Abstract
The role of bacteria in the occasional emergence of red water, which has been documented worldwide, has yet to be determined. To better understand the mechanisms that drive occurrences of red water, the bacterial community composition and the relative abundance of several functional bacterial groups in a water distribution system of Beijing during a large-scale red water event were determined using several molecular methods. Individual clone libraries of the 16S rRNA gene were constructed for three red water samples and one sample of normal water. Beta-, Alpha-, and Gammaproteobacteria comprised the major bacterial communities in both red water and normal water samples, in agreement with previous reports. A high percentage of red water clones (25.2 to 57.1%) were affiliated with or closely related to a diverse array of iron-oxidizing bacteria, including the neutrophilic microaerobic genera Gallionella and Sideroxydans, the acidophilic species Acidothiobacillus ferrooxidans, and the anaerobic denitrifying Thermomonas bacteria. The genus Gallionella comprised 18.7 to 28.6% of all clones in the three red water libraries. Quantitative real-time PCR analysis showed that the 16S rRNA gene copy concentration of Gallionella spp. was between (4.1 ± 0.9) × 107 (mean ± standard deviation) and (1.6 ± 0.3) × 108 per liter in red water, accounting for 13.1% ± 2.9% to 17.2% ± 3.6% of the total Bacteria spp. in these samples. By comparison, the percentages of Gallionella spp. in the normal water samples were 0.1% or lower (below the limit of detection), suggesting an important role of Gallionella spp. in the formation of red water.
On occasion, extensive precipitation of iron oxides in drinking water distribution systems manifests as red water at the tap and results in serious deterioration of water quality, with undesirable esthetic and health effects (18, 40, 46). The abundance of ferrous iron in source water or the acceleration of corrosion of iron pipelines after the loosening of chemical and microbial films from the interior surfaces of distribution systems might be the sources of iron oxides in red water. Switching of water sources has been observed to be associated with red water due to disruption of the delicate chemical equilibrium in water supply systems (18). High concentrations of anions, particularly sulfate ions, have been recognized as a causative agent of red water in many cases, reflected in high values on indices such as the Larson-Skold index (18, 29). Other physicochemical factors, such as insufficient disinfection residue, extended hydraulic retention time, low levels of dissolved oxygen, high temperature, low alkalinity, and high chloride concentration, have also been implicated in the emergence of red water (18, 46).
In addition to physicochemical factors, microorganisms may also participate in the unique phenomenon of red water. Drinking water distribution systems are a unique niche for microorganisms, despite oligotrophic conditions and the presence of free or combined chlorine (3, 18). Phylogenetically diverse bacterial groups can inhabit the bulk water or biofilms attached to pipes. Culture-based and independent analyses have revealed that members of the class Proteobacteria, including the Alpha-, Beta-, and Gammaproteobacteria, are typically the most abundant bacterial group in water distribution systems, followed by bacterial phyla such as Actinobacteria, Firmicutes, and Bacteroidetes (13, 38). Bacteria inhabiting distribution systems mainly fill functions of diverse carbon source utilization and nitrification, as well as microbial corrosion (3). Meanwhile, during periods of red water, abundant ferrous iron in the bulk water creates favorable conditions for the growth of bacteria in the distribution systems, as this iron scavenges residual chlorine and serves as an energy source for iron-oxidizing bacteria. Some neutrophilic iron oxidizers, such as Gallionella spp. and Leptothrix ochracea, which have occasionally been observed in association with red water events because of their distinct morphology, can promote the precipitation of iron oxides by converting ferrous iron to ferric iron (9, 46). As very little energy can be generated during the oxidation of ferrous to ferric iron, a large quantity of iron needs to be oxidized to support the growth of lithotrophic iron oxidizers. It has been calculated that the ratio of iron to the weight of bacterial cell material could be up to approximately 450 to 500, assuming that the oxidation of ferrous iron provides the sole energy for the synthesis of cell material (9). Emerson et al. have found that the oxidation rate of ferrous iron could be up to 600 to 960 nmol per h per cm3 of mat material that contained up to 109 bacterial cells, most of which were iron oxidizers like Gallionella spp. and Leptothrix ochracea, and the oxidation rate of ferrous iron by iron oxidizers could be as high as four times that of dissolved oxygen (15). These neutrophilic iron oxidizers have been even utilized to remove iron from groundwater by passage of preaerated water through sand filters during drinking water treatment (24, 36). Thus, iron-oxidizing species might play an important role in red water events. With the exception of specific neutrophilic iron oxidizers (e.g., Gallionella spp. and Leptothrix ochracea), the whole microbial community composition in red water and the presence of potentially functional groups, including neutrophilic iron-oxidizing bacteria in red water, is poorly defined, possibly because the appearance of this unique phenomenon in real distribution systems is so irregular. To better understand the mechanisms that drive the emergence of red water, the bacterial community composition and the relative abundance of several functional bacterial groups in a water distribution system of Beijing during a large-scale red water event were determined using several molecular methods. The results of this comprehensive investigation of the biological component of red water will provide valuable information for those managing red water events in water distribution systems.
MATERIALS AND METHODS
Study site and sampling.
Red water from a drinking water supply company occurred in large areas of Beijing, China, soon after 80% of the source water was switched in steps from local surface water to water from a neighboring province in late September, 2008. The percentage of water from the new source was soon decreased to approximately 30% after the appearance of red water. However, the phenomenon of red water still persisted for nearly 3 months. The water supply company has an average output of about 1.5 million m3 of drinking water per day. Raw water undergoes conventional and enhanced treatments, including chemical precipitation and flocculation, sedimentation, coal and sand filtration, biological activated carbon filtration, and finally, chloramination. The drinking water distribution system was mainly made of cast iron pipes. Although the majority of the areas supplied by this company reported the phenomenon of red water, the water quality in some places was still quite normal, without perceptible color or turbidity. Forty-liter tap water samples were individually obtained in autoclaved glass bottles from four endpoints of the same distribution system, including three red water points, Z, J, and S, and a normal point, D, located in different downtown areas on 11, 18, 22, and 26 October 2008, respectively. The distance between different sampling points ranged from 1.0 km to 5.7 km. Finished water was also sampled from the waterworks. Water samples were kept at 4°C in darkness for at most 2 h before analysis. Twenty-liter water samples were used for water quality analysis, and the remaining 20-liter amounts were used for microbial analysis. The detailed water quality parameters analyzed and the ranges of values are listed in Table 1.
