Abstract
It has been suggested that Mycobacterium avium subspecies paratuberculosis has a role in Crohn's disease. The organism may be acquired but is difficult to culture from the environment. We describe a quantitative PCR (qPCR) method to detect M. avium subsp. paratuberculosis in drinking water and the results of its application to drinking water and faucet biofilm samples collected in the United States.
Mycobacterium avium subspecies paratuberculosis is a member of the Mycobacterium avium complex. M. avium subsp. paratuberculosis causes Johne's disease in bovine and ovine animals and has been hypothetically linked to Crohn's disease in humans. Several review articles have been written describing the association between M. avium subsp. paratuberculosis and Crohn's disease (1, 2, 10, 11, 16, 23). Most mycobacterial infections are acquired from the environment; however, M. avium subsp. paratuberculosis can elude laboratory culture from environmental samples (28). M. avium subsp. paratuberculosis has been cultured only once from drinking water in the United States; therefore, its occurrence in drinking water is unknown (17). There are several reasons one could expect to find M. avium subsp. paratuberculosis in drinking water. The bacterium has been isolated from surface water used as a source of drinking water (19, 20, 24, 26). It is resistant to chlorine disinfection (25). Also, other subspecies of M. avium have been detected in biofilms obtained from drinking water pipes in the United States (8, 22, 27).
Due to the potential for waterborne transmission of mycobacteria and the association of M. avium subsp. paratuberculosis with human illness, the focus of this study was to estimate the organism's occurrence in drinking water in the United States using quantitative PCR (qPCR) (15). A comprehensive method was developed for detection of M. avium subsp. paratuberculosis in drinking water and biofilms that includes the concentration of microorganisms from samples using membrane filtration, total DNA extraction and purification, and detection of two targets unique to this bacterium: IS900 and target 251. IS900 is a common target used to identify M. avium subsp. paratuberculosis, and the average number of copies per genome is 14 to 18 (13). Target 251 qPCR analysis, which corresponds to the M. avium subsp. paratuberculosis gene 2765c (David Alexander, personal communication), was developed by Rajeev et al. (21). Samples positive for both targets are considered positive for M. avium subsp. paratuberculosis. TaqMan primer and probe sequences and qPCR assay characteristics are described in Table 1. The complete method is described in Fig. S1 in the supplemental material.
TABLE 1.
qPCR assay primers, probes, DNA targets, and assay characteristicsa
| DNA target | Primer or probe (sequence, 5′→3′) | Product (bp) | Reference | ||
|---|---|---|---|---|---|
| LODb | LOQc | ||||
| IS900 | IS900F (CCGCTAATTGAGAGATGCGATTGG) | 230 | 1.8 | 1.8 | 13 |
| IS900R (ATTCAACTCCAGCAGCGCGGCCTC) | |||||
| IS900P (6-FAM-TCCACGCCCGCCCAGACAGG-TAMRA) | |||||
| Target 251 | 251F (GCAAGACGTTCATGGGAACT) | 200 | ND | ND | 21 |
| 251R (GCGTAACTCAGCGAACAACA) | |||||
| 251P (6-FAM-CTGACTTCACGATGCGGTTCTTC-TAMRA) | |||||
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine; ND, not determined.
The limit of detection (LOD) of the IS900 qPCR assay was defined as the lowest copy number resulting in a CT of <40, determined from six independent dilution series.
The limit of quantification (LOQ) was defined as the lowest copy number per assay yielding a coefficient of variation (CV) of less than 25% (33).
A master standard curve was generated from six series of 10-fold dilutions of genomic DNA from M. avium subsp. paratuberculosis strain 49164 for quantification of IS900 target copies (see Fig. S2A in the supplemental material). Each dilution series contained eight standards run in triplicate for a total of 18 threshold cycle (CT) measurements per standard. A linear regression was performed on CT versus log IS900 copy number and R2 was 0.997. The standard error of y was used to create two equations to estimate the upper and lower concentration, or range, of M. avium subsp. paratuberculosis IS900 copy number.
