccumulating ability of P. vittata is due to the interactions among plants, rhizobacteria, and arsenic. A study of microbial communities present in the plant rhizosphere is important to illustrate the mechanisms of arsenic hyperaccumulation in P. vittata. Thus, the objectives of this research were to understand how microbial metabolic diversities, communities, and functional genes/relative abundances were affected by soil arsenic contamination and the P. vittata rhizosphere environment. To determine the soil microbial metabolic diversity, the Biolog system (Biolog, Carlsbad, CA) was used to analyze the sole-carbon-source-utilizing capabilities of the soil microbial communities. For functional gene analysis, a high-density, sensitive, oligonucleotide-based microarray (GeoChip 3.0) was used. GeoChip-based technologies have revealed the structure, metabolic activity, and dynamics of microbial communities from complex environments, such as soil, sediments, and groundwater (10, 38, 39, 45, 48). Our results provide evidence that changes of microbial community structure, functional gene distribution, and microbial metabolic diversity are associated with the soil arsenic level and the rhizosphere effect of P. vittata and suggest that plant phytoremediation is an interactive process among plants, microorganisms, and soil contaminants.
MATERIALS AND METHODS
Site description and soil sample collection.
Soil samples were collected from an arsenic phytoremediation experimental station located at Dengjiatang village, Chenzhuo city, Hunan province, Central South China (25°48′N and 113°02′E, with an elevation of 185 m), where the soil was contaminated by arsenic due to the waterfall wastes from an adjacent arsenic-smelting factory at a higher elevation. Rice was cultivated in this area until the soil was highly contaminated with arsenic in 1999, when the rice field was abandoned. In order to remediate the arsenic-contaminated soil, P. vittata was planted in 2002 with plants at distances of 40 cm by 40 cm. Before phytoremediation, the concentrations of arsenic were about 300 mg/kg in the highly arsenic-contaminated soil and 200 mg/kg in the moderately contaminated soils. From 2002 to the end of 2008, the leaves and shoots of P. vittata were harvested each year, and the roots were left in the soil, which regrew in the following year. After continual phytoremediation for 6 years, the soil arsenic level was low enough for plant cultivation (Table 1). In October 2008, five soil samples representing highly arsenic-contaminated P. vittata rhizosphere (HR), highly arsenic-contaminated nonrhizosphere (HA), moderately arsenic-contaminated P. vittata rhizosphere (MR), moderately arsenic-contaminated nonrhizosphere (MA), and uncontaminated control soil (CK) were collected for this study. P. vittata rhizosphere soils were the soils attached to the roots of P. vittata (5- to 20-cm depth), while the nonrhizosphere soil samples were taken from the area between two P. vittata plants (5- to 20-cm depth). The control soil was taken near the experimental station without obvious arsenic contamination and plants. To prevent the P. vittata plants from being damaged, it was not permitted to take a sufficient amount of soil for the rhizosphere samples, so five subsamples (heterogeneity within a certain arsenic contamination range) from each soil site were collected, mixed thoroughly, and stored at 4°C for soil characterization and Biolog analyses and at −80°C for genomic DNA extraction.
TABLE 1.
Selected physical and chemical properties of the soilsa
| Sample | Soil texture | O-M concn (mg/kg) | pH | Concn of: |
||||
|---|---|---|---|---|---|---|---|---|
| As (μg/kg) | N (mg/kg) | P (mg/kg) | S (mg/kg) | Fe (mg/kg) | ||||
| CK | Silty clay | 73.75 | 7.57 | 39.76 | 102.80 | 13.15 | 5.54 | 7.59 |
| MA | Silty clay | 40.52 | 7.63 | 303.52 | 119.14 | 3.35 | 6.30 | 8.60 |
| MR | Silty clay | 42.02 | 7.69 | 356.56 | 112.23 | 4.47 | 6.38 | 10.11 |
| HA | Silty clay | 27.96 | 7.58 | 344.52 | 87.60 | 6.20 | 9.69 | 9.26 |
| HR | Silty clay | 28.84 | 7.52 | 415.26 | 90.22 | 8.81 | 12.54 | 11.84 |
O-M, organic matter. pH, 1:1 soil-H2O suspension. As, 0.05 M (NH4)2 SO4 soluble concentration.
Soil physical and chemical features, including pH, soil texture, organic matter (O-M), N, P, S, Fe, Zn, Cd, and Cu, were analyzed as described previously by Liao et al. (21). Arsenic analysis was performed by using an atomic fluorescence spectrometer (AFS-830; Titan Instruments, Beijing, China).
Biolog analysis of different soils.
Sole-carbon-source utilization by soil microbial communities was analyzed by using a Biolog system, which contains 31 individual carbon sources in triplicate and three negative controls (without a carbon source) in a 96-well-plate format. Five grams of soil (field moist weight) of each sample was added to 45 ml double-distilled water (ddH2O) and incubated at 4°C with shaking (200 rpm) for 45 min and then standing for 30 min. The samples were then serially diluted to 10−3 based on a pilot experiment, and 100 μl of each sample was inoculated into each well of an EcoPlate (Biolog) and incubated at 25°C for 168 h. The metabolic diversity of the soil samples was measured every 15 min by recording the absorbance of each well at 590 nm. The community metabolic diversity (CMD) was calculated as the sum of positive carbon responses (purple wells with threshold optical densities [ODs] of 0.25) observed over the incubation time (30).
Soil community DNA extraction, amplification, labeling, and GeoChip microarray hybridization.
Soil community DNA extraction and purification were performed as described previously (47). The DNA quantity was detected with an ND-1000 spectrophotometer (Nanodrop Inc., Wilmington, DE). Purified DNA was stored at −20°C until use.