TABLE 1.
Summary of water quality parameter valuesa for normal and red water samples
| Siteb | Temp (°C) | Residual chlorine (mg/liter) | Turbidity (NTUc) | Color (color units) | pH | Total iron (mg/liter) | Dissolved iron (mg/liter) | Sulfate (mg/liter) | Chloride (mg/liter) | Conductivity (μS/cm) | Dissolved oxygen (mg/liter) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| R1 | NDd | ND | 0.56-3.30 | ND | 7.66-8.34 | ND | <0.05 | 23.7-43.6 | 14.2-18.7 | ND | ND |
| R2 | ND | ND | ND | ND | ND | ND | ND | 200-230 | 41.0-44.7 | ND | ND |
| Z | 20-22 | 0.03-0.08 | 1.60-27.5 | <5-35 | 7.59-7.69 | 0.39-1.39 | <0.05-0.17 | 63.0-68.3 | 23.4-24.7 | 393-472 | 6.73-7.41 |
| J | 20-22 | 0.03-0.05 | 0.45-31.9 | <5-28 | 7.56-7.60 | 0.12-1.61 | <0.05-0.07 | 58.8-64.2 | 23.2-23.6 | 403-463 | 7.02-7.50 |
| S | 20-22 | 0.03-0.2 | 0.98-12.3 | <5-12 | 7.59-7.73 | 0.15-0.67 | <0.05-0.15 | 59.1-70.7 | 23.4-26.0 | 440-487 | 7.02-7.48 |
| D | 20-22 | 0.1-0.6 | 0.09-0.63 | <5 | 7.50-7.84 | <0.05-0.07 | <0.05 | 54.1-89.0 | 22.0-25.5 | 390-470 | 8.01-8.62 |
| Inlet | 17-20 | 0.7-0.8 | 0.09-0.12 | <5 | 7.68-7.74 | <0.05 | <0.05 | 57.8-95.7 | 24.3-30.4 | 400-460 | 8.14-8.55 |
The ranges of values are shown.
R1 represents raw water before the switch to a new source of water; R2 represents the new source of water. The four sampling points (Z, J, S, and D) represent endpoints of the drinking water distribution system located in different downtown areas; samples were taken during the red water event. Inlet represents inlet water of the drinking water distribution system during the red water event.
NTU, nephelometric turbidity units.
ND, not determined.
Heterotrophic bacterium counts.
To roughly estimate heterotrophic bacterium counts, the bacteria in 2 liters of the water samples were harvested on 0.22-μm-pore-size Millipore GSWP filters by filtration and subsequently resuspended from the filter surface with 2 to 10 ml of physiological saline by vortexing, and then 0.05- to 0.1-ml aliquots were inoculated onto LB agar medium. The plates were incubated aerobically at 37°C for 24 h to roughly determine heterotrophic bacterium counts according to the drinking water standards of China (GB5749-85 [34]).
DNA extraction and PCR analysis.
Bacteria from 3 liters of water of each sample were harvested by membrane filtration with 0.22-μm-pore-size Millipore GSWP filters and then suspended in 10 ml of physiological saline. After centrifugation, DNA was extracted using a FastDNA spin kit for soil (Qbiogene, Solon, OH) facilitated with the FastPrep-24 bead beater system, following the manufacturer's instructions, and then quantified with a Nanodrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE).
Nearly the entire bacterial 16S rRNA gene was amplified using bacterial universal primers 27f and 1492r (Table 2). The 16S rRNA gene fragment of ammonia-oxidizing bacteria (AOB) affiliating with the Betaproteobacteria was amplified using the primers CTO189fA/B, CTO189fC, and CTO654r. The first two forward primers were used in a 2:1 ratio as described before (26). The partial dsrB gene of sulfate-reducing bacteria (SRB) was amplified with the primers DSRp2060f and DSR4r. The standard 50-μl PCR mixture (Takara, Dalian, China) included 1× PCR buffer containing 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 10 pmol each primer, 1.25 U of TaKaRa rTaq polymerase, and approximately 50 ng of template DNA. The PCR conditions for the amplification of the bacterial 16S rRNA gene using universal primers 27f and 1492r were as follows: 95°C for 10 min, followed by 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min 30 s and a final extension at 72°C for 15 min. For the amplification of AOB, the PCR conditions were 95°C for 10 min, followed by 35 cycles of 95°C for 1 min, 57°C for 1 min, and 72°C for 1 min and a final extension at 72°C for 10 min. The amplification conditions for SRB were the same as for AOB except that the annealing temperature was 55°C. PCR products were confirmed by electrophoresis in 1.2% (wt/vol) agarose gel.
TABLE 2.
Oligonucleotide primers and probes used in this study
| Primer or probe | Specificity | Sequence (5′-3′) | Target genes | Base position | Application | Reference |
|---|---|---|---|---|---|---|
| 27f | Bacteria | AGAGTTTGATCCTGGCTCAG | 16S rRNA | 8-27 | PCR | 28 |
| 1492r | Universal | TACGGYTACCTTGTTACGACTT | 16S rRNA | 1492-1513 | PCR | 28 |
| CTO189fA/B | AOB | GGAGRAAAGCAGGGGATCG | 16S rRNA | 190-208 | PCR | 26 |
| CTO189fC | AOB | GGAGGAAAGTAGGGGATCG | 16S rRNA | 190-208 | PCR | 26 |
| CTO654r | AOB | CTAGCYTTGTAGTTTCAAACGC | 16S rRNA | 633-654 | PCR | 26 |
| DSRp2060f | SRB | CAACATCGTYCAYACCCAGGG | dsrB | PCR | 17 | |
| DSR4r | SRB | GTGTAGCAGTTACCGCA | dsrB | PCR | 17 | |
| GAL1f | Gallionella genus | CGAAAGTTACGCTAATACCGCATA | 16S rRNA | 158-181a | qPCR | This study |
| GAL1r | Gallionella genus | CTCAGACCAGCTACGGATCGT | 16S rRNA | 279-299a | qPCR | This study |
| GAL1p | Gallionella genus | CCTCTCGCTTTCGGAGTGGCCG | 16S rRNA | 214-235a | qPCR | This study |
| GAL2f | Gallionella genus | AAGCGGTGGATTATGTGGATT | 16S rRNA | 937-957a | qPCR | This study |
| GAL2r | Gallionella genus | ACAAGGGTTGCGCTCGTT | 16S rRNA | 1101-1118a | qPCR | This study |
| GAL2p | Gallionella genus | CCAGGAAGATTTCAGAGATGAGATTGTGCC | 16S rRNA | 999-1028a | qPCR | This study |
| UNI331f | Bacteria | TCCTACGGGAGGCAGCAGT | 16S rRNA | 340-358 | qPCR | 37 |
| UNI797r | Bacteria | GGACTACCAGGGTATCTAATCCTGTT | 16S rRNA | 781-806 | qPCR | 37 |
| UNIp | Bacteria | CGTATTACCGCGGCTGCTGGCAC | 16S rRNA | 515-537 | qPCR | 37 |
| NSO190 | AOB | CGATCCCCTGCTTTTCTCC | 16S rRNA | 190-208 | FISH | 35 |
According to Escherichia coli numbering (4).