The specificities of the IS900 and target 251 primer/probe sets were evaluated by Rajeev et al. (21) on 211 M. avium subsp. paratuberculosis and 38 non-M. avium subsp. paratuberculosis isolates, and each assay was 100% specific for M. avium subsp. paratuberculosis. We further evaluated specificity using 22 M. avium subsp. paratuberculosis isolates from animals and 10 non-M. avium subsp. paratuberculosis ATCC reference strains (see Table S1 in the supplemental material) (18). Target 251 was 100% specific; however, one M. avium subsp. paratuberculosis isolate (3063) repeatedly produced a negative result by IS900 qPCR. Results suggest that a small subset of M. avium subsp. paratuberculosis isolates may not contain the IS900 element or may have a sequence that differs from that of the IS900 primer/probe set.
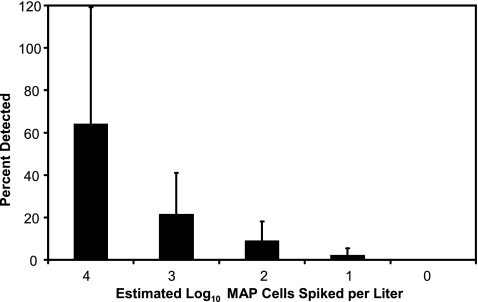
The sensitivity of the method for detection of M. avium subsp. paratuberculosis in different drinking water matrices was evaluated by spiking serial dilutions of strain 1112 cells, ranging from 104 cells to no addition of cells, into 1-liter tap water samples obtained from five locations in the United States. The number of M. avium subsp. paratuberculosis cell equivalents was estimated by dividing the IS900 copy number obtained from the master standard curve by 18 (mean, 18 IS900 copies/M. avium subsp. paratuberculosis genome). The method provided consistent detection (5/5 samples) in a spiked sample of 100 cells/liter. In a spiked sample of 10 cells/liter, the IS900 target was detected 40% (2/5 samples) of the time, and at 1 cell/liter we did not detect the target in any spiked sample. Percent recovery was variable and decreased as the number of spiked cells decreased (Fig. 1). At a spike level of 1 × 104 cells/liter, the average percent recovery was 64%; this decreased to 9.2% at 1 × 102 cells/liter. Cell surface hydrophobicity, a property of mycobacteria, may have influenced clumping of the spiked sample or partitioning of M. avium subsp. paratuberculosis onto the sample bottle or filtration unit, affecting recovery of the bacterium (3).
FIG. 1.
Average percent recovery of M. avium subsp. paratuberculosis spiked into drinking water collected from five sites in the United States. Error bars denote standard deviation. MAP, M. avium subsp. paratuberculosis.
Midwest and temporal study.
Two liters of cold drinking water and a biofilm sample were collected from 33 homes or commercial buildings in two metropolitan areas in the Midwest from May to November 2007. The first liter, or “first-pull” sample, was collected immediately upon turning on the tap. The second liter, or “standard methods” sample, was collected according to sections 9060A and B of Standard Methods for the Examination of Water and Wastewater (7). Biofilm samples were collected by swabbing the surface of the faucet grating at the end of the tap with a sterile cotton swab. The swab was broken off in a tube containing 10 ml sterile molecular biology-grade water and transported back to the lab. Samples were analyzed by the method described in Fig. S1 in the supplementary material. No template controls and standards were included with every set of qPCR analyses. Method controls were prepared by filtering sterile molecular biology-grade water and processed with every set and on the same day as that for water and biofilm samples.