The methods for whole-soil genomic DNA amplification, labeling, and microarray hybridization were based on a previously established method (44). The amplified DNA (triplicates) was quantified with a Quant-It PicoGreen kit (Invitrogen, Carlsbad, CA) using a FLUOStar Optima plate reader (BMG Labtech, San Francisco, CA). Three micrograms of product DNA from each soil sample was used for labeling to ensure that similar amounts of DNA were used for all hybridizations. Fluorescent dye Cy5-labeled DNA was purified, dried, resuspended in 130 μl hybridization solution containing a Cy3 normalization standard (19), denatured at 98°C for 3 min, and then hybridized with a GeoChip 3.0 on an HS4800 Pro hybridization station (Tecan US, Durham, NC) at 42°C for 10 h in a dark room. The GeoChip 3.0 is fabricated with 50-mer oligonucleotides and contains 27,812 probes covering 56,990 coding sequences (CDS) of 292 functional genes involved in carbon, nitrogen, phosphorus, and sulfur cycles; energy metabolism; metal resistance (including arsenic resistance); and organic contaminant degradation as well as phylogenetic (e.g., gyrB and 16S rRNA) markers as positive controls (15).
GeoChip data analysis.
Hybridized microarrays were scanned by using a ScanArray 5000 (Perkin-Elmer, Wellesley, MA) system at 95% laser power and 85% photomultiplier tube gain to scan Cy5 and Cy3 at their respective wavelengths. The images were analyzed by quantifying the pixel density (intensity) of each spot using ImaGene 6.1 (Biodiscovery Inc., Los Angeles, CA). Empty and poor spots were removed before the signal intensities were normalized by the mean signal across the slide. A signal-to-noise ratio (SNR) [SNR = (signal mean − background mean)/background standard deviation] of >2 was used as the cutoff for positive spots (14). For each sample, a gene was considered positive if it was detected in two or more of three replicates and was used for further statistical analysis (13).
Cluster analysis was performed by using the pairwise average linkage hierarchical clustering algorithm (6) and visualized by using Treeview software (http://rana.stanford.edu). The correlation coefficients of the samples and detected genes were analyzed by using same program (6) and were shown on the tree branches. BioEnv was used to determine the best correlation between the community structure and the measured geochemical variables (5). Canonical correspondence analysis (CCA) and principal-components analysis (PCA) were performed by using the Canoco program for Windows 4.5 (Biometris, Wageningen, Netherlands). A Mantel test was used to examine the correlations between the differences of soil chemical concentrations and those of various functional gene abundances (35).
RESULTS AND DISCUSSION
Characterization of soil samples.
Several physical and chemical parameters that play an important role in controlling arsenic transformation and microbial metabolism were analyzed. The five soils were all silty clay with similar pH values (pH 7.52 to 7.69) (Table 1), which could be due to the high buffering capacity of clay soils. Other studies have also shown that induced pH changes of the P. vittata rhizosphere were not associated with arsenic phytoremediation in clay soil (9). The Cd, Cu, and Zn concentrations in the five soils were low, with maximum concentrations of 3.416 mg/kg, 6.38 mg/kg, and 0.368 mg/kg, respectively.
The arsenic concentration was significantly lower in the control soil sample (CK) than in the arsenic-contaminated soil samples (Table 1). Even though the soluble arsenic concentrations of the four contaminated soil samples (303.52 to 415.26 μg/kg) were still higher than the Chinese national standard (50 μg/kg), those concentrations were about 700 times lower than those before planting P. vittata, indicating the success of the phytoremediation. In contrast, the control soil had the highest O-M and P concentrations (73.35 mg/kg and 13.15 mg/kg, respectively) compared to the contaminated soils, indicating that arsenic contamination had an adverse effect on soil features.
The concentrations of As, Fe, S, and P were all higher in the rhizosphere soil samples (HR and MR) than in the corresponding nonrhizosphere soil samples (HA and MA). It was reported previously that iron oxides/hydroxides are the primary compounds associated with arsenic solubility in clay soils (9, 43) and that available P increases arsenic accumulation efficiency by P. vittata (42). The increased Fe along with higher As concentrations observed in the rhizosphere samples (Table 1) indicate that the rhizosphere may assist with the dissolution of arsenic to accelerate arsenic accumulation by P. vittata.
Sole-carbon-source-utilizing capabilities among the five soils.
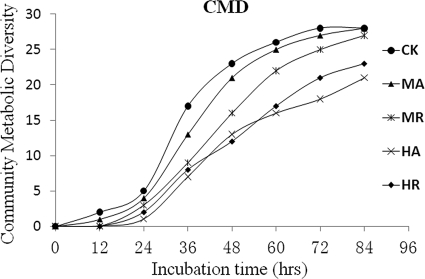
Using the Biolog system, a total of 31 sole-carbon-source utilization profiles of the five soil samples were measured. The community metabolic diversity (CMD) curve showed that the control soil harbored the highest diversity in sole-carbon utilization capabilities (Fig. 1), whereas those of the highly arsenic-contaminated soil samples (HA and HR) were the lowest. In addition, the MR and HR soil samples had higher CMD values than did the nonrhizosphere MA and HA soil samples (Fig. 1), indicating that the rhizosphere microorganisms of P. vittata were more metabolically activated than the nonrhizosphere ones. Such results suggest that the community metabolic diversity was adversely affected by arsenic contamination levels and that the rhizosphere of P. vittata may provide the degradable carbons and energy sources needed to increase rhizosphere microbial metabolic diversities and mitigate the toxic effects of arsenic.
FIG. 1.
Community metabolic diversity (CMD) of the soil samples analyzed with the Biolog system (threshold OD of 0.25).