Cloning and sequencing of 16S rRNA genes.
Three separate reactions for the bacterial 16S rRNA gene using the universal primers 27f and 1492r were run for each sample to minimize PCR bias in subsequent cloning steps, and all PCR products of water samples from the same point were further pooled together. The amplification products were purified with a QIAquick PCR cleanup kit (Qiagen, Inc., Chatsworth, CA) and cloned into the TOPO TA cloning vector pCR2.1, with TOP10 Escherichia coli transformants further selected according to the manufacturer's instructions (Invitrogen). Cloned inserts were amplified from lysed colonies by PCR with plasmid vector-specific primers M13F and M13R under the same conditions as for the 16S rRNA gene listed above. Positive clones were sequenced with an ABI 3730 automated sequencer (Invitrogen, Shanghai, China).
Phylogenetic and statistical analysis.
The detailed phylogenetic and statistical analyses were generally the same as described before (30). DNA sequences were assembled with the Phred/Phrap/Consed package (www.phred.org), and possible chimeras were checked with Bellerophon version 3 (http://greengenes.lbl.gov/cgi-bin/nph-bel3_interface.cgi). The most similar reference sequences were retrieved from RDP and the GenBank database (1, 6), and then phylogenetic trees were constructed using MEGA 4 (27). The operational taxonomic unit (OTU) number was determined using DOTUR by defining the sequences sharing 97% or greater similarity as one OTU (42). OTU richness values SChao1 and SACE, as well as the Shannon diversity index (H), were calculated using EstimateS version 8.0 (7). Evenness (E) indices were calculated as follows: E = H/lnn, where n is the number of OTUs. Coverage (C) was calculated as follows: C = 1 − (n1/N), where n1 is the number of OTUs that occurred once and N is the total number of clones (44). Rarefaction curves were constructed using DOTUR. UniFrac computational analysis was performed to compare clone libraries from different sampling sites (32). All statistical analyses were performed by using the SPSS version 16.0 release.
Primer and probe design and quantitative real-time PCR (qPCR).
All available 16S rRNA sequences of Gallionella cultured and as-yet-uncultured strains were retrieved from RDP release 10, Greengenes (10), and the GenBank database. These sequences were added to the ARB database ssujun02.arb together with the sequences affiliated with this group in our study (33). Possible TaqMan probes specific for Gallionella spp. were designed using the ARB probe design and probe match programs and Primer Express software version 3.0 (Applied Biosystems, Foster City, CA). The specificity of the probes was evaluated in silico with the Probe Match tool in RDP, ARB Probe Match Online, the Probe tool in Greengenes, and the BLAST search at the National Center for Biotechnology Information. The primers that were combined with the probes were designed using Primer Express software 3.0. Two sets of primers and probes for Gallionella bacteria were designed, including one set applicable for almost all Gallionella sequences available now and one set specific for partial sequences obtained from the red water of this study (Table 2). A universal probe and primer set described before was used for the quantification of the total bacteria. The probes were 5′-end labeled with 6-carboxyfluorescein (FAM) as the reporter and 3′-end labeled with 6-carboxytetramethylrhodamine (TAMRA) as a quencher (Takara, Dalian, China).
qPCR was performed in a 25-μl final reaction mixture volume consisting of 12.5 μl of Premix Ex Taq (perfect real time) (Takara, Dalian, China), 0.5 μl of 10 μM forward and reverse primers, 1.0 μl of 3 μM TaqMan probe, 8.5 μl of distilled water, 0.5 μl of ROX reference dye (50×), and 2.0 μl of DNA template. PCR amplifications were carried out in 96-well optical plates on an Applied Biosystems 7300 qPCR system with 7300 SDS 1.4 software (Applied Biosystems) using the following protocols: 95°C for 30 s, followed by 45 cycles of 95°C for 30 s and 58°C for 40 s. Clones harboring 16S rRNA sequences of Gallionella spp. were selected, and the carried plasmids were used as standard template DNA for both Gallionella spp. and total Bacteria spp. after extraction with a Tianprep mini plasmid kit (Tiangen Biotech, China) and purification with a QIAquick PCR cleanup kit (Qiagen, Inc., Chatsworth, CA). Standard curves were generated with serial dilutions (10 to 108 copies per microliter) of the plasmids, and the threshold cycle values of unknown samples were plotted on the standard curves to determine the copy numbers of target sequences. All qPCRs were performed in triplicate.
FISH.
The bacteria in 2 liters of the water samples were collected by filtration and resuspension as described above. Then, fluorescence in situ hybridization (FISH) was performed according to the standard procedures of Amann (2). The Cy3-labeled probe NSO190 was applied to enumerate the target AOB group of Betaproteobacteria in red water samples. Samples were counterstained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) at 4°C in darkness for 5 min prior to microscopy. A negative control (lacking a probe) was prepared to monitor autofluorescence. Microscopy counts of hybridized and DAPI-stained cells were performed with an epifluorescence microscope (Axioskop2 mot plus; Zeiss, Germany) equipped with a cooled charge-coupled device camera (AxioCam MRm; Zeiss, Germany) by using the software provided by Zeiss (Axio Vision 4.1). The final results were determined from 20 views with a minimum of 50 cells per view counted.
Nucleotide sequence accession numbers.
The 16S rRNA nucleotide sequence data from this study were deposited in the GenBank database under the accession numbers GQ388775 to GQ389207.
RESULTS
Water quality and total cell counts.