Eighty-one percent of first-pull water samples, 88% of standard methods samples, and 76% of the biofilm samples were positive for both IS900 and target 251 in the qPCR assays (n = 33). IS900 copy concentrations varied widely among sites (see Table S2 in the supplemental material). IS900 copy concentrations in first-pull and standard methods samples from the same site were not significantly different (α = 0.05; P > 0.095). Additionally, 82% of IS900-positive samples contained <100 target copies per liter of water or biofilm sampled, and 68% contained <500 target copies/liter. These results are similar to what Hilborn et al. (12) found in a geographically localized survey of home taps in the Pacific Northwest, where greater than 50% of water samples were positive for M. avium. Five M. avium subsp. paratuberculosis-positive biofilm samples had negative results for water samples. Conversely, three M. avium subsp. paratuberculosis-positive water samples had negative results for biofilm samples. Little is currently known about the ability of M. avium subsp. paratuberculosis to survive or grow in drinking water biofilms (9).
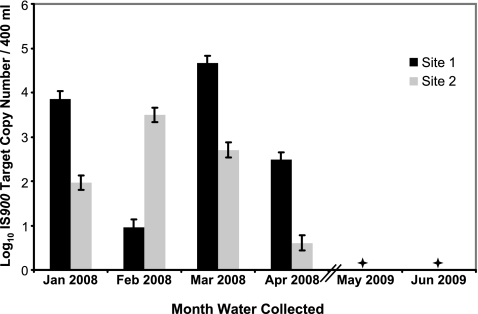
In order to determine if M. avium subsp. paratuberculosis persists in drinking water, 1 liter of water was collected from two homes in the Midwest once a month for 4 months (January to April 2008). The homes were positive for M. avium subsp. paratuberculosis in 2007. M. avium subsp. paratuberculosis was consistently detected at both taps, though the quantity was highly variable during the period sampled (Fig. 2). Repeated detection at drinking water taps supports the findings of Hilborn et al. (12), who isolated the same M. avium genotypes from three taps during a 2-year period.
FIG. 2.
Results of the temporal study showing the change in log10 estimated M. avium subsp. paratuberculosis IS900 target copy number over time between sites. Error bars denote the range between high and low estimates. ✦, negative detection for IS900 on those dates; Jan, January; Feb, February; Mar, March; Apr, April; Jun, June.
National study.
Drinking water samples (n = 238) were collected from January to December 2009 from taps in homes or commercial buildings at 41 sites in 25 states, one district, and one U.S. territory. A spectrum of source waters (surface and ground water), system sizes (large and small utilities, private wells), and disinfection types (chlorine, chloramine, and chlorine dioxide) was included. Twenty-nine samples were analyzed using TaqMan universal master mix, and 209 samples were analyzed using TaqMan environmental master mix 2.0. All reaction mixtures included the TaqMan exogenous internal positive-control reagent to detect PCR inhibition. A standard curve using the new reagents was generated (see Fig. S2B in the supplemental material) and was not significantly different from the curve obtained with the original reagents (α = 0.05; P > 0.31).
In contrast to the Midwest study, no samples in the national study were positive for M. avium subsp. paratuberculosis, though five were positive for IS900 but not the target 251 qPCR assay. Additionally, five Midwestern sites positive for M. avium subsp. paratuberculosis in 2007 were negative for that bacterium in 2009. Such disparate findings between the two studies were unexpected because a large number of Midwest samples were positive for M. avium subsp. paratuberculosis in the Midwest. In a similar survey of the United States, Covert et al. (5) measured the occurrence of nontuberculous mycobacteria (NTM) in drinking water from geographically dispersed sites. In that study, only one sample (<1%) was positive for M. avium. The studies of Hilborn et al. (12) and Covert et al. (5) demonstrate the variability of detecting M. avium in a localized versus national survey of drinking water. The cause of the variability observed in these studies is unknown, though it is important to note that the Midwest experienced a severe drought in 2007 but not in 2009, as reported by the National Climatic Data Center. Furthermore, the area sampled in the Midwest was in the top 10th percentile for temperatures in August 2007 (http://www.ncdc.noaa.gov/oa/climate/research/2007/ann/drought-summary.html#regdrot). Only one of five IS900-positive samples from the national study was from a state experiencing a drought in 2009. Additional abiotic and biotic factors which could influence the occurrence of M. avium subsp. paratuberculosis in drinking water include water temperature, the effect of drinking water disinfection, and the source of drinking water and its proximity to M. avium subsp. paratuberculosis-infected herds. Further research is needed to understand the geographical and temporal differences in the occurrence of this bacterium, as has been described for other NTM (4, 8, 14). This is the first report on the occurrence of M. avium subsp. paratuberculosis in drinking water in the United States using a molecular method that may prove useful for understanding the ecology and epidemiology of the organism.