Additionally, the CMD analysis showed the time points when microorganisms started to be active. In Fig. 1, the microbial community of the control soil reached stationary phase in a shorter time than did the microbial communities of the other soil samples, which further indicated that uncontaminated soil possessed a higher metabolic potential. The metabolic diversity might be positively correlated with O-M and P, with higher concentrations in the control soil. The arsenic concentration was much lower in the control soil than in the arsenic-contaminated soil samples (Table 1), suggesting that arsenic may be a key factor in reducing the microbial sole-carbon-source utilization capability.
Functional gene diversity in the soils.
DNA from the five soil samples was hybridized to the GeoChip 3.0, which contains 27,812 probes, and a total of 2,594 functional genes showed positive hybridization signals for at least one of the five soil samples. Among these, 1,567 genes were unique and were detected in only a single sample. Even though the Simpson evenness values were very similar among the five soils (Table 2), only 172 (172/2,594; 6.63%) genes were shared among the five soil samples, indicating that microbial community structures were quite different from each other. Furthermore, a pairwise comparison showed that only a small proportion of genes overlapped (16.52% to 45.75%) and that the percentage of unique genes of each sample ranged from 13.38% to 48.09% (Table 2). The number of functional genes detected varied considerably among the five soil samples. In each gene category, more genes were detected in the control soil than in the arsenic-contaminated soils, indicating the significant influence of arsenic on the reduction of microbial community diversity. The highest number of functional genes (1,620) was detected in the control soil, 48.09% of which were unique, indicating that there is a high level of heterogeneity in the uncontaminated soil. The microbial community composition in the control soil was significantly different from those in the arsenic-contaminated soil samples (P = 0.019) based on Adonis tests (permutational multivariate analysis of variance). A Mantel test also revealed that there was a significant correlation between the whole-gene structure and the arsenic concentration (r = −0.68; P = 0.036). A Simpson reciprocal diversity index also indicated much higher levels of microbial diversity in the control soil (1,123.8) than in the arsenic-contaminated soils (737.9, 467.0, 460.5, and 319.3 for MR, MA, HR, and HA, respectively) (Table 2). The same diversity trend was also obtained in the order of CK > MR > MA > HR > HA, using a Shannon-Weaver H index (Table 2). The CK and HA soil samples shared the lowest proportion of overlapping genes (16.52%), indicating that a high level of arsenic contamination significantly reduced microbial survival, thereby reducing community richness and altering the microbial functional structure. Furthermore, the highly arsenic-contaminated rhizosphere sample HR shared similar numbers of genes detected and the highest number of overlapping functional genes with MA (45.75%) (Table 2). Principal-component analysis (PCA) also showed that rhizosphere samples were much closer to the CK sample than to the corresponding nonrhizosphere samples (data not shown). This indicated that the rhizosphere probably reduced arsenic toxicity and increased microbial activity (18). These results showed that both As and the rhizosphere exerted selection effects.
TABLE 2.
GeoChip hybridization data, including percentages of gene overlap, gene uniqueness, and diversity indices for each soil sample
| Variable | Value for groupa |
||||
|---|---|---|---|---|---|
| CK | MA | MR | HA | HR | |
| % Gene overlap | |||||
| CK | 48.09 | 29.40 | 26.29 | 16.52 | 28.17 |
| MA | 13.38 | 31.36 | 24.85 | 45.75 | |
| MR | 38.99 | 19.74 | 29.58 | ||
| HA | 24.95 | 25.30 | |||
| HR | 14.56 | ||||
| No. of genes detected | 1,620 | 792 | 1,185 | 489 | 769 |
| Diversity indices | |||||
| Simpson index | 1,123.8 | 467.0 | 737.9 | 319.3 | 460.5 |
| Simpson evenness | 0.694 | 0.581 | 0.622 | 0.652 | 0.607 |
| Shannon index | 7.195 | 6.417 | 6.839 | 5.979 | 6.399 |
Values in boldface type represent unique genes in each sample. Values in italic type represent overlap genes between two samples.
Comparing functional gene numbers between P. vittata rhizosphere and nonrhizosphere soils, there were more genes detected in the rhizosphere soil samples (769 genes in the HR sample versus 489 in the HA sample, and 1,185 genes in the MR sample versus 792 in the MA sample) (Table 2), even though the arsenic concentrations of the rhizosphere soils were higher (Table 1). Rhizosphere soil samples had much higher levels of microbial diversity than did nonrhizosphere soil samples with similar arsenic contamination levels; for example, the MR soil sample had 38.99% unique genes. This phenomenon implied that the rhizosphere could affect the dynamics of certain microbes and their genes (see details in “Arsenic resistance genes” below).
Arsenic resistance genes.
The arsenic resistance genes were well represented in the five soil samples, with a total of 44 arsCBA gene sequences detected (see Fig. S1 in the supplemental material). Those gene sequences were retrieved from 37 bacterial genera, which reflected the high level of diversity of arsenic-resistant bacteria to some extent. The gene diversity was significantly correlated with the arsenic concentration (P = 0.014; r = 0.575). Only one gene sequence was detected across all five soil samples, which was that of the arsC sequence from Parvularcula bermudensis HTCC2503 (GI 84691528; GI protein sequence in the NCBI database, the same as that described below), a strictly aerobic, chemoheterotrophic strain.
Arsenic contamination appeared to be the primary factor in the selection of arsenic-resistant microorganisms since the functional gene cluster of the uncontaminated soil sample (CK) (see Fig. S1 in the supplemental material) was well separated from that of the arsenic-contaminated soil samples. It was shown previously that As contamination led to the selection of diverse arsenic-resistant bacteria and activated arsenic metabolic systems, such as the arsenite efflux pump (arsB and acr3) (2).