Following the switch to a new water source in a water distribution system in Beijing, the new raw source water contained significantly higher concentrations of sulfate and chloride than the local source (Table 1). After the appearance of red water, the water quality of the control samples (collected from point D) was similar to the effluent from the waterworks (inlet), with the exception of residual chlorine, which is normally consumed during the distribution process. In contrast, the red water samples (collected from points Z, J, and S) differed significantly in several respects from the control samples. The residual chlorine and dissolved oxygen (DO) levels were lower in the red water samples than in the controls (Mann-Whitney U test, P < 0.02). Turbidity, color, total iron, and dissolved iron levels were higher in the red water samples than in the control samples (Mann-Whitney U test, P < 0.03 for turbidity and total iron). The levels of turbidity, color, and total iron in the red water samples were above permissible levels according to the drinking water standards of China (GB5749-85). Other parameters, including pH, sulfate, chloride, and conductivity were not statistically different between red water and control samples (Mann-Whitney U test, all P > 0.4). The heterotrophic bacterium counts were (2.73 ± 0.27) × 104 (mean ± standard deviation), (1.07 ± 0.37) × 104, (1.43 ± 0.21) × 104, and (3.91 ± 0.47) × 102 CFU/liter, respectively. The bacterial cell numbers were significantly higher in red water samples than in the controls, by at least an order of magnitude (Mann-Whitney U test, P < 0.02). No distinct trends in water quality parameters were evident in water samples collected at different times during the red water event.
Bacterial community composition.
For each of the water samples (Z, J, S, and D), a 16S rRNA gene library was constructed. A total of 433 sequences were obtained and grouped into 200 OTUs. Possible chimeras were discarded. UniFrac metric analysis showed that the three red water bacterial communities (Z, J, and S) were more similar to each other than to the control bacterial community (D), and this result was confirmed by principal component analysis (data not shown).
Sequence analysis of clones derived from the control water samples (D) indicated that the majority (64.1%) were affiliated with the phylum Proteobacteria, including the classes Betaproteobacteria, Gammaproteobacteria, and Alphaproteobacteria, followed by the phyla Bacteroidetes (32.8%) and Actinobacteria (3.1%) (Table 3). The bacterial genera that were identified were common residents of potable water distribution systems or fresh water and included Pseudomonas spp., Flavobacterium spp., Propionivibrio spp., Sphingomonas spp., Comamonas spp., Rhodoferax spp., Ferribacterium spp., Dyadobacter spp., and Pedobacter spp. One clone, D42, which was grouped into Rhodoferax spp., showed 97.6% similarity to R. ferrireducens type strain T118 (GenBank accession no. AF435948), and clones D14 and D48 within the genus Ferribacterium showed moderate similarity (94.0 to 97.2%) to F. limneticum type strain cda-1 (GenBank accession no. Y17060) (Fig. 1). These two species are both dissimilatory ferric iron reducers, which are a phylogenetically diverse group usually falling into the Gamma- and Deltaproteobacteria (31).
TABLE 3.
Distribution of phylogenetic groups among bacterial 16S rRNA gene clone libraries for three red water samples and a normal water samplea
| Phylum | Class | Order | Family | Genus | No. of clones (no. of OTUs) in sampleb: |
|||
|---|---|---|---|---|---|---|---|---|
| Z | J | S | D | |||||
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | Caulobacter | 1 (1) | 1 (1) | ||
| Brevundimonas | 1 (1) | |||||||
| Rhodospirillales | Unclassified | 1 (1) | 1 (1) | |||||
| Sphingomonadales | Sphingomonadaceae | Sphingomonas | 4 (3) | 2 (2) | 7 (5) | 1 (1) | ||
| Novosphingobium | 2 (1) | |||||||
| Sphingopyxis | 2 (1) | 1 (1) | 3 (2) | |||||
| Unclassified | 1 (1) | 3 (3) | ||||||
| Rhodobacterales | Rhodobacteraceae | Rhodobacter | 1 (1) | |||||
| Rhizobiales | Bradyrhizobiaceae | Afipia | 1 (1) | |||||
| Hyphomicrobiaceae | Hyphomicrobium | 1 (1) | ||||||
| Brucellaceae | Ochrobactrum | 1 (1) | ||||||
| Unclassified | 10 (3) | 2 (2) | 2 (1) | |||||
| Unclassified | 2 (2) | 4 (2) | 5 (5) | |||||
| Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | Rhodocyclus | 2 (1) | 2 (1) | |||
| Ferribacterium | 3 (1) | 2 (2) | ||||||
| Propionivibrio | 4 (1) | |||||||
| Unclassified | 40 (7) | 8 (5) | 16 (9) | 5 (5) | ||||
| Nitrosomonadales | Gallionellaceae | Gallionella | 42 (15) | 27 (13) | 20 (11) | |||
| Burkholderiales | Comamonadaceae | Curvibacter | 1 (1) | |||||
| Rhodoferax | 1 (1) | 1 (1) | ||||||
| Comamonas | 1 (1) | |||||||
| Simplicispira | 1 (1) | |||||||
| Unclassified | 1 (1) | 3 (3) | ||||||
| Oxalobacteraceae | Herbaspirillum | 14 (2) | 2 (2) | 6 (4) | ||||
| Unclassified | 10 (7) | 2 (2) | 2 (2) | 1 (1) | ||||
| Gammaproteobacteria | Legionellales | Legionellaceae | Legionella | 1 (1) | ||||
| Coxiellaceae | Coxiella | 1 (1) | 1 (1) | |||||
| Pseudomonadales | Pseudomonadaceae | Pseudomonas | 1 (1) | 57 (11) | 14 (6) | |||
| Xanthomonadales | Xanthomonadaceae | Nevskia | 4 (2) | |||||
| Hydrocarboniphaga | 1 (1) | |||||||
| Thermomonas | 2 (2) | |||||||
| Unclassified | 1 (1) | 3 (2) | 1 (1) | |||||
| Unclassified | 3 (1) | 1 (1) | ||||||
| Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Desulfovibrio | 1 (1) | ||||
| Unclassified | 1 (1) | |||||||
| Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | Sulfuricurvum | 2 (1) | 16 (6) | |||
| Unclassified | 2 (2) | 5 (1) | 5 (5) | 2 (2) | ||||
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | Empedobacter | 1 (1) | |||
| Flavobacterium | 9 (7) | |||||||
| Cloacibacterium | 1 (1) | |||||||
| Unclassified | 1 (1) | |||||||
| Sphingobacteria | Sphingobacteriales | Sphingobacteriaceae | Sphingobacterium | 3 (2) | ||||
| Pedobacter | 1 (1) | |||||||
| Flexibacteraceae | Dyadobacter | 1 (1) | ||||||
| Unclassified | 1 (1) | 5 (4) | ||||||
| Actinobacteria | Actinobacteria | Actinomycetales | Actinomycetaceae | Actinomyces | 1 (1) | |||
| Micrococcaceae | Arthrobacter | 1 (1) | ||||||
| Microbacteriaceae | Microbacterium | 1 (1) | ||||||
| Planctomycetes | Planctomycetacia | Planctomycetales | Planctomycetaceae | Unclassified | 1 (1) | |||
| Unclassified | 4 (4) | |||||||
| Total | 147 (57) | 115 (45) | 107 (72) | 64 (48) | ||||
Classification was based on match results of RDP and the GenBank database.