Supplementary Material
Acknowledgments
We thank Srinand Sreevatsan, College of Veterinary Medicine at the University of Minnesota, for the generous gift of M. avium subsp. paratuberculosis isolates. We also acknowledge the help of Stephen Vesper, Manju Varma, and Jeff Hester from the U.S. Environmental Protection Agency for their assistance with the lysate recovery portion of the M. avium subsp. paratuberculosis qPCR method.
This research was supported in part by an appointment to the Postgraduate Research Participation Program administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the EPA. The EPA, through its Office of Research and Development, funded and managed the research described here. This research has been subjected to the agency's administrative review and has been approved for publication.
Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print on 3 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abubakar, I., D. Myhill, S. H. Aliyu, and P. R. Hunter. 2008. Detection of Mycobacterium avium subspecies paratuberculosis from patients with Crohn's disease using nucleic acid-based techniques: a systematic review and meta-analysis. Inflamm. Bowel Dis. 14:401-410. [DOI] [PubMed] [Google Scholar]
- 2.Behr, M. A., and V. Kapur. 2008. The evidence for Mycobacterium paratuberculosis in Crohn's disease. Curr. Opin. Gastroenterol. 24:17-21. [DOI] [PubMed] [Google Scholar]
- 3.Bendinger, B., H. H. M. Rijnaarts, K. Altendor, and A. J. B. Zehnder. 1993. Physiochemical cell surface and adhesive properties of coryneforms bacteria related to the presence and chain length of mycolic acids. Appl. Environ. Microbiol. 59:3973-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks, R. W., B. C. Parker, H. Gruft, and J. O. Falkinham III. 1984. Epidemiology of infection by nontuberculous mycobacteria. V. Numbers in eastern United States soils and correlation with soil characteristics. Am. Rev. Respir. Dis. 130:630-633. [DOI] [PubMed] [Google Scholar]
- 5.Covert, T. C., M. R. Rodgers, A. L. Reyes, and G. N. Stelma, Jr. 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl. Environ. Microbiol. 65:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reference deleted.
- 7.Eaton, A. D., L. S. Clesceri, E. W. Rice, and A. E. Greenberg (ed.). 2005. Standard methods for the examination of water and wastewater, 21st ed. American Public Health Association, Washington, DC.