The ars genes between rhizosphere and nonrhizosphere soil samples were quite different and seldom overlapped (HR versus HA and MR versus MA) (see Fig. S1 in the supplemental material). More genes involved in arsenic resistance and arsenate reduction, with higher relative abundances, were detected in the rhizosphere samples MR and HR than in the corresponding nonrhizosphere samples HA and MA (Fig. 2 and Fig. S1), whereas genes in other categories did not show this trend, even for other metal resistance genes (data not shown). Although the DNA-based microarray hybridization data could not evaluate the gene expression level directly, the relative abundance of detected ars genes might provide a quantitative measurement of the environmental microbial gene densities in the samples.
FIG. 2.
Proportion of functional gene categories detected. The percentages were calculated by the total signal intensity values of each gene family divided by the total signal intensity values of all genes detected on the array after combination and normalization. C deg and C fix represent carbon degradation and carbon fixation, respectively.
Comparing rhizosphere and nonrhizosphere soil samples, there were several genes detected only in the rhizosphere (Fig. S1, red), such as arsB from Herminiimonas arsenicoxydans (an arsenite-oxidizing bacterium isolated from a heavily arsenic-contaminated sludge) (GI 133740356), which was strongly hybridized in the HR and MR samples. More importantly, the detected ars genes in nonrhizosphere soil samples were observed for at least one of the rhizosphere soil samples, further indicating higher levels of microbially diverse populations in the rhizosphere (Fig. S1). This result indicated that the rhizosphere might be able to maintain certain microorganisms to interact with P. vittata roots.
Interestingly, the MA and HR samples clustered together (see Fig. S1 in the supplemental material). One possibility is that P. vittata may reduce the arsenic toxicity to the rhizosphere microorganisms since it is extremely efficient in extracting arsenic from soils (22), resulting in a similar microbial community structure with high levels of overlapping genes (45.75%) between those two samples. It was reported previously that plant roots exude carboxylate, which could play a crucial role in arsenic metabolism in the rhizosphere by dissolving iron-bound or desorbing arsenic (8), which may result in a higher local arsenic concentration in the rhizosphere (Table 1). In return, the increase in levels of soluble arsenic may promote its translocation and uptake by P. vittata roots (37), ultimately reducing the local toxicity to the microorganisms, which is in line with a previous study that showed that an increase in available As concentrations in soil resulted in more arsenic accumulation by P. vittata (4). The above-described results indicate that arsenic and the rhizosphere are two crucial factors resulting in a greater diversity of arsenic-resistant microorganisms that may promote soil arsenic uptake by P. vittata.
Phosphorus-utilizing genes.
Available P is positively correlated with the arsenic accumulation efficiency of P. vittata (42). However, P is often limited in terrestrial environments. Mineralization is a major pathway in phosphorus cycling to produce bioavailable P, which is accomplished mainly through microbial phosphatase (26). In addition, arsenate is taken up via the phosphate uptake system of P. vittata (41). Thus, the bacterial phosphorus-utilizing genes may have an effect not only on P metabolism but also on As bioavailability in soil systems. Therefore, the genes in phosphorus cycling were analyzed and evaluated for their effects on arsenic dynamics.
A total of eight genes (five exopolyphosphatase ppk genes and three exopolyphosphatase ppx genes) belonging to Agrobacterium, Bordetella, Gluconobacter, Idiomarina, Mycobacterium, Pseudomonas, Sodalis, and Streptomyces spp. were detected in these five soil samples (see Fig. S2 in the supplemental material). Furthermore, their signal intensities were high and did not change too much, indicating that these populations were probably the dominant community members and maintained their functions in this soil ecosystem. Different from the arsenic resistance gene category, the detected relative abundances of the phosphorus-utilizing genes were similar among the five soil samples (Fig. 2); however, changes in community structure and diversity were still observed. Cluster analysis revealed four different phylogenetic phosphorus-utilizing gene groups. Group 1 contained mostly unique genes from the HA sample and was quite different from the other groups, which might maintain phosphorus metabolism in a highly arsenic-contaminated environment. Unique genes in the control soil sample were located mainly in group 2, and those in the MR and MA samples were grouped mainly in groups 3 and 4, respectively. Most genes in HR were shared with MA, and no unique genes were detected in HR (Fig. S2).
There were two exopolyphosphatase (ppx) genes, GI 115371933 from Stigmatella aurantiaca DW4/3-1 and GI 83846326 from Sulfitobacter sp. strain EE-36, which were detected only in the rhizosphere among the As-contaminated soils (Fig. S2, red). The nonrhizosphere MA and HA samples each harbored some unique genes that were not detected in the rhizosphere samples (Fig. S2, red). Microbial exopolyphosphatase is able to degrade long-chain inorganic polyphosphates, which may result in a higher available P concentration in the rhizosphere samples than in the nonrhizosphere samples (Table 1). It was reported previously that plants showed an enhanced exudation of carboxylic acids to dissolve metal-chelate complexes with P, resulting in an enhanced availability of P in the rhizosphere (26). Even though arsenate and phosphate have similar chemical structures and update systems by plants, the available P showed a positive correlation with the arsenic accumulation efficiency of P. vittata (41). Increased levels of available P could exchange arsenic from the soil binding sites and enhance the mobility of As, thus significantly increasing As uptake efficiency by P. vittata (3), which may explain why the rhizosphere had higher As and P concentrations than the respective nonrhizosphere soil samples, even for the soil samples that were phytoremediated by P. vittata for several years.
Sulfate-reducing genes.
Some sulfate-reducing bacteria (SRB) were previously reported to be able to reduce arsenate (23). Sulfite-reducing genes (dsrA and dsrB), indicators of SRB, were detected across the five soil samples. This phenomenon may suggest that the SRB were active in the soil system, even though the available soil sulfate concentrations were very low (Table 1), although this would have to be confirmed with mRNA.