A blank indicates that no related clones were obtained.
FIG. 1.
Phylogenetic relationships of representative bacterial 16S rRNA gene sequences from four clone libraries of this study determined by the neighbor-joining method. Bootstrap values of >50% (obtained with 1,000 resamplings) are shown at nodes. The scale bar indicates 0.05 nucleotide substitutions per site. Reference sequences were obtained from RDP release 10 or GenBank. Methanobrevibacter ruminantium was used as an outgroup. GenBank accession numbers are in parentheses.
The vast majority of the clones (93.5 to 99.3%) derived from the three red water samples were affiliated with the phylum Proteobacteria, primarily comprising the classes Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria (Table 3). The remaining clones were classified into the phyla Bacteroidetes, Actinobacteria, and Planctomycetes, with only one clone grouped into each. The dominant bacterial group in the Z library was Betaproteobacteria (74.8% of all sequences), followed by Alphaproteobacteria (17.0% of all sequences). Similar results were observed for the S library, with Betaproteobacteria as the dominant class (46.7%), and Alphaproteobacteria as the second most abundant (20.6%). In the J library, Gammaproteobacteria was the dominant class (51.3%), with abundant Pseudomonas sequences, and the class Betaproteobacteria was the second largest group (33.9%). There were also 2 and 16 sequences of the J and S libraries, respectively, that aligned with the class Epsilonproteobacteria and were grouped into the sulfur-oxidizing genus Sulfuricurvum. Sulfur-oxidizing acidophiles could convert ferrous sulfide to sulfuric acid, releasing ferrous iron in the process, which in turn could be used by lithoautotrophic iron-oxidizing acidophiles, such as Acidithiobacillus ferrooxidans and Leptospirillum ferrooxidans (5, 20). Two sequences of the S library were classified into the class Deltaproteobacteria, with one sequence, S112, further grouped into the sulfate-reducing genus Desulfovibrio. Sulfate-reducing bacteria are usually associated with anaerobic iron corrosion by producing hydrogen sulfide as a corrosive agent and consuming cathodic hydrogen or a hydrogen film on iron in aqueous solutions (11). Overall, most of the bacterial genera in the three red water samples were also common residents of drinking water distribution systems, like Gallionella spp., Herbaspirillum spp., Sphingopyxis spp., Novosphingobium spp., Nevskia spp., Caulobacter spp., Hyphomicrobium spp., and Rhodocyclus spp.
There were several notable characteristics of the three red water libraries. The first was the abundance of sequences of the neutrophilic iron-oxidizing genus Gallionella (the phylogenetic details of these clones are shown in Fig. 2). The percentage of Gallionella sequences in the three red water libraries ranged from 18.7% to 28.6%. Second, 39, 2, and 13 sequences of the Z, J, and S libraries, respectively, (i.e., Z123, S1, and J78) (Fig. 1) were grouped into the family Rhodocyclaceae and showed various levels of similarity (92.9 to 98.2%) to the circumneutral, microaerobic, lithotrophic, iron-oxidizing bacteria Sideroxydans lithotrophicus strain ES-1 (GenBank accession no. DQ386264) and S. lithotrophicus strain LD-1 (GenBank accession no. DQ386859). Most of these clones (87.0%) also showed moderate similarity (94.1 to 97.1%) to Gallionella sp. clones MWE_N10 and A531 (GenBank accession no. FJ391503 and EU283473, respectively) (Fig. 1). Furthermore, four sequences in the Z and S libraries (including Z124 and S71; see Fig. 1) showed similarity (92.4 to 92.8%) to Acidithiobacillus ferrooxidans strain DSM 2392 and iron-oxidizing acidophile m-1 (GenBank accession no. AJ459800 and AF387301, respectively). One clone, D38 of the D library, was 96.0% similar to the anaerobic, denitrifying, iron-oxidizing bacterial strain BrG3 (GenBank accession no. U51103). Anaerobic oxidizers of ferrous iron, including anoxygenic phototrophic bacteria and several nitrate reducers coupling ferrous iron oxidation to nitrate reduction, have recently been isolated (12, 45). Several clones related to the dissimilatory ferric iron-reducing bacteria were also identified. In the S library, one clone belonging to the genus Rhodoferax showed 94.7% similarity to R. ferrireducens type strain T118, and three clones showed high similarity (99.3%) to Ferribacterium limneticum type strain cda-1.
FIG. 2.
Phylogenetic relationships of representative bacterial 16S rRNA gene sequences of Gallionella spp. retrieved from RDP release 10, Greengenes, and the GenBank database, as well as four clone libraries of this study determined by the neighbor-joining method. Filled triangles indicate sequence matches using the qPCR TaqMan probe GAL1p, open triangles indicate sequence matches using the TaqMan probe GAL2p, and filled squares indicate sequence matches using both probes. The sources of the reference Gallionella sequences obtained from the public databases are shown. Bootstrap values of >50% (obtained with 1,000 resamplings) are shown at the nodes. The scale bar indicates 0.02 nucleotide substitutions per site. Several reference sequences of the other bacterial genera that are phylogenetically close to the genus Gallionella were also obtained from RDP release 10 or GenBank. Sphingomonas asaccharolytica was used as an outgroup. GenBank accession numbers are in parentheses.
As shown in Table 4, the Shannon diversity indices for all drinking water samples ranged from 3.15 to 4.02, comparable to those of river water or even soil (8, 19). These results indicated that the bacterial species diversity was high in this oligotrophic potable water distribution system, although the bacterial genera were not so diverse in all red and normal water samples (Table 3). Furthermore, despite the fact that drinking water samples from four different sampling points (Z, J, S, and D) were supplied by the same company, there were only 2 OTUs in common among the four samples, with approximately 50 or more bacterial OTUs obtained for each sampling point. Among the red water libraries, 6 OTUs were shared by all three, and 10 to 17 OTUs were shared by two of the three. Nearly a third of the shared OTUs were grouped into Gallionella spp.