- 8.Falkinham, J. O., III, C. D. Norton, and M. W. LeChevallier. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 67:1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feazel, L. M., K. L. Peterson, D. N. Frank, J. K. Harris, and N. R. Pace. 2009. Opportunistic pathogens enriched in showerhead biofilms. Proc. Natl. Acad. Sci. U. S. A. 106:16393-16399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feller, M., K. Huwiler, R. Stephan, E. Altpeter, A. Shang, H. Furrer, G. E. Pfyffer, T. Jemmi, A. Baumgartner, and M. Egger. 2007. Mycobacterium avium subspecies paratuberculosis and Crohn's disease: a systematic review and meta-analysis. Lancet Infect. Dis. 7:607-613. [DOI] [PubMed] [Google Scholar]
- 11.Hermon-Taylor, J., T. J. Bull, J. M. Sheridan, J. Cheng, M. L. Stellakis, and N. Sumanr. 2000. Causation of Crohn's disease by Mycobacterium avium subspecies paratuberculosis. Can. J. Gastroenterol. 14:521-539. [DOI] [PubMed] [Google Scholar]
- 12.Hilborn, E. D., T. C. Covert, M. A. Yakrus, S. I. Harris, S. F. Donnelly, E. W. Rice, S. Toney, S. A. Bailey, and G. N. Stelma, Jr. 2006. Persistence of nontuberculous mycobacteria in a drinking water system after addition of filtration treatment. Appl. Environ. Microbiol. 72:5864-5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, S. G., S. J. Shin, R. H. Jacobson, L. J. Miller, P. R. Harpending, S. M. Stehman, C. A. Rossiter, and D. A. Lein. 2002. Development and application of quantitative polymerase chain reaction assay based on the ABI 7700 system (TaqMan) for detection and quantification of Mycobacterium avium subsp. paratuberculosis. J. Vet. Diagn. Invest. 14:126-131. [DOI] [PubMed] [Google Scholar]
- 14.Kirschner, R. A., Jr., B. C. Parker, and J. O. Falkinham III. 1992. Epidemiology of infection by nontuberculous mycobacteria. Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum in acid, brown-water swamps of the southeastern United States and their association with environmental variables. Am. Rev. Respir. Dis. 145:271-275. [DOI] [PubMed] [Google Scholar]
- 15.LeChevalier, M. W. (ed.). 2006. Mycobacterium avium complex, p. 125-127. In Waterborne pathogens, 2nd ed. American Public Health Association, Denver, CO.
- 16.Mendoza, J. L., R. Lana, and M. Diaz-Rubio. 2009. Mycobacterium avium subspecies paratuberculosis and its relationship with Crohn's disease. World J. Gastroenterol. 15:417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishina, D., P. Katsel, S. T. Brown, E. C. Gilberts, and R. J. Greenstein. 1996. On the etiology of Crohn disease. Proc. Natl. Acad. Sci. U. S. A. 93:9816-9820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motiwala, A. S., M. Strother, A. Amonsin, B. Byrum, S. A. Naser, J. R. Stabel, W. P. Shulaw, J. P. Bannantine, V. Kapur, and S. Sreevatsan. 2003. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: evidence for limited strain diversity, strain sharing, and identification of unique targets for diagnosis. J. Clin. Microbiol. 41:2015-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickup, R. W., G. Rhodes, S. Arnott, K. Sidi-Boumedine, T. J. Bull, A. Weightman, M. Hurley, and J. Hermon-Taylor. 2005. Mycobacterium avium subsp. paratuberculosis in the catchment area and water of the River Taff in South Wales, United Kingdom, and its potential relationship to clustering of Crohn's disease cases in the city of Cardiff. Appl. Environ. Microbiol. 71:2130-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickup, R. W., G. Rhodes, T. J. Bull, S. Arnott, K. Sidi-Boumedine, M. Hurley, and J. Hermon-Taylor. 2006. Mycobacterium avium subsp. paratuberculosis in lake catchments, in river water abstracted for domestic use, and in effluent from domestic sewage treatment works: diverse opportunities for environmental cycling and human exposure. Appl. Environ. Microbiol. 72:4067-4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajeev, S., Y. Zhang, S. Sreevatsan, A. S. Motiwala, and B. Byrum. 2005. Evaluation of multiple genomic targets for identification and confirmation of Mycobacterium avium subsp. paratuberculosis isolates using real-time PCR. Vet. Microbiol. 105:215-221. [DOI] [PubMed] [Google Scholar]
- 22.Torvinen, E., M. J. Lehtola, P. J. Martikainen, and I. T. Miettinen. 2007. Survival of Mycobacterium avium in drinking water biofilms as affected by water flow velocity, availability of phosphorus, and temperature. Appl. Environ. Microbiol. 73:6201-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waddell, L