Some of these genes were derived from typical sulfate-reducing bacterial genera, including Chlorobium, Desulfovibrio, Desulfarculus, Desulfofaba, Desulforhabdus, Desulfotomaculum, Pyrobaculum, and Syntrophobacter, but most of them were retrieved from uncultured bacteria (see Fig. S3 in the supplemental material). Although the relative abundances were similar among the five samples (Fig. 2), only two dsrA genes, from Desulfovibrio aerotolerans and Pyrobaculum calidifontis JCM 11548, hybridized across the five soil samples. The number of detected genes decreased sharply with the aggregative arsenic contamination level. With a higher sulfate concentration, the sulfate-reducing gene numbers decreased, whereas the relative abundance of dsr genes increased, especially in HA samples (Fig. 2 and Fig. S3).
The gene diversities among the five soils were obviously different since each sample had several unique dsr genes from different uncultured SRB (see Fig. S3 in the supplemental material). There was one dsrA gene (GI 15055576), derived from Desulfotomaculum ruminis, that preferentially respires arsenate over sulfate, which was detected only in the rhizosphere samples HR and MR. One dsrA gene (GI 46307986) and three dsrB genes (GI 118424524, GI 109290262, and GI 14389215), all derived from uncultured sulfate-reducing bacteria, were not detected in the highly As-contaminated soils (HR and HA), which might be due to the inhibition of sulfate reduction by the high level of arsenic (28).
The dsrAB cluster patterns were very similar to the “arsenic resistance genes.” HR and MA were grouped together, and CK was well separated from them (see Fig. S3 in the supplemental material). The Mantel test showed that there was a significant correlation between the sulfate-reducing gene structure and the combination of sulfate and arsenic concentrations (r = −0.826; P = 0.032), further indicating an interaction of the arsenic level with sulfate reduction. Sulfur often coexists with arsenic, and some microorganisms are capable of reducing both arsenic and sulfate and then precipitating the arsenic as arsenic trisulfide (27), which may be important for arsenic transformation and bioavailability. The homeostasis of dissolved arsenic may be controlled by the solubility of sulfide phases, which depends on microbe-mediated sulfate reduction (16). Scherer (32) reported previously that the rhizosphere possesses a higher level of activity of arylsulfatase, which mineralizes sulfur from organic matter and provides a sulfur source for plants to synthesize phytochelatins (PCs). The As-PC complexes may increase with the arsenic accumulation ability of P. vittata (12).
Genes related to nitrogen cycles, carbon degradation, organic contaminant degradation, and metal resistance.
Large numbers of functional genes detected by the GeoChip 3.0 were involved in carbon degradation, organic remediation, and metal resistance (Fig. 2). Little is known about the contribution of organic remediation to arsenic cycling and hyperaccumulation processes in the rhizosphere of P. vittata. For carbon degradation and organic remediation genes, no obvious changes in relative abundance were detected between arsenic-contaminated and uncontaminated soil samples and rhizosphere and nonrhizosphere soil samples (data not shown), although such microorganisms may be metabolically active in those soils.
Moderately As-contaminated soil samples had the highest relative abundance of carbon degradation genes, whereas highly arsenic-contaminated soil samples harbored more of the denitrification genes nirS and nirK than the moderately contaminated samples (Fig. 2). However, whether these differences in primary energy metabolic pathways were caused by arsenic contamination levels still needs direct experimental evidence, such as that obtained by radiolabeled carbon/nitrogen source uptake assays. The rhizosphere samples contained more of the NH4+ transformation-related genes nifH and ureC than did the nonrhizosphere samples (data not shown). The nitrogen fixation gene nifH plays a role in reducing N2 to NH4+. NH4+-N was reported to be more efficient than other nitrogen sources in stimulating arsenic accumulation by P. vittata (20). The ureC gene encodes urease, which plays a key role in transferring organic N to NH4+, which was highly abundant, especially in the HR sample (Fig. 2). Such processes may promote the growth of P. vittata and/or stimulate arsenic hyperaccumulation in P. vittata.
The most frequently detected metal resistance genes were those for resistance to Cr6+, Hg2+, Cu2+, and Zn2+. The relative abundance and diversity of those metal resistance genes were similar across the five soil samples (data not shown), indicating that metal resistance mechanisms contributed less to the change in the microbial community.
Correlations between microbial community structure and environmental factors.
Arsenic and organic matter were identified as the combination of geochemical variables that provided the best correlation (P = 0.014; r = 0.600) with the microbial community structure using the BioEnv program. The top three (As, O-M, and P) geochemical variables identified by automatic selection were analyzed by canonical correspondence analysis (CCA) for correlation with community structure (Fig. 3). Due to multicollinearity, other variables were removed based on the VIF (variable inflation factor).
FIG. 3.
Canonical correspondence analysis (CCA) of GeoChip 3.0 hybridization signal intensities and soil geochemistry variables. The percentage of variation explained by each axis is shown.
The CCA biplot delineates the effect of selected soil geochemical properties on the microbial communities, and the results revealed a significant correlation between microbial community structure and environmental factors. The total canonical eigenvalue was 1.306. The first canonical axis explained 35.0% of the microbial diversity detected, and the second axis represented 31.4% variance. A total of 66.4% variation was found (Fig. 3).
The microbial community of the control soil was quite different, which was well separated from those of the arsenic-contaminated soils, mainly by the second axis that positively correlated with the P concentration and negatively correlated with the As concentration (Fig. 3). The arsenic-contaminated soil communities spread along the second axis, showing that the amount of O-M was an obvious factor affecting the microbial communities in the arsenic-contaminated soils, although O-M itself may have originally been affected by the arsenic contamination level. The HA and MR samples were well separated, which was potentially caused by differences in their soil geochemical features. The HR and MA samples shared similar microbially diverse populations, even though their geochemical features were quite different. Such a phenomenon indicated that the P. vittata rhizosphere might improve microbial survival. As a result, the highly arsenic-contaminated rhizosphere sample (HR) and moderately arsenic-contaminated soil sample (MA) shared similar community structures.