TABLE 4.
Coverage and diversity indices of bacterial 16S rRNA gene libraries for three red water samples and a normal water sample
| Sample | No. of clones | No. of OTUs | SChao1 value | SACE value | Shannon index | Evenness index | % Coverage |
|---|---|---|---|---|---|---|---|
| Z | 147 | 57 | 121 | 188 | 3.33 | 0.824 | 74.1 |
| J | 115 | 45 | 154 | 172 | 3.15 | 0.827 | 73.0 |
| S | 107 | 72 | 249 | 277 | 4.02 | 0.940 | 46.7 |
| D | 64 | 48 | 212 | 329 | 3.67 | 0.948 | 35.9 |
Quantification of Gallionella bacteria.
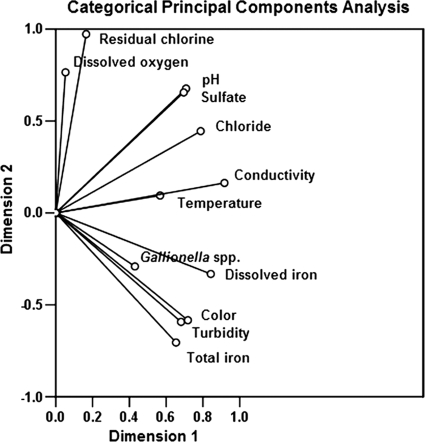
Because of the high percentage of Gallionella sequences in all three red water libraries, we developed a qPCR assay to investigate the detailed 16S rRNA gene copy numbers of Gallionella bacteria and the total Bacteria spp. in normal and red water samples. Two sets of primers and TaqMan probes were designed for the analysis. Probe GAL1p was specific for nearly all Gallionella sequences obtained from public databases and this study, whereas probe GAL2p was specific for approximately half of the Gallionella sequences obtained from this study (Fig. 2). Based on qPCR using probe GAL1p, the 16S rRNA gene copy number of Gallionella spp. ranged from (4.1 ± 0.9) × 107 to (1.6 ± 0.3) × 108 copies per liter in red water samples Z, J, and S (Table 5), indicating that the cell number of this genus had a magnitude of 107 to 108 per liter (25). By comparison, in control samples taken from point D, the copy number reached 2.2 × 104 per liter at its highest and, in several samples, was below the limit of detection (20 copies per liter). Using probe GAL2p, the 16S rRNA gene copy number of Gallionella bacteria ranged from (1.9 ± 0.5) × 107 to (4.5 ± 0.8) × 107 copies per liter in the three red water samples and from undetectable to 8.4 × 103 copies per liter in normal water samples. The total bacterial copy number in the three red water samples ranged from (3.1 ± 0.9) × 108 to (9.3 ± 4.6) × 108 per liter, an order of magnitude higher than the copy number in the control samples [(4.5 ± 2.0) × 107 per liter]. The Gallionella copy numbers (based on probe GAL1p) as the percentage of total bacteria were 17.2 ± 3.6%, 13.1 ± 2.9%, and 17.1 ± 2.6% for red water samples Z, J, and S, respectively, whereas in the control samples, the highest percentage obtained was 0.1%. These percentages, based on the qPCR results, were roughly concordant with the results obtained by sequence analysis of the clone libraries, in which Gallionella sequences accounted for 28.6%, 23.5%, and 18.7%, respectively, of all sequences in libraries Z, J, and S, whereas no sequences of this genus were present in the D library (Table 3). Furthermore, the ratio of Gallionella copy numbers obtained using GAL1p versus GAL2p in all red water samples was 2.7 ± 0.8, which was similar to the results of in silico matching of the clone libraries using the two probes (2.3 ± 1.1). As shown in Fig. 3, categorical principal component analysis of water quality parameters and 16S rRNA gene copy number of Gallionella bacteria (based on probe GAL1p) of all water samples from points Z, J, S, and D showed that the appearance of Gallionella bacteria correlated closely with total iron, dissolved iron, turbidity, and color and was negatively associated with residual chlorine and DO. These results were generally concordant with those of previous reports of conditions that favor the emergence of red water (18, 46).
TABLE 5.
Comparison of 16S rRNA gene copy numbers of Gallionella bacteria and total Bacteria spp. in three red water samples and a normal water sample
| Sample | Value (mean ± SD) for Gallionella spp. obtained using probe: |
Total Bacteria copy no. | |||
|---|---|---|---|---|---|
| GAL1p |
GAL2p |
||||
| Copy no. per liter | % Relative abundancea | Copy no. per liter | % Relative abundance | ||
| Z | 8.0 × 107 ± 1.6 × 107 | 17.2 ± 3.6 | 2.9 × 107 ± 0.4 × 107 | 6.2 ± 0.9 | 4.6 × 108 ± 1.1 × 108 |
| J | 4.1 × 107 ± 0.9 × 107 | 13.1 ± 2.9 | 1.9 × 107 ± 0.5 × 107 | 6.1 ± 1.2 | 3.1 × 108 ± 0.9 × 108 |
| S | 1.6 × 108 ± 0.3 × 108 | 17.1 ± 2.6 | 4.5 × 107 ± 0.8 × 107 | 4.9 ± 0.8 | 9.3 × 108 ± 4.6 × 108 |
| D | NDb to 2.2 × 104 | ND, 0.1 | ND to 8.4 × 103 | ND, 0 | 4.5 × 107 ± 2.0 × 107 |
Abundance of the 16S rRNA gene copy numbers of Gallionella bacteria relative to the total Bacteria copy number.
ND, not detected. The low detection limit is 20 copies per liter.
FIG. 3.
Categorical principal components analysis of water quality parameters and 16S rRNA gene copy number of Gallionella bacteria of all water samples (Z, J, S, and D). Dimension 1 explained 42.5% of the observed variation; dimension 2 explained 33.6% of the variation.
A partial dsrB gene sequence of SRB was detected by PCR in all water samples (red water and normal samples; data not shown), whereas only two Deltaproteobacteria sequences were detected in all four libraries. The presence of AOB in all four water samples was confirmed by PCR, with the quantity of less than 1% of the total cells further determined by FISH (data not shown).
