Abstract
This study investigated the effect of bacteriophages (phages) e11/2 and e4/1c against Escherichia coli O157:H7 in an ex vivo rumen model and in cattle in vivo. In the ex vivo rumen model, samples were inoculated with either 103 or 106 CFU/ml inoculum of E. coli O157:H7 and challenged separately with each bacteriophage. In the presence of phage e11/2, the numbers of E. coli O157:H7 bacteria were significantly (P < 0.05) reduced to below the limit of detection within 1 h. Phage e4/1c significantly (P < 0.05) reduced E. coli O157:H7 numbers within 2 h of incubation, but the number of surviving E. coli O157:H7 bacteria then remained unchanged over a further 22-h incubation period. The ability of a phage cocktail of e11/2 and e4/1c to reduce the fecal shedding of E. coli O157:H7 in experimentally inoculated cattle was then investigated in two cattle trials. Cattle (yearlings, n = 20 for trial one; adult fistulated cattle, n = 2 for trial two) were orally inoculated with 1010 CFU of E. coli O157:H7. Animals (n = 10 for trial one; n = 1 for trial two) were dosed daily with a bacteriophage cocktail of 1011 PFU for 3 days postinoculation. E. coli O157:H7 and phage numbers in fecal and/or rumen samples were determined over 7 days postinoculation. E. coli O157:H7 numbers rapidly declined in all animals within 24 to 48 h; however, there was no significant difference (P > 0.05) between the numbers of E. coli O157:H7 bacteria shed by the phage-treated or control animals. Phages were recovered from the rumen but not from the feces of the adult fistulated animal in trial two but were recovered from the feces of the yearling animals in trial one. While the results from the rumen model suggest that phages are effective in the rumen, further research is required to improve the antimicrobial effectiveness of phages for the elimination of E. coli O157:H7 in vivo.
Escherichia coli O157:H7 has become a worldwide public health concern since it was first identified as a human pathogen in 1982 (31). This pathogen has a very low infectious dose (approximately 10 cells) in humans, and symptoms of infection range from watery diarrhea to hemorrhagic colitis and hemolytic uremic syndrome, and in some cases, death (22, 39). Ruminants are recognized as reservoirs for this pathogen and are the most common sources for food-borne outbreaks (8, 13, 25). It has been reported that the occurrence of E. coli O157:H7 in the feces and, in particular, the hide of cattle is a significant source of the pathogen on the carcass and in derived meat products (11, 12, 25). The control of this pathogen within the animal is difficult, because carriage in ruminants is asymptomatic and shedding can be intermittent and seasonal (12, 19).
Research has highlighted the necessity for preharvest intervention strategies to control or reduce E. coli O157:H7 in the food chain (17, 18). Successful strategies to reduce the carriage of E. coli O157:H7 in ruminant animals could potentially reduce the risk of human exposure to this pathogen. There are currently no effective and reliable commercially available intervention strategies to control the carriage of E. coli O157:H7 in ruminants. However, research in this area is increasing, and numerous agents, such as vaccines, probiotics, and bacteriophages (phages), are being evaluated (15, 17, 18). The use of phages for the control of food-borne pathogens in the food chain is desirable, as they are natural, nontoxic viruses that target only specific bacteria (2) and are already being used in human and veterinary medicine, particularly prior to antibiotics (6, 14, 15, 30, 37). Many studies have investigated the use of different phages for the control of E. coli O157:H7 in various animals, including mice, calves, and sheep (4, 5, 35, 37, 41). Although the results between studies vary, some have reported the successful reduction of E. coli O157:H7 levels in animals (4), and one study has resulted in a U.S. patent (41). There are very few commercially available phage products to date, but research indicates promising outcomes for the use of phages for the control of E. coli O157:H7 within the food chain.
The E. coli O157:H7-specific phages e11/2 and e4/1c were isolated from bovine slurry in a previous study (26) and have the potential to be used as biocontrol agents for E. coli O157:H7. Both phages have been found to be active against E. coli O157:H7 in a number of relevant test conditions involving different pHs, water activity, and temperatures (B. Coffey, L. Rivas, G. Duffy, A. Coffey, R. P. Ross, and O. McAuliffe, unpublished data). In addition, whole-genome sequencing revealed that neither phage encodes undesirable properties, such as virulence factors, that would hinder its use as a biocontrol agent for E. coli O157:H7 (B. Coffey, G. O'Flynn, A. Coffey, O. O'Sullivan, O. McAuliffe, and R. P. Ross, unpublished data). The objective of the present study was, first, to evaluate the effect of phages e4/1c and e11/2 against inoculated E. coli O157:H7 in an ex vivo model rumen system, and second, to assess the ability of a phage cocktail (e11/2 and e4/1c) to reduce the shedding of E. coli O157:H7 in experimentally inoculated cattle. Findings from ex vivo studies determined our phages to be effective against E. coli O157:H7 in a model rumen system; however, complete eradication of E. coli O157:H7 from cattle was not achieved.
MATERIALS AND METHODS
Bacterial cultures.
A toxigenic E. coli O157:H7 isolate (J21) was used in the model rumen system, and three nontoxigenic but eae- and hylA-positive E. coli O157:H7 strains (NCTC 12900, DAF454, and 13C1T3) were used in the animal trials. Nontoxigenic strains of E. coli O157:H7 were used in the animal trials as a safety precaution for staff exposed to these animals. To aid detection, the strains were made resistant to streptomycin sulfate (1,000 μg/ml) and nalidixic acid (50 μg/ml) (-nas) as described by Park (28). For phage propagation and enumeration of phages in all assays, the nontoxigenic host strain E. coli O157:H7 P1432 was used. Cultures for the experiments were prepared from Protect beads (Technical Service Consultants, Lancashire, United Kingdom) stored at −20°C in 10 ml of brain heart infusion (BHI) broth (Oxoid, Basingstoke, United Kingdom) and incubated overnight at 37°C without shaking.
Phage propagation.