Variation partitioning analysis (VPA) was performed to determine the proportion of variation in the microbial communities affected by the geochemical variables identified by CCA (Fig. 4). Three variables, As, P, and O-M, explained a major part of the variation observed, leaving 24.9% of the variation unexplained by those factors. Arsenic alone explained 20.1% (P = 0.043), P explained 19.1% (P = 0.108), and O-M explained 22.0% (P = 0.068) of the variation, respectively. The interactions between As and P and between P and O-M accounted for 14.7% and 2.3% of the variation, respectively. These results indicate that As, P, and O-M concentrations greatly influenced the microbial functional gene structure. The unexplained amount of variation in this study (24.9%) was considerably less than those observed for a Tc/U-contaminated groundwater sample (45.1% [39]) and in situ U(VI) biostimulation soil samples (35.9% [38]). Also, unexplained variation may be the result of other geochemical factors, such as carbonate, enhancing or suppressing arsenic-adsorptive characteristics (1), and oracetate, stimulating arsenite release (17).
FIG. 4.
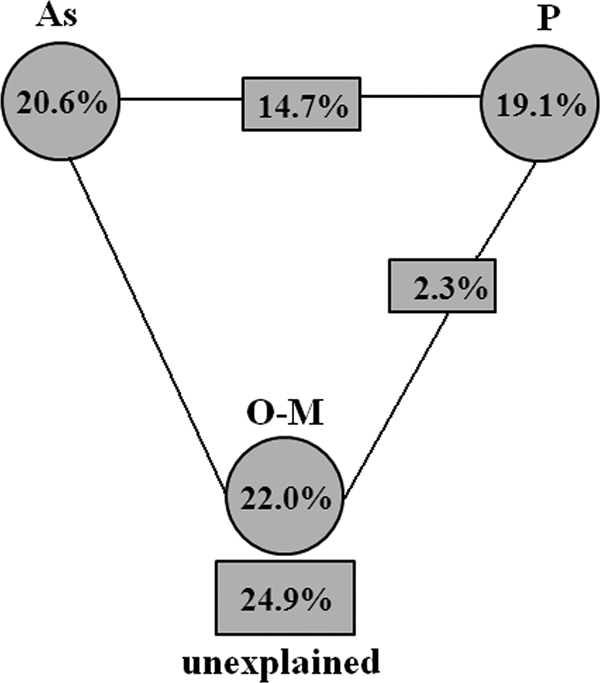
Variation partitioning analysis (VPA) of the variance in microbial diversity among important geochemical variables, As, O-M, and P, and their interactions.
Summary.
Since the collection area shared similar features before arsenic mining, any later changes in microbial properties in the arsenic-polluted soils could be attributed to the effects of arsenic contamination levels and phytoremediation treatment using P. vittata. Our results indicated that the microbial metabolic diversity, functional gene diversity, and structure were affected by both the arsenic contamination level and the rhizosphere of P. vittata. The soil physical and chemical properties, microbial sole-carbon-source utilization, functional processes, and microbial community structure were varied among the five selected soil samples. Arsenic resistance, sulfur reduction, phosphorus utilization, and denitrification genes were very different between rhizosphere and nonrhizosphere soils, as were the arsenic contamination levels. Microbial community structure analysis provided the evidence for a strong linkage among the level of arsenic contamination, the rhizosphere, and functional gene distribution. Arsenic was the main driver in reducing the soil functional gene diversity; however, organic matter and phosphorous also exhibited significant effects on the soil microbial community structures. Arsenic contamination impacted the microbial community functional structure by the selection of arsenic-resistant populations, whereas the rhizosphere of the arsenic accumulator P. vittata appeared to mitigate this toxicity to microbes and maintained high-diversity microbial species that most probably provided some dissolution and transformation functions to promote the P. vittata arsenic uptake ability. Thus, this study provides a sketch on how microbial community and metabolic activity interact with arsenic and the rhizosphere of P. vittata and provides valuable information necessary to understand the interactive phytoremediation processes among plants, microorganisms, and soil contaminants.
Supplementary Material
Acknowledgments
J.X. is supported by the exchanging Ph.D. student scholarship of the Ministry of Education, People's Republic of China. This work was supported by the National Natural Science Foundation of China (grant 30970075), the Chinese 863 project (grant 2007AA06Z332), and the Virtual Institute for Microbial Stress and Survival (http://VIMSS.lbl.gov), supported by the U.S. Department of Energy Office of Biological and Environmental Research Genomics Program, GTL, through contract DE-AC02-05CH11231 between the Lawrence Berkeley National Laboratory and the U.S. Department of Energy.
We thank Ping Zhao and Qian Yang for sample collection.