DISCUSSION
Several factors could have influenced the heterotrophic bacterium levels in this study, including the adoption of activated carbon filtration in the waterworks which could reduce nutrient levels in the drinking water, the inefficient extraction of bacterial cells from filter membranes before plating onto agar medium, and the presence of numerous large iron oxide particles which might absorb many bacterial cells. Distinct differences in the values of several physicochemical and microbial parameters were observed between the red water and the control water samples. The most notable microbial characteristic was the abundance of iron-oxidizing bacteria, particularly Gallionella bacteria, in the red water samples. Neutrophilic iron-oxidizing bacteria like Gallionella spp. are difficult to obtain in pure culture, presumably because of the delicate requirement of DO and ferric iron concentrations in isolation (16). As a result, the presence of Gallionella strains in various aquatic environments is usually determined morphologically, by virtue of their unique helical stalks with bean-shaped cells attached at the termini (21). Using a gradient growth method, Gallionella strains have been obtained in pure culture (21), which has facilitated the identification of this bacterial species in various environmental samples through constructing bacterial 16S rRNA gene clone libraries and phylogenetic analyses. As shown in Fig. 2, 16S rRNA gene sequences of Gallionella spp. from diverse niches, mainly aquatic environments such as springs, mineral water, mine water, groundwater, rivers, lakes, and drinking water facilities, have been identified. Gallionella as well as Leptothrix strains have occasionally been identified in red water but are rarely quantified under these conditions. The ratio of iron-oxidizing bacteria (Gallionella and Leptothrix strains) to total bacteria in a circumneutral iron seep was approximately 10%, based on the most probable number method (16), which is comparable to the ratio of Gallionella spp. in red water in the current study, based on qPCR analysis using two sets of primers and probes specific for Gallionella spp. in aquatic samples. Gallionella bacteria in all three red water samples were significantly more abundant than in the control water samples, based on qPCR and sequence analysis of 16S rRNA libraries derived from each water sample, demonstrating the close relationship between this particular bacterial genus and the phenomenon of the red water event.
Many neutrophilic iron-oxidizing bacteria are still poorly defined due to the lack of pure cultures and nonspecific morphologies, e.g., many species in the Siderocapsaceae family (22, 23, 43). A number of the sequences (1.73 to 26.5%) derived from the three red water libraries in the current study showed moderate similarity to both Sideroxydans strains and Gallionella sequences, suggesting that these sequences may be representative of novel circumneutral-pH, microaerobic, iron-oxidizing bacterial species within this particular water distribution system. A more definitive affiliation of these sequences awaits the availability of pure cultures of more iron-oxidizing bacteria. Several clones appeared to be related to the iron-oxidizing acidophile Acidithiobacillus ferrooxidans and anaerobic, denitrifying, iron-oxidizing bacteria. In contrast, although Leptothrix ochracea strains are frequently observed in different iron-oxidizing environments (16) and this species of bacteria can grow actively in oxygen concentrations approaching 50% (4.2 mg/liter) of the ambient water (14), no L. ochracea-related sequences were identified in any of the red water or control samples. Overall, the presence of Gallionella and many other iron-oxidizing bacterial sequences indicates that a highly diverse iron-oxidizing bacterial niche exists in this drinking water supply system.
Previous studies have shown that iron-oxidizing bacteria are ubiquitous in drinking water supply systems (13, 39). The abundance of ferrous iron in source water or the release of ferrous iron into the bulk water due to chemical corrosion of iron pipelines would provide a permissive environment for heavy growth of iron-oxidizing bacteria in the bulk water. In the current study, except for the possible ferrous iron in source water, high sulfate concentrations in the new water source would also lead to depletion of the uniform calcium carbonate scales on the interior surfaces of iron pipes with the formation of calcium sulfate, resulting in significant acceleration of chemical iron corrosion and the release of ferrous iron into the bulk water. In any case, high levels of Gallionella spp., as well as of other neutrophilic iron-oxidizing bacteria in the bulk water, could facilitate the precipitation of iron oxides by converting ferrous to ferric iron, thus contributing to the formation of a red water event. Control of these iron-oxidizing bacteria, therefore, might be important in mitigating the deleterious effects of red water events. It should be still noted that only one SRB clone, which is often associated with iron corrosion in water pipes, was identified in the water libraries. Furthermore, sequences of ferric iron reduction bacteria, such as Rhodoferax ferrireducens and Ferribacterium limneticum, were not very abundant in the red water samples. The lack of SRB and ferric iron reduction bacteria suggests that biocorrosion is not a major factor in the emergence of red water.
In the current study, we identified few clones that were related to human-health-associated bacterial genera, indicating that the presence of pathogenic organisms in red water may not be a significant issue. However, the exhaustion of residual disinfectant during red water events could promote the growth of certain opportunistic pathogens. Furthermore, disinfectants such as chloramines are not effective against iron-oxidizing bacteria (41). Thus, proper disinfection measures should be considered both in terms of mitigating the effects of red water events and the overall microbial safety of drinking water (46).
Acknowledgments
This work was financially supported by the Ministry of Science and Technology of China (grant no. 2007AA06A414 and 2006DFA91870) and the National Natural Science Foundation of China (grant no. 50921064).
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Altschul, F. S., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I. 1995. In situ identification of microorganisms by whole cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkerman, D. J. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 3.Berry, D., C. Xi, and L. Raskin. 2006. Microbial ecology of drinking water distribution systems. Curr. Opin. Biotechnol. 17:297-302. [DOI] [PubMed] [Google Scholar]
- 4.Brosius, J., T. L. Dull, D. D. Steeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 5.Bruneel, O., R. Duran, C. Casiot, F. Elbaz-Poulichet, and J.-C. Personné. 2006. Diversity of microorganisms in Fe-As-rich acid mine drainage waters of Carnoulès, France. Appl. Environ. Microbiol. 72:551-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, and A. M. Bandela. 2007. The ribosomal database project (RDP-II): introducing My RDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colwell, R. K. 2005. EstimateS 8.0 user's guide. http://purl.oclc.org/estimates.
- 8.Cottrell, M. T., L. A. Waidner, L. Yu, and D. L. Kirchman. 2005. Bacterial diversity of metagenomic and PCR libraries from the Delaware River. Environ. Microbiol. 7:1883-1895. [DOI] [PubMed] [Google Scholar]
- 9.Cullimore, D. R., and A. E. McCann. 1978. The identification, cultivation and control of iron bacteria in ground water, p. 219-261. In F. A. Skinner and M. J. Shewan (ed.), Aquatic microbiology, Academic Press, London, England.