Phages e11/2 and e4/1c were propagated separately using the host strain E. coli O157:H7 P1432. In brief, an overnight culture of E. coli O157:H7 P1432 was inoculated into 1 liter of Luria-Bertani (LB; Merck KGaA, Darmstadt, Germany) broth containing 10 mM CaCl2. Following incubation at 37°C for 2 h with shaking, the 1-liter E. coli O157:H7 culture was divided into two aliquots. Phage was added to one of the E. coli O157:H7 culture aliquots at a multiplicity of infection (MOI; defined as the ratio of phages to target host bacteria) of 100, together with 200 ml of fresh LB broth. All samples were incubated at 37°C with shaking for a further 2 h. Subsequently, phage-containing and non-phage-containing samples were pooled and were reincubated for a final 2 h to increase the phage titer. A final concentration of 1 M NaCl was added, and the sample was stirred at 4°C for 1 h. The sample was then centrifuged at 6,000 × g for 2 h at 4°C, and the supernatant (lysate containing the phage) was filtered through a 0.45-μm sterile filter (Sarstedt, Nümbrecht, Germany). Polyethylene glycol 8000 (PEG 8000; Sigma-Aldrich, St. Louis, MO) was added to a make a final concentration of 15% (wt/vol), and the mixture was stirred at 4°C. Highly concentrated phage preparations were obtained by the CsCl density gradient ultracentrifugation method as previously described by Sambrook and Russell (33). The phage titer was calculated as PFU per ml by plaque assay as described previously (27).
Ex vivo model rumen assay with E. coli O157:H7 and bacteriophages.
Ruminal fluid was obtained from three fistulated steers fed a grass silage and concentrate (60:40) diet. The rumen fluid was collected prior to morning feeding and strained through four layers of cheesecloth before use. The assays were carried as described by Tilley and Terry (40) with modifications. Large glass test tubes (100 ml) sealed with rubber stoppers fitted with a one-way gas release valve were used for the assay. The assay system consisted of 10 ml of rumen fluid and 40 ml of rumen buffer (26 mM Na2HPO4·12H2O, 117 mM NaHCO3, 8 mM NaCl, 7.65 mM KCl, 0.63 mM MgCl2·6H2O, 0.54 mM CaCl2·6H2O). Each test tube contained 0.5 g of feed substrate composed of a concentrate-grass silage mixture at 1:4. The percentage of dry matter of the substrate was 90.4%. An overnight culture of E. coli O157:H7 (J21-nas) was prepared as described above, serially diluted in 9-ml volumes of maximum recovery diluent (MRD; Merck), and inoculated into the rumen assay at either 103 or 106 CFU/ml. The two phages were tested separately in each assay; the MOI values chosen were based on previous experiments that showed this level of phage to be effective at reducing E. coli O157:H7 levels in the presence of an artificial cocktail of microorganisms at 39°C (data not shown). Phage e11/2 was added at MOIs of 100 and 1, while the e4/1c phage was added at MOIs of 10 and 1,000. CaCl2 (10 mM) was also added to each test tube prior to the flushing of each tube with CO2 for approximately 2 min. Tubes were then incubated at 39°C without agitation.
Samples were taken from each tube over a 24-h incubation period. For spread plating, 1-ml aliquots taken from each tube were serially diluted in 9 ml MRD, and 0.1-ml aliquots of each dilution were spread onto tryptone soy agar (TSA) plates, which were first incubated at 37°C for 2 h. Sorbitol MacConkey agar (Oxoid) containing nalidixic acid at 50 μg/ml and streptomycin sulfate at 1,000 μg/ml (SMAC-nas) was then poured over the plates, which were reincubated for a further 48 h to recover injured cells. Another 1-ml aliquot from each sample tube was taken at each time point and added to a 9-ml modified tryptone soy broth (Oxoid) containing nalidixic acid at 50 μg/ml and streptomycin sulfate at 1,000 μg/ml (mTSB-nas) and incubated at 37°C for 24 h. If no E. coli O157:H7 colonies were recovered from either medium, then the enriched broth samples were processed by immunoseparation (IMS) using anti-E. coli O157 beads (Dynal, Oslo, Norway) and a BeadRetriever (Dynal) according to the manufacturers' instructions. Collected beads were plated onto SMAC-nas, and the plates were incubated at 37°C for 24 to 48 h. Suspected E. coli O157:H7-positive colonies were confirmed by latex agglutination (Oxoid). Samples with zero E. coli O157:H7 counts on plating media were assigned the arbitrary value of 1.00 log10 CFU/ml, while positive IMS samples, which indicated the presence of E. coli O157:H7 following enrichment, were assigned an arbitrary value of 1.00 CFU/g or CFU/ml (0 log10 CFU/g or CFU/ml), respectively. These values are the levels of detection of direct plating and IMS for this assay. After sampling at each time point, all tubes were flushed with CO2 for approximately 1 min before reincubation at 39°C. Sampling at each time point took approximately 3 min, and the final pH of each tube after 24 h of incubation was determined using a pH meter (Orion 260A pH meter). The assay was performed in triplicate using fresh rumen fluid for each assay. The assay was also repeated using the cocktail of nontoxigenic E. coli O157:H7 strains (NCTC 12900-nas, DAF454-nas, and 13C1T3-nas) used in the animal trials outlined below.
Animals and housing facilities used for the animal trials.
A license for the animal trials was obtained under the Cruelty to Animals Act 1876, as amended by the European Communities (Amendment of Cruelty to Animals Act 1876), regulations 2002 and 2005. For trial one, 20 animals, aged approximately 16 months, were selected by weight and randomly assigned to one of two (phage or control) groups. All animals were adapted to a grass silage-concentrate diet for 3 weeks. The experiment was performed indoors in a large shed containing 60 pens (the approximate size of each pen was 10 m2). The shed was divided into two halves, whereby one half of the shed contained the phage-treated animals and the other half contained the control animals. Each animal was placed in an individual pen with no animals in adjacent pens, to avoid cross contamination. A row of pens and plywood was also used to divide the two treatment groups. None of the animals had contact with any other animal during the trial. The pens had concrete slatted flooring, and feed was offered on the concrete solid passage in front of each pen every day.
For trial two, two ruminally fistulated animals, aged 8 and 9 years, were selected. These animals were housed in a metabolism housing unit in separate pens. Pens were divided by steel gates to avoid the occurrence of cross contamination. Pens had concrete flooring and separate steel feeding boxes which were cleaned out each day.
In both trials, each pen had a separate automatic water supply. A plastic tag on the right ear individually identified all animals. One week before the start of the trials, feces and/or rumen fluid from each animal was screened to ensure that the animals were not shedding enteric bacteria that possess the same antibiotic resistance profile as the marked E. coli O157:H7 strains and for other naturally occurring phages that could potentially lyse the marked E. coli O157:H7 strains. Both trials were performed once.
Preparation of the E. coli O157:H7 inoculum and phage doses used in the animal trials.
Overnight cultures of the three nontoxigenic E. coli O157:H7 strains (NCTC 12900-nas, DAF454-nas, and 13C1T3-nas) were prepared as outlined above. One ml of each culture was aseptically transferred to 100 ml BHI broth and incubated at 37°C for 18 h to achieve a stationary-phase culture. The cultures were centrifuged at 3,000 × g for 10 min, and pellets from the three strains were pooled into 50-ml sterile distilled water to achieve an inoculum of 1010 CFU of E. coli O157:H7 per animal.
To obtain sufficient volumes of phages for dosing, each phage was propagated as described above with slight modifications. Ten milliliters of an overnight culture of E. coli O157:H7 (P1432) was added to 500 ml LB broth containing 10 mM CaCl2 in a 1-liter Schott bottle and incubated for 1 h at 37°C with shaking. The relevant phage was added at an MOI of 100 and incubated at 37°C with shaking until clearing was observed. The sample was then centrifuged at 6,000 × g for 90 min, and the supernatant was filter sterilized using 0.45-μm Stericup filters (Millipore, Billerica, MA). Phage lysates of e11/2 and e4/1c were combined to make a phage cocktail comprising approximately 1011 PFU per dose. The use of a phage cocktail of the two phages as well as the amounts of E. coli O157:H7 and phage used per dose were based on results obtained from other studies (9, 30, 32).
Inoculation and sampling procedures used for the animal trials.
For trial one, on day 0, all 20 animals were inoculated orally by syringe (Plastitek, 60 ml; Becton Dickinson, Oxford, United Kingdom) with 50-ml sterile distilled water that contained approximately 1010 CFU of E. coli O157:H7. Immediately after inoculation, each animal was dosed with 100 ml of sterile distilled water to wash down the culture. On days 1, 2, and 3, 10 animals on one side of the shed were inoculated orally by syringe with 100 ml of bacteriophage cocktail containing approximately 1011 PFU phages, followed immediately by a dose of 100 ml of sterile distilled water with 10 mM CaCl2 to wash down the phages. The same inoculation procedure was performed for trial two, but only one animal was used for each treatment.
For trial one, fecal samples were taken via rectal palpation prior to all dosing on days 0, 1, 2, 3, 4, and 7. For trial two, rumen fluid was taken via the fistulation, and fecal samples were taken via rectal palpation prior to all dosing; samples were taken twice per day (morning and afternoon) for 7 days. All fecal and rumen samples were placed into sterile jars (Medfor Products Ltd., Hampshire, United Kingdom) and transported to the laboratory within 1 h.
To avoid cross-contamination among pens during sampling, the researchers wore separate disposable laboratory coats while in each pen. Latex gloves were worn and changed following collection of each sample. Boots were disinfected after exiting each pen by using a boot dip consisting of a large (50-liter) container with water and detergent (Osmodex detergent; Osmonds Broomhill, Ireland). Sampling of all the control animals was performed before that of the phage-treated animals each day to avoid cross contamination of phage to the control animals. None of the protective equipment or boot dip used during sampling was tested for E. coli O157:H7 or phage.
Processing of fecal and rumen samples during the animal trials.
For the enumeration of E. coli O157:H7, 1 g of feces or 1 ml of rumen fluid was placed into 9 ml of mTSB-nas and vortexed for 1 min. The samples were prepared in duplicate; one set of broth samples was used for direct counts, while the other set was enriched overnight at 37°C. On days 1 and 2, serial dilutions in 9 ml MRD were performed before direct plating. For all sample days, a volume of 1 ml of the sample was spread in duplicate onto plates of TSA and allowed to dry in a laminar hood before incubation at 37°C for 2 h, and SMAC-nas was poured over the plate before reincubation for a further 48 h to recover injured cells. If neither of the spread-plated media resulted in the recovery of E. coli O157:H7 colonies, then the enriched broth samples were processed by IMS as outlined above. For the animal trials, fecal (trials 1 and 2) and rumen samples with zero E. coli O157:H7 counts were assigned the arbitrary value of 0.7 log10 CFU/g (feces) or CFU/ml (rumen fluid). Samples positive for IMS, which indicated the presence of E. coli O157:H7 following enrichment, were assigned an arbitrary value of 1.00 CFU/g or CFU/ml (0 log10 CFU/g or CFU/ml), respectively. These values are the levels of detection of direct plating and IMS for this assay.
For phage enumeration, 1 g of feces was placed in 9 ml of MRD and vortexed for 1 min. Samples were then centrifuged at 7,000 × g for 10 min, and approximately 4 to 5 ml of the supernatant was transferred into 1.5-ml sterile Eppendorf tubes before centrifugation at 10,000 × g for 10 min. The supernatant was then filtered using a 0.45-μm filter (Sartorius AG, Göttingen, Germany), and a plaque assay was performed. The plaque assay involved adding 1 ml of filtrate to 5 ml of BHI soft-top agar (0.7% [wt/wt]) containing 100 μl of overnight-grown E. coli O157:H7 (P1432) and 10 mM CaCl2. The soft-top agar was then mixed by vortexing and poured over a BHI agar plate. All plates were incubated at 37°C for 24 h, and plaques were enumerated. If no plaques were found following incubation, a pre-enrichment step was performed. Basically, 450 μl of filtrate was enriched with 50 μl of E. coli O157:H7 (P1432) at 37°C for 2 h, 100 μl of the enriched filtrate was included in the plaque assay, and plaques were observed following incubation of plates at 37°C for 24 h. The limit for detection for phage enumeration was 1.00 log10 PFU/ml.
Processing of hide, pen, and feed samples during the animal trials.
For trial one, swabs of hides (approximately 900 cm2 of the rump area on one side of the animal) and pen barriers (unlimited areas) were taken using a sterile cellulose sponge (Sydney Heath and Son, Ltd., Staffordshire, United Kingdom) moistened with 10 ml of MRD and placed into sterile stomacher bags (Seward Laboratory, London, United Kingdom). Hide swabs were taken on days 2, 4, and 7, and pen barrier samples were taken on days 4 and 7. Upon return to the laboratory, sponges were placed in 90 ml of mTSB-nas and stomached in a Colworth stomacher (model BA 6024; A. J. Steward and Co., Ltd, London, United Kingdom) for 1 min and enriched overnight at 37°C prior to undergoing IMS as outlined above.
For feed samples, approximately 25 g of feed was collected from each pen (feed remaining from the previous day) on day 4 and placed into sterile stomacher bags. In addition, fresh silage and concentration samples were tested prior to the feeding of the animals. Samples were added to 100-ml volumes of mTSB-nas, stomached for 1 min, and enriched and then underwent IMS detection of E. coli O157 as outlined above. The presence or absence of phage on the hide or in environmental or feed samples was not determined.
Statistical analysis.
All counts were log10 transformed, and one-way analysis of means and comparison of means (Tukey's method) was performed for all ex vivo model rumen data sets (SAS Institute, Inc., Cary, NC). For the animal trials, analysis of variance was performed using the SAS mixed-model procedure using the spatial model for the covariance structure. The experimental unit was the individual animal in each pen. The least significant difference (LSD) test was used to determine the differences among means, where significance was determined at a P value of <0.05. Time/treatment interactions were discounted due to the natural reduction of inoculated E. coli O157:H7 populations in the animals; therefore, only pointwise comparisons were performed.
RESULTS
Ex vivo model rumen system.
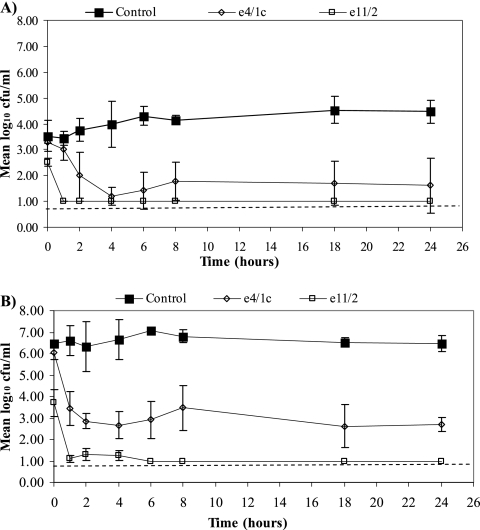
Figure 1 shows the mean number of surviving E. coli O157:H7 bacteria (A, starting inoculum, 103 CFU/ml; B, starting inoculum, 106 CFU/ml) following exposure to either e11/2 or e4/1c in the model rumen assay. The numbers of E. coli O157:H7 bacteria surviving exposure to either phage, e11/2 or e4/1c, were found to be significantly (P < 0.05) lower than the numbers recovered from the controls (no phage) after 24 h of incubation (Fig. 1A and B). Although the same E. coli O157:H7 inoculum levels were used for the e4/1c and e11/2 assays, e11/2 was found to significantly (P < 0.05) reduce E. coli O157:H7 levels within minutes of addition of the phage (time zero; sampling time was approximately 3 min), and after 1 h of incubation, the E. coli O157:H7 counts were significantly (P < 0.05) reduced to below the level of detection for direct plating and remained undetected with direct plating after 24 h of incubation. However, E. coli O157:H7 was found to be present following enrichment and IMS. In the presence of e4/1c, E. coli O157:H7 levels were significantly (P < 0.05) reduced within 2 h, and counts did not significantly change throughout the remaining incubation period. In all control samples (no phage), the numbers of E. coli O157:H7 inoculated did not significantly (P > 0.05) change throughout the incubation period. The presence of neither phage nor E. coli O157:H7 strains significantly (P > 0.05) altered the final pH (∼6.8) of the assay in comparison to that of the controls. Both phages were also found to significantly (P < 0.05) reduce a cocktail of nontoxigenic E. coli O157:H7 strains that were subsequently used in the animal trials within 1 h, and E. coli O157:H7 counts remained significantly (P < 0.05) lower than in the controls after 24 h (data not shown).
FIG. 1.
E. coli O157:H7 (J21-nas) numbers following exposure to bacteriophages e4/1c and e11/2 in a model rumen system. The assay used E. coli O157:H7 inocula of 103 (A) and 106 (B) CFU/ml. Phages e11/2 and e4/1c were added at MOIs of 100 and 1,000, respectively (A), and at MOIs of 1 and 100, respectively (B). A control underwent the same assay with the E. coli O157:H7 inoculum but without the addition of phage (▪). E. coli O157:H7 counts obtained following exposure to e4/1c (⋄) and e11/2 (□) over 24 h at 39°C were determined. All assays were replicated at least three times. The dotted line represents the limit of detection for direct plating.
Fecal results for animal trial one.
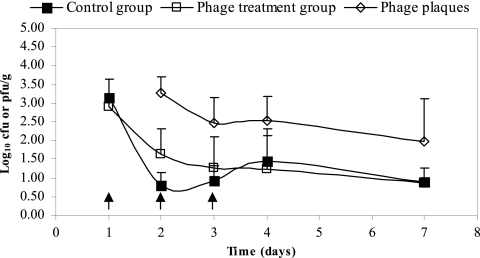
In both animal trials, all animals remained healthy for the duration of the experiment. Preinoculation fecal samples taken from all animals were negative for E. coli O157:H7 or other bacteria with antibiotic resistance profiles similar to those of the marked E. coli O157 strains. Figure 2 shows the mean numbers of E. coli O157:H7 bacteria recovered in the feces of animals with and without phage treatment and the mean numbers of phages recovered from the feces of phage-treated animals in trial one. The numbers of E. coli O157:H7 bacteria recovered in the feces of phage-treated animals were not statistically (P > 0.05) different from those of the untreated control group. The numbers of E. coli O157:H7 shed in the feces in animals in both the control and phage-treated groups rapidly decreased within 48 h of inoculation, with mean E. coli O157:H7 counts of 3.14 and 2.90 log10 CFU/g of feces, respectively. Following 48 h, the majority of fecal samples from both groups required enrichment for E. coli O157:H7; however, 5 out of 10 control animals and all phage-treated animals were enrichment positive after 7 days postinoculation. The numbers of phages shed in the feces did not significantly differ (P > 0.05) for days 2, 3, 4, and 7. Phage plaques in the feces of some of the control animals were sporadically observed following enrichment. These plaques were similar to plaques produced by phages e11/2 and/or e4/1c used in the trial; however, further analysis, such as enzyme restriction of the phage DNA, is required for confirmation.
FIG. 2.
Mean log10 counts of E. coli O157:H7 shed in the feces of cattle with (□) and without (▪) bacteriophage treatment and the mean plaque counts of bacteriophages (⋄) shed in the feces of animals dosed with bacteriophages. Twenty animals were dosed with 1010 CFU of E. coli O157:H7 cocktail (NCTC 12900-nas, DAF454-nas, and 13C1T3-nas) on day 0. Ten animals were dosed with a 1011 PFU cocktail of e11/2 and e4/1c phages on days 1, 2, and 3 (indicated by the arrows).
Hide, pen, and feed sample results for animal trial one.
Between 8 to 10 out of 10 animal hides in each group were E. coli O157:H7 positive on days 2 and 4. However, on day 7, hide samples for the control animals were all negative, while 8 out of 10 hide samples from the phage treatment group were positive. All pen barriers (10 of 10) for the phage-treated group and 9 out of 10 pens for the control group were positive for E. coli O157:H7 on day 2; however, on day 7, only 1 and 3 out of 10 pens each were E. coli O157:H7 positive for the control and phage-treated groups, respectively. On day 4, feed collected from the front of each pen (feed remaining from the previous day) was collected. Four out of 10 feed samples from the control group and 2 out of 10 feed samples from the phage treatment group were positive for E. coli O157:H7. Fresh silage and concentrate samples were negative for E. coli O157:H7 prior to distribution to the pens and feeding.
Fecal and rumen results for animal trial two.
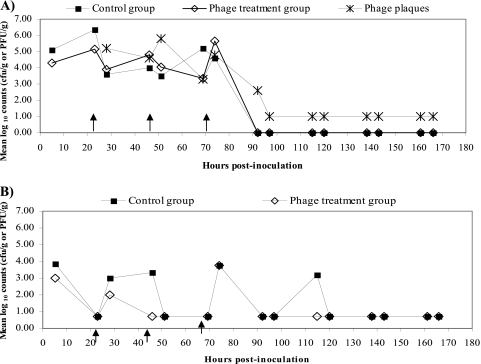
Figure 3 shows the numbers of E. coli O157:H7 and phages recovered in the rumen (A) and fecal (B) samples for trial two. E. coli O157:H7 was recovered from the rumen 5 h postinoculation in both the phage-treated and untreated control animals (Fig. 3A). However, E. coli O157:H7 numbers significantly (P < 0.05) decreased over time and were not recovered by enrichment after 92 h. The numbers of E. coli O157:H7 bacteria recovered from the feces were significantly (P < 0.05) lower than those from the rumen samples (Fig. 3B). E. coli O157:H7 counts, particularly in the control animal, were unstable over the first 120 h of the experiment but were then recovered only by enrichment of the feces after 120 h. Phage was also recovered from the rumen 5 h after dosing. The numbers of phages in the rumen were found to gradually decrease following dosing, but numbers would again peak after subsequent dosing and then decline rapidly after the completion of all doses. Bacteriophage was not recovered from the feces of the animals during trial two and therefore could not be enumerated.
FIG. 3.
Mean number of E. coli O157:H7 bacteria in the rumen (A) and feces (B) of cattle inoculated with (⋄) and without (▪) phage treatment and mean plaque counts of bacteriophages (*) recovered in the rumens of animals dosed with bacteriophages. Two fistulated animals were dosed with 1010 CFU of E. coli O157:H7 cocktail (NCTC 12900-nas, DAF454-nas, and 13C1T3-nas) on day 0. One animal was dosed with a 1011 PFU cocktail of e11/2 and e4/1c phages on days 1, 2, and 3 (indicated by the arrows). Plaques were not found in fecal samples (B) directly or following enrichment and are not shown in the figure. This experiment was performed once with two animals.
DISCUSSION
This study successfully demonstrated that bacteriophages e11/2 and e4/1c at MOI values of 1 to 1,000 PFU/CFU reduced inoculated E. coli O157:H7 levels in an ex vivo model rumen system within 1 to 4 h. One of the few published studies that have used an ex vivo rumen system to evaluate the activity of phage against E. coli O157:H7 is a study by Bach et al. (5). This study reported that phage D22 required MOI values of 1,000 to 10,000 PFU/CFU to significantly reduce levels of inoculated E. coli O157:H7 in an ex vivo Rusitec system. Variations in MOI values between the studies may be attributed to the different phages evaluated as well as the different ex vivo systems used. Other studies using ex vivo broth techniques have also suggested that aeration, high MOIs, and the use of phage cocktails are critical for rapid cell lysis and complete elimination of the pathogen (16). While low MOI values of each individual phage in the present study were successful in rapidly reducing E. coli O157:H7 levels in the ex vivo rumen system, mixing during the flushing of tubes with CO2 may have increased the opportunity for phage-bacterium interaction (16). A cocktail of the two phages was used in the in vivo animal trials to prevent the emergence of phage-resistant cells, as noted by other studies (38). Previous work with e11/2 and e4/1c found an emergence of bacteriophage-insensitive mutants following challenge, but their formation was at a low level, and the mutants commonly reverted to a bacteriophage-sensitive form (26).
In vivo application of the phages to cattle artificially inoculated with E. coli O157:H7 showed no effect on the fecal shedding of E. coli O157:H7. Studies showing successful application of bacteriophages to decrease E. coli O157:H7 shedding in animals have been published (4, 35, 41). Differences between studies may explain why the bacteriophages in the present study did not reduce E. coli O157:H7 carriage in cattle. The most likely explanation could be the lack of exposure of the phages to the inoculated E. coli O157:H7, whereby the concentration of the phages did not reach a sufficient concentration in close proximity to the E. coli O157:H7 population colonizing the animal (32). E. coli O157:H7 and phage numbers were also found to be significantly lower in the feces than in the rumen samples at similar time points, indicating that E. coli O157:H7 and the phages may have been inactivated by various physiological conditions or experienced nonspecific binding within the gastrointestinal tract (GIT) of the animal (1, 5).
In the present study, E. coli O157:H7 was found to independently survive the ex vivo model rumen system for over 24 h, whereas Bach et al. (5) reported a natural decline of E. coli O157:H7 in a continuous Rusitec fermentation system over a longer incubation time (192 h). Indeed, E. coli O157:H7 and the bacteriophages in the present study were also found to be recovered in vivo in the rumen but were found to decline over time (∼92 h). Shedding of E. coli O157:H7 also appeared to vary between animals in the trials, but the numbers of E. coli O157:H7 shed usually decreased over time, and most animals remained E. coli O157:H7 enrichment positive through to the end time of the trials. Other researchers have reported a heterogeneity of E. coli O157:H7 carriage and excretion rates between animals and studies (10). The factors that contribute to the persistence of E. coli O157:H7 in ruminants are unclear, but it has been suggested that the rumen may be a transient site for E. coli O157:H7 and that the microorganism primarily colonizes at the recto-anal junction (RAJ) of the animal (23, 42). It was previously reported that the application of phage directly to the RAJ, as well as the addition of the phage to the drinking water, significantly reduced E. coli O157:H7 populations but did not eliminate the organism in cattle (35). It was encouraging to find that the phages in the present study were recoverable from the feces of the yearling animals, as it demonstrated that the phages can survive transit through the GITs of these animals. It may be possible in the future to incorporate the phages into protective delivery systems, such as microencapsulation, which could aid in increasing the protection of the phages from inhibition and successfully transferring higher numbers of phages to target areas within the GIT, such as the RAJ (4).
For practical reasons, the bacteriophages in the present study were applied as a therapeutic method (phages administered 24 h after inoculation with E. coli O157:H7) to represent a method that could be used preslaughter on infected animals. A greater reduction in numbers of E. coli O157:H7 has been achieved through the administration of bacteriophages prior to or together with the target bacteria as a preventative application (35, 36, 41). A study by Waddell et al. (41) reported the successful reduction of E. coli O157:H7 fecal shedding in calves by using a mixture of E. coli O157-specific phages as a preventative treatment (administered before E. coli O157:H7 inoculation) as well as after inoculation of E. coli O157:H7 (days 0 and 1). It has been suggested that bacterial reductions are achieved through the use of large or repeated doses of bacteriophages associated with no actual replication of bacteriophages in the host (passive therapy), although sometimes active phage replication (active therapy) has been seen in vivo (29). Self-replication would make bacteriophage therapy a far more viable option for the control of E. coli O157:H7 within a livestock production environment. In contrast, high phage levels have been found to be detrimental to efficacy (9), whereby adding too much phage interferes with subsequent phage penetration to bacteria, such as by effecting lysis from without (which is a phage-mediated bacterial killing without corresponding phage release) (1).
In general, greater success has been achieved when experimental trials are carried out with smaller animals such as mice, sheep, and calves (4, 9, 30, 35, 37, 41). In the present study, yearling cattle (240 kg, average weight) were used in one trial, while adult (fistulated) cattle (750 kg, average weight) were used in another trial. Both E. coli O157:H7 and phage were recovered from the feces of yearling cattle, but phage was not recovered from the feces of the adult fistulated cattle. The adult animals were much larger than the yearling cattle, which could indicate a difference in phage and E. coli O157:H7 ratios present in the GITs of these animals, as well as the influence of other factors, such as the ability of the phage and/or E. coli O157:H7 to survive and attach to the RAJ or differences between the GIT environments of these animals. Also, the age-related difference in development of the GITs and the immune systems may explain the difference in efficacy in calves and older animals (15, 35).
In addition, studies that have reported a reduction in E. coli O157:H7 numbers usually find that the organism is never eliminated (35). Ideally, the reduction of the pathogen load in the animals would potentially reduce food-borne illness risk without a reduction in prevalence of the organism (7). E. coli O157:H7 is easily transferred between animals and their surrounding environments (21, 34). Sources such as water, soil, and manure can be long-term reservoirs for E. coli O157:H7, and reinfection can occur. In the present study, the hides and pens of many animals (trial one) were positive for E. coli O157:H7 within 2 days after E. coli O157:H7 inoculation. Contamination of the hide can also contribute to the contamination of the carcass during preslaughter and is one factor increasing risk of infection (20). Feed was also often found to become contaminated by the animals following feeding and could possibly contribute to reinfection of the animal.
Bacteriophage plaques were also found in the feces (following enrichment) of some of the animals in the control group. The plaques produced by these phages were similar to those produced by e11/2 and e4/1c; however, further analysis, such as enzyme digestion of the phage DNA, is required for confirmation. Every effort was made to avoid cross-contamination between the two treatment groups; however, it is possible that the phages used in the study could have become airborne and contaminated the control animals. Alternatively, these plaques may also represent other naturally occurring phages that were not detected in our initial screening procedures or lysogenic phages propagated from the host. This is not unusual, as phages have been found to be ubiquitous within feedlots and have been commonly isolated from animal feces and slurry (24, 26). Although these plaques were so few and sporadic, it is unknown whether their numbers could have influenced the E. coli O157:H7 numbers shed by the control animals.
Conclusions.
This study shows that bacteriophages e11/2 and e4/1c were able to reduce E. coli O157:H7 CFU inoculated in an ex vivo model rumen system but in the cattle trials showed no effect on the fecal shedding of E. coli O157:H7. There is a considerable potential for development and inclusion of phage therapies in preharvest intervention strategies. However, further research is required to improve the activities and/or survival of the phages in vivo as well as to optimize delivery systems and route and timing of phage administration. The dose and route of administration of the phages to animals in particular are very important factors to consider, as it must be economical and practical for agricultural use. Although the effectiveness of other preharvest interventions (for example, vaccination and probiotic feeds) varies, no intervention is completely effective in reducing the fecal prevalence of E. coli O157:H7. It would be beneficial to incorporate phage application into a number of strategies to successfully reduce the prevalence of E. coli O157:H7 throughout the beef chain (3).
Acknowledgments
This project was funded by the Department of Agriculture, Fisheries and Food (DAFF) under the Food Institutional Research Measure (FIRM), Ireland, and Dairy Levy.
We thank colleagues from Teagasc, Beef Research Centre, Grange, and Moorepark Food Research Centre for all their assistance with the rumen model and animal experiments.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Abedon, S. T. 2009. Kinetics of phage-mediated biocontrol of bacteria. Foodborne Pathog. Dis. 6:807-815. [DOI] [PubMed] [Google Scholar]
- 2.Alisky, J., K. Iczkowski, A. Rapoport, and N. Troitsky. 1998. Bacteriophages show promise as antimicrobial agents. J. Infect. 36:5-15. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, T. M., J. E. Keen, J. M. Bosilevac, D. M. Brichta-Harhay, N. Kalchayanand, S. D. Shackelford, T. L. Wheeler, X. W. Nou, and M. Koohmaraie. 2009. Longitudinal study of Escherichia coli O157:H7 in a beef cattle feedlot and role of high-level shedders in hide contamination. Appl. Environ. Microbiol. 75:6515-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bach, S. J., R. P. Johnson, K. Stanford, and T. A. McAllister. 2009. Bacteriophages reduce Escherichia coli O157:H7 levels in experimentally inoculated sheep. Can. J. Anim. Sci. 89:285-293. [Google Scholar]
- 5.Bach, S. J., T. A. McAllister, D. M. Veira, V. P. J. Gannon, and R. A. Holley. 2003. Effect of bacteriophage DC22 on Escherichia coli O157:H7 in an artificial rumen system (Rusitec) and inoculated sheep. Anim. Res. 52:89-101. [Google Scholar]
- 6.Barrow, P., M. Lovell, and A. Berchieri. 1998. Use of lytic bacteriophage for control of experimental Escherichia coli septicemia and meningitis in chickens and calves. Clin. Diagn. Lab. Immunol. 5:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brichta-Harhay, D. M., T. M. Arthur, J. M. Bosilevac, M. N. Guerini, N. Kalchayanand, and M. Koohmaraie. 2007. Enumeration of Salmonella and Escherichia coli O157:H7 in ground beef, cattle carcass, hide and faecal samples using direct plating methods. J. Appl. Microbiol. 103:1657-1668. [DOI] [PubMed] [Google Scholar]
- 8.Callaway, T. R., M. A. Carr, T. S. Edrington, R. C. Anderson, and D. J. Nisbet. 2009. Diet, Escherichia coli O157:H7, and cattle: a review after 10 years. Curr. Issues Mol. Biol. 11:67-79. [PubMed] [Google Scholar]
- 9.Callaway, T. R., T. S. Edrington, A. D. Brabban, R. C. Anderson, M. L. Rossman, M. J. Engler, M. A. Carr, K. J. Genovese, J. E. Keen, M. L. Looper, E. M. Kutter, and D. J. Nisbet. 2008. Bacteriophage isolated from feedlot cattle can reduce Escherichia coli O157:H7 populations in ruminant gastrointestinal tracts. Foodborne Pathog. Dis. 5:183-191. [DOI] [PubMed] [Google Scholar]
- 10.Chase-Topping, M., D. Gally, C. Low, L. Matthews, and M. Woolhouse. 2008. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat. Rev. Microbiol. 6:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duffy, G., F. Butler, E. Cummins, S. O'Brien, P. Nally, E. Carney, M. Henchion, D. Mahon, and C. Cowan. 2006. E. coli O157:H7 in beefburgers produced in the Republic of Ireland: a quantitative microbial risk assessment. Teagasc, Ashtown Food Research Centre, Dublin, Ireland.
- 12.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Brarkocy-Gallagher, M. Koohmaraie, and W. W. Lagreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. U. S. A. 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Food Safety Authority. 2007. Monitoring of verotoxigenic Escherichia coli (VTEC) and identification of human pathogenic VTEC types: scientific opinion of the panel on biological hazards. EFSA J. 591:1-61. [Google Scholar]
- 14.Greer, G. G. 2005. Bacteriophage control of foodborne bacteria. J. Food Prot. 68:1102-1111. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, R. P., C. L. Gyles, W. E. Huff, S. Ojha, G. R. Huff, N. C. Rath, and A. M. Donoghue. 2008. Bacteriophages for prophylaxis and therapy in cattle, poultry and pigs. Anim. Health Res. Rev. 9:201-215. [DOI] [PubMed] [Google Scholar]
- 16.Kudva, I. T., S. Jelacic, P. I. Tarr, P. Youderian, and C. J. Hovde. 1999. Biocontrol of Escherichia coli O157 with O157-specific bacteriophages. Appl. Environ. Microbiol. 65:3767-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeJeune, J. T., and A. N. Wetzel. 2007. Preharvest control of Escherichia coli O157 in cattle. J. Anim. Sci. 85:E73-E80. [DOI] [PubMed] [Google Scholar]
- 18.Loneragan, G. H., and M. M. Brashears. 2005. Pre-harvest interventions to reduce carriage of E. coli O157 by harvest-ready feedlot cattle. Meat Sci. 71:72-78. [DOI] [PubMed] [Google Scholar]
- 19.Matthews, L., J. C. Low, D. L. Gally, M. C. Pearce, D. J. Mellor, J. A. Heesterbeek, M. Chase-Topping, S. W. Naylor, D. J. Shaw, S. W. Reid, G. J. Gunn, and M. E. Woolhouse. 2006. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc. Natl. Acad. Sci. U. S. A. 103:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McEvoy, J. M., A. M. Doherty, J. J. Sheridan, F. M. Thomson-Carter, P. Garvey, L. McGuire, I. S. Blair, and D. A. McDowell. 2003. The prevalence and spread of Escherichia coli O157:H7 at a commercial beef abattoir. J. Appl. Microbiol. 95:256-266. [DOI] [PubMed] [Google Scholar]
- 21.McGee, P., L. Scott, J. J. Sheridan, B. Earley, and N. Leonard. 2004. Horizontal transmission of Escherichia coli O157:H7 during cattle housing. J. Food Prot. 67:2651-2656. [DOI] [PubMed] [Google Scholar]
- 22.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naylor, S. W., J. C. Low, T. E. Besser, A. Mahajan, G. J. Gunn, M. C. Pearce, I. J. McKendrick, D. G. Smith, and D. L. Gally. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu, Y. D. Xu, Y. McAllister, T. A. Rozema, E. A. Stephens, T. P. Bach, S. J. Johnson, and R. P. Stanford. 2008. Comparison of fecal versus rectoanal mucosal swab sampling for detecting Escherichia coli O157:H7 in experimentally inoculated cattle used in assessing bacteriophage as a mitigation strategy. J. Food Prot. 71:691-698. [DOI] [PubMed] [Google Scholar]
- 25.O'Brien, S. B., G. Duffy, E. Carney, J. J. Sheridan, D. A. McDowell, and I. S. Blair. 2005. Prevalence and numbers of Escherichia coli O157 on bovine hides at a beef slaughter plant. J. Food Prot. 68:660-665. [DOI] [PubMed] [Google Scholar]
- 26.O'Flynn, G., R. P. Ross, G. F. Fitzgerald, and A. Coffey. 2004. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 70:3417-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Sullivan, D., R. P. Ross, D. P. Twomey, G. F. Fitzgerald, C. Hill, and A. Coffey. 2001. Naturally occurring lactococcal plasmid pAH90 links bacteriophage resistance and mobility functions to a food-grade selectable marker. Appl. Environ. Microbiol. 67:929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, R. W. A. 1978. The isolation and use of streptomycin resistant mutants for following development of bacteria in mixed populations, p. 107-112. In D. Lovelock and R. Davis (ed.), Techniques for the study of mixed populations. Academic Press, London, United Kingdom.
- 29.Payne, R. J. H., and V. A. A. Jansen. 2001. Understanding bacteriophage therapy as a density-dependent kinetic process. J. Theor. Biol. 208:37-48. [DOI] [PubMed] [Google Scholar]
- 30.Raya, R. R., P. Varey, R. A. Oot, M. R. Dyen, T. R. Callaway, T. S. Edrington, E. M. Kutter, and A. D. Brabban. 2006. Isolation and characterization of a new T-even bacteriophage, CEV1, and determination of its potential to reduce Escherichia coli O157:H7 levels in sheep. Appl. Environ. Microbiol. 72:6405-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. Mcgee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 32.Rozema, E. A., T. P. Stephens, S. J. Bach, E. K. Okine, R. P. Johnson, K. Stanford, and T. A. McAllister. 2009. Oral and rectal administration of bacteriophages for control of Escherichia coli O157:H7 in feedlot cattle. J. Food Prot. 72:241-250. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Schouten, J. M., E. A. M. Graat, K. Frankena, F. Van Zijderveld, and M. C. M. De Jong. 2009. Transmission and quantification of verocytotoxin-producing Escherichia coli O157 in dairy cattle and calves. Epidemiol. Infect. 137:114-123. [DOI] [PubMed] [Google Scholar]
- 35.Sheng, H., H. J. Knecht, I. T. Kudva, and C. J. Hovde. 2006. Application of bacteriophages to control intestinal Escherichia coli O157:H7 levels in ruminants. Appl. Environ. Microbiol. 72:5359-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, H. W., M. B. Huggins, and K. M. Shaw. 1987. The control of experimental Escherichia coli diarrhea in calves by means of bacteriophages. J. Gen. Microbiol. 133:1111-1126. [DOI] [PubMed] [Google Scholar]
- 37.Tanji, Y., T. Shimada, H. Fukudomi, K. Miyanaga, Y. Nakai, and H. Unno. 2005. Therapeutic use of phage cocktail for controlling Escherichia coli O157:H7 in gastrointestinal tract of mice. J. Biosci. Bioeng. 100:280-287. [DOI] [PubMed] [Google Scholar]
- 38.Tanji, Y., T. Shimada, M. Yoichi, K. Miyanaga, K. Hori, and H. Unno. 2004. Toward rational control of Escherichia coli O157:H7 by a phage cocktail. Appl. Microbiol. Biotech. 64:270-274. [DOI] [PubMed] [Google Scholar]
- 39.Teunis, P., K. Takumi, and K. Shinagawa. 2004. Dose response for infection by Escherichia coli O157:H7 from outbreak data. Risk Anal. 24:401-407. [DOI] [PubMed] [Google Scholar]
- 40.Tilley, J. M., and R. A. Terry. 1963. A two stage technique for the in vitro digestion of forage crops. Grass Forage Sci. 18:104-111. [Google Scholar]
- 41.Waddell, T. E., A. Mazzocco, J. Pacan, R. Ahmed, R. Johnson, C. Poppe, and R. Khakhria. 26 November 2002. Use of bacteriophages for control of Escherichia coli O157. U.S. patent 6,485,902 B2.
- 42.Walker, C., X. Shi, M. Sanderson, J. Sargeant, and T. G. Nagaraja. 2010. Prevalence of Escherichia coli O157:H7 in gut contents of beef cattle at slaughter. Foodborne Pathog. Dis. 7:249-255. [DOI] [PubMed] [Google Scholar]