Footnotes
Published ahead of print on 10 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arai, Y., D. L. Sparks, and J. A. Davis. 2004. Effects of dissolved carbonate on arsenate adsorption and surface speciation at the hematite-water interface. Environ. Sci. Technol. 38:817-824. [DOI] [PubMed] [Google Scholar]
- 2.Cai, L., G. Liu, C. Rensing, and G. Wang. 2009. Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol. 9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao, X., L. Q. Ma, and A. Shiralipour. 2003. Effects of compost and phosphate amendments on arsenic mobility in soils and arsenic uptake by the hyperaccumulator, Pteris vittata L. Environ. Pollut. 126:157-167. [DOI] [PubMed] [Google Scholar]
- 4.Cao, X., L. Q. Ma, and C. Tu. 2004. Antioxidative responses to arsenic in the arsenic-hyperaccumulator Chinese brake fern (Pteris vittata L.). Environ. Pollut. 128:317-325. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, K. R., and M. Ainsworth. 1993. A method of linking multivariate community structure to environmental variables. Mar. Ecol. Prog. Ser. 92:205-219. [Google Scholar]
- 6.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan, H., C. Su, Y. Wang, J. Yao, K. Zhao, and G. Wang. 2008. Sedimentary arsenite-oxidizing and arsenate-reducing bacteria associated with high arsenic groundwater from Shanyin, Northwestern China. J. Appl. Microbiol. 105:529-539. [DOI] [PubMed] [Google Scholar]
- 8.Fitz, W. J., and W. W. Wenzel. 2002. Arsenic transformations in the soil-rhizosphere-plant system: fundamentals and potential application to phytoremediation. J. Biotechnol. 99:259-278. [DOI] [PubMed] [Google Scholar]
- 9.Fitz, W. J., W. W. Wenzel, H. Zhang, J. Nurmi, K. Stipek, Z. Fischerova, P. Schweiger, G. Kollensperger, L. Q. Ma, and G. Stingeder. 2003. Rhizosphere characteristics of the arsenic hyperaccumulator Pteris vittata L. and monitoring of phytoremoval efficiency. Environ. Sci. Technol. 37:5008-5014. [DOI] [PubMed] [Google Scholar]
- 10.Gentry, T. J., G. S. Wickham, C. W. Schadt, Z. He, and J. Zhou. 2006. Microarray applications in microbial ecology research. Microb. Ecol. 52:159-175. [DOI] [PubMed] [Google Scholar]
- 11.Gihring, T. M., G. K. Druschel, R. B. McCleskey, R. J. Hamers, and J. F. Banfield. 2001. Rapid arsenite oxidation by Thermus aquaticus and Thermus thermophilus: field and laboratory investigations. Environ. Sci. Technol. 35:3857-3862. [DOI] [PubMed] [Google Scholar]
- 12.Hartley-Whitaker, J., G. Ainsworth, R. Vooijs, W. M. Ten Bookum, H. Schat, and A. A. Meharg. 2001. Phytochelatins are involved in differential arsenate tolerance in Holcus lanatus L. Plant Physiol. 126:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He, Z., T. J. Gentry, C. W. Schadt, L. Wu, J. Liebich, S. C. Chong, Z. Huang, W. Wu, B. Gu, P. Jardine, C. Criddle, and J. Zhou. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1:67-77. [DOI] [PubMed] [Google Scholar]
- 14.He, Z., and J. Zhou. 2008. Empirical evaluation of a new method for calculating signal-to-noise ratio for microarray data analysis. Appl. Environ. Microbiol. 74:2957-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He, Z., Y. Deng, J. D. Van Nostrand, Q. Tu, M. Xu, C. L. Hemme, X. Li, L. Wu, T. C. Hazen, and J. Zhou. 2010. GeoChip 3.0 as a high throughput tool for analyzing microbial community structure, composition, and functional activity. ISME J. 4:1167-1179. [DOI] [PubMed] [Google Scholar]
- 16.Inskeep, W. P., T. R. McDermott, and S. Fendorf. 2002. Arsenic (V)/(III) cycling in soils and natural waters: chemical and microbiological processes, p. 183-215. In W. T. Frankenberger (ed.), Environmental chemistry of arsenic. Marcel Dekker, New York, NY.
- 17.Islam, F. S., A. G. Gault, C. Boothman, D. A. Polya, J. M. Charnock, D. Chatterjee, and J. R. Lloyd. 2004. Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature 430:68-71. [DOI] [PubMed] [Google Scholar]
- 18.Joner, E. J., A. Johansen, A. P. Loibner, M. A. Cruz, O. H. Szolar, J. M. Portal, and C. Leyval. 2001. Rhizosphere effects on microbial community structure and dissipation and toxicity of polycyclic aromatic hydrocarbons (PAHs) in spiked soil. Environ. Sci. Technol. 35:2773-2777. [DOI] [PubMed] [Google Scholar]
- 19.Liang, Y., Z. He, L. Wu, Y. Deng, G. Li, and J. Zhou. 2010. Development of a common oligonucleotide reference standard for microarray data normalization and comparison across different microbial communities. Appl. Environ. Microbiol. 76:1088-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao, X. Y., T. B. Chen, X. Y. Xiao, H. Xie, X. L. Yan, L. M. Zhai, and B. Wu. 2007. Selecting appropriate forms of nitrogen fertilizer to enhance soil arsenic removal by Pteris vittata: a new approach in phytoremediation. Int. J. Phytorem. 9:269-280. [DOI] [PubMed] [Google Scholar]
- 21.Liao, X. Y., T. B. Chen, H. Xie, and Y. R. Liu. 2005. Soil as contamination and its risk assessment in areas near the industrial districts of Chenzhou City, Southern China. Environ. Int. 31:791-798. [DOI] [PubMed] [Google Scholar]
- 22.Ma, L. Q., K. M. Komar, C. Tu, W. Zhang, Y. Cai, and E. D. Kennelley. 2001. A fern that hyperaccumulates arsenic. Nature 409:579. [DOI] [PubMed] [Google Scholar]