- 10.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinh, H. T., J. Kuever, M. Mußmann, A. W. Hassel, M. Stratmann, and F. Widdel. 2004. Iron corrosion by novel anaerobic microorganisms. Nature 427:829-832. [DOI] [PubMed] [Google Scholar]
- 12.Ehrenreich, A., and F. Widdel. 1994. Anaerobic oxidation of ferrous iron by purple bacteria, a new type of phototrophic metabolism. Appl. Environ. Microbiol. 60:4517-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichler, S., R. Christen, C. Höltje, P. Westphal, J. Bötel, I. Brettar, A. Mehling, and M. G. Höfle. 2006. Composition and dynamics of bacterial communities of a drinking water supply system as assessed by RNA- and DNA-based 16S rRNA gene fingerprinting. Appl. Environ. Microbiol. 72:1858-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emerson, D., and N. P. Revsbech. 1994. Investigation of an iron-oxidizing microbial mat community located near Aarhus, Denmark: field studies. Appl. Environ. Microbiol. 60:4022-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emerson, D., and N. P. Revsbech. 1994. Investigation of an iron-oxidizing microbial mat community located near Aarhus, Denmark: laboratory studies. Appl. Environ. Microbiol. 60:4032-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emerson, D., and J. V. Weiss. 2004. Bacterial iron oxidation in circumneutral freshwater habitats: findings from the field and the laboratory. Geomicrobiol. J. 21:405-414. [Google Scholar]
- 17.Geets, J., B. Borremans, L. Diels, D. Springael, J. Vangronsveld, D. van der Lelie, and K. Vanbroekhoven. 2006. DsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. J. Microbiol. Methods 66:194-205. [DOI] [PubMed] [Google Scholar]
- 18.Geldreich, E. E. 1996. Microbial quality of water supply in distribution systems, p. 20-27 and 104-144. CRC Lewis Publishers, Boca Raton, FL.
- 19.Hackl, E., S. Zechmeister-Boltenstern, L. Bodrossy, and A. Sessitsch. 2004. Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl. Environ. Microbiol. 70:5057-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallberg, K. B., K. Coupland, S. Kimura, and D. B. Johnson. 2006. Macroscopic streamer growths in acidic, metal-rich mine waters in North Wales consist of novel and remarkably simple bacterial communities. Appl. Environ. Microbiol. 72:2022-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanert, H. H. 2006. The genus Gallionella, p. 990-995. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 7. Springer-Verlag, New York, NY. [Google Scholar]
- 22.Hanert, H. H. 2006. The genus Siderocapsa (and other iron- and manganese-oxidizing Eubacteria), p. 1005-1015. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 7. Springer-Verlag, New York, NY. [Google Scholar]
- 23.Hirsch, P. 2006. The genus Toxothrix, p. 986-989. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 7. Springer-Verlag, New York, NY. [Google Scholar]
- 24.Katsoyiannis, I. A., and A. I. Zouboulis. 2006. Use of iron- and manganese-oxidizing bacteria for the combined removal of iron, manganese and arsenic from contaminated groundwater. Water Qual. Res. J. Can. 41:117-129. [Google Scholar]
- 25.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar, S., J. Dudley, M. Nei, and K. Tamura. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley, Chichester, United Kingdom.
- 29.Larson, T. E., and R. V. Skold. 1958. Laboratory studies relating mineral quality of water to corrosion of steel and cast iron. Corrosion 14:285-288. [Google Scholar]
- 30.Li, D., M. Yang, Z. Li, R. Qi, J. He, and H. Liu. 2008. Change of bacterial communities in sediments along Songhua River in northeastern China after a nitrobenzene pollution event. FEMS Microbiol. Ecol. 65:494-503. [DOI] [PubMed] [Google Scholar]
- 31.Lonergan, D. J., H. L. Jenter, J. D. Coates, E. J. P. Phillips, T. M. Schmidt, and D. R. Lovley. 1996. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J. Bacteriol. 178:2402-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ministry of Public Health of the People's Republic of China. 1985. National standard of the People's Republic of China sanitary standards for drinking water. GB5749-85. Ministry of Public Health, Beijing, People's Republic of China.
- 35.Mobarry, B. K., M. Wagner, V. Urbain, B. E. Rittmann, and D. A. Stahl. 1996. Phylogenetic probes for analysing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mouchet, P. 1992. From conventional to biological removal of iron and manganese in France. J. Am. Water Works Assoc. 84:158-167. [Google Scholar]
- 37.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 38.Norton, C. D., and M. W. Lechevallier. 2000. A pilot study of bacteriological population changes through potable water treatment and distribution. Appl. Environ. Microbiol. 66:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridgway, H. F., E. G. Means, and B. H. Olson. 1981. Iron bacteria in drinking-water distribution systems: elemental analysis of Gallionella stalks, using X-ray energy-dispersive microanalysis. Appl. Environ. Microbiol. 41:288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarin, P., V. L. Snoeyink, J. Bebee, K. K. Jim, M. A. Beckett, W. M. Kriven, and J. A. Clement. 2004. Iron release from corroded iron pipes in drinking water distribution systems: effect of dissolved oxygen. Water Res. 38:1259-1269. [DOI] [PubMed] [Google Scholar]
- 41.Satpathy, K. K., T. S. Rao, V. P. Venugopalan, K. V. K. Nait, and P. K. Mathur. 1994. Studies on the response of iron oxidising and slime forming bacteria to chlorination in a laboratory model cooling tower, p. 373-383. In A. K. Tiller and C. A. C. Sequeira (ed.), Microbial corrosion. The Institute of Metals, London, United Kingdom.
- 42.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt, J. M., and G. A. Zavarzin. 2006. The genera Caulococcus and Kusnezovia, p. 996-997. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 7. Springer-Verlag, New York, NY. [Google Scholar]
- 44.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Straub, K. L., W. A. Schonhuber, B. E. E. Buchholz-Cleven, and B. Schink. 2004. Diversity of ferrous iron-oxidizing, nitrate-reducing bacteria and their involvement in oxygen-independent iron cycling. Geomicrobiol. J. 21:371-378. [Google Scholar]
- 46.Volk, C., E. Dundore, J. Schiermann, and M. Lechevallier. 2000. Practical evaluation of iron corrosion control in a drinking water distribution system. Water Res. 34:1967-1974. [Google Scholar]