- 23.Macy, J. M., J. M. Santini, B. V. Pauling, A. H. O'Neill, and L. I. Sly. 2000. Two new arsenate/sulfate-reducing bacteria: mechanisms of arsenate reduction. Arch. Microbiol. 173:49-57. [DOI] [PubMed] [Google Scholar]
- 24.Mandal, B. K., and K. T. Suzuki. 2002. Arsenic round the world: a review. Talanta 58:201-235. [PubMed] [Google Scholar]
- 25.Meharg, A. A., and J. Hartley-Whitaker. 2002. Arsenic uptake and metabolism in arsenic resistant and nonresistant plant species. New Phytol. 154:29-43. [Google Scholar]
- 26.Neumann, G., and V. Römheld. 1999. Root excretion of carboxylic acids and protons in phosphorous-deficient plants. Plant Soil 211:121-130. [Google Scholar]
- 27.Newman, D. K., E. K. Kennedy, J. D. Coates, D. Ahmann, D. J. Ellis, D. R. Lovley, and F. M. Morel. 1997. Dissimilatory arsenate and sulfate reduction in Desulfotomaculum auripigmentum sp. nov. Arch. Microbiol. 168:380-388. [DOI] [PubMed] [Google Scholar]
- 28.Nicholas, D. R., S. Ramamoorthy, V. Palace, S. Spring, J. N. Moore, and R. F. Rosenzweig. 2003. Biogeochemical transformations of arsenic in circumneutral freshwater sediments. Biodegradation 14:123-137. [DOI] [PubMed] [Google Scholar]
- 29.Oremland, R. S., J. F. Stolz, and J. T. Hollibaugh. 2004. The microbial arsenic cycle in Mono Lake, California. FEMS Microbiol. Ecol. 48:15-27. [DOI] [PubMed] [Google Scholar]
- 30.Preston-Mafham, J., L. Boddy, and P. F. Randerson. 2002. Analysis of microbial community functional diversity using sole-carbon-source utilisation profiles—a critique. FEMS Microbiol. Ecol. 42:1-14. [DOI] [PubMed] [Google Scholar]
- 31.Qin, J., B. P. Rosen, Y. Zhang, G. Wang, S. Franke, and C. Rensing. 2006. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 103:2075-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scherer, H. W. 2009. Sulfur in soils. J. Plant Nutr. Soil Sci. 172:326-335. [Google Scholar]
- 33.Silver, S., and T. Phungle. 2005. A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 32:587-605. [DOI] [PubMed] [Google Scholar]
- 34.Smith, A. H., E. O. Lingas, and M. Rahman. 2000. Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bull. World Health Organ. 78:1093-1103. [PMC free article] [PubMed] [Google Scholar]
- 35.Smouse, P. E., J. C. Long, and R. R. Sokal. 1986. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Biol. 35:627-632. [Google Scholar]
- 36.Sun, G. 2004. Arsenic contamination and arsenicosis in China. Toxicol. Appl. Pharmacol. 198:268-271. [DOI] [PubMed] [Google Scholar]
- 37.Tu, C., and L. Q. Ma. 2002. Effects of arsenic concentrations and forms on arsenic uptake by the hyperaccumulator ladder brake. J. Environ. Qual. 31:641-647. [PubMed] [Google Scholar]
- 38.Van Nostrand, J. D., W. M. Wu, L. Wu, Y. Deng, J. Carley, S. Carroll, Z. He, B. Gu, J. Luo, C. S. Criddle, D. B. Watson, P. M. Jardine, T. L. Marsh, J. M. Tiedje, T. C. Hazen, and J. Zhou. 2009. GeoChip-based analysis of functional microbial communities during the reoxidation of a bioreduced uranium-contaminated aquifer. Environ. Microbiol. 11:2611-2626. [DOI] [PubMed] [Google Scholar]
- 39.Waldron, P. J., L. Wu, J. D. Van Nostrand, C. W. Schadt, Z. He, D. B. Watson, P. M. Jardine, A. V. Palumbo, T. C. Hazen, and J. Zhou. 2009. Functional gene array-based analysis of microbial community structure in groundwaters with a gradient of contaminant levels. Environ. Sci. Technol. 43:3529-3534. [DOI] [PubMed] [Google Scholar]
- 40.Wang, G., S. P. Kennedy, S. Fasiludeen, C. Rensing, and S. DasSarma. 2004. Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. J. Bacteriol. 186:3187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, J., F. J. Zhao, A. A. Meharg, A. Raab, J. Feldmann, and S. P. McGrath. 2002. Mechanisms of arsenic hyperaccumulation in Pteris vittata. Uptake kinetics, interactions with phosphate, and arsenic speciation. Plant Physiol. 130:1552-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei, C. Y., X. Sun, C. Wang, and W. Y. Wang. 2006. Factors influencing arsenic accumulation by Pteris vittata: a comparative field study at two sites. Environ. Pollut. 141:488-493. [DOI] [PubMed] [Google Scholar]
- 43.Wenzel, W. W., N. Kirchbaumer, T. Prohaska, G. Stingeder, E. Lombi, and D. C. Adriano. 2001. Arsenic fractionation in soils using an improved sequential extraction procedure. Anal. Chim. Acta 436:309-323. [Google Scholar]
- 44.Wu, L., X. Liu, C. W. Schadt, and J. Zhou. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl. Environ. Microbiol. 72:4931-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, L., L. Kellogg, A. H. Devol, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2008. Microarray-based characterization of microbial community functional structure and heterogeneity in marine sediments from the Gulf of Mexico. Appl. Environ. Microbiol. 74:4516-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, Q., S. Tu, G. Wang, X. Liao, and X. Yan. Effectiveness of applying of arsenate reducing bacteria to enhance arsenic removal from polluted soils by Pteris vittata L. Int. J. Phytorem., in press. [DOI] [PubMed]
- 47.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, J., S. Kang, C. W. Schadt, and C. T. Garten. 2008. Spatial scaling of functional gene diversity across various microbial taxa. Proc. Natl. Acad. Sci. U. S. A. 105:7768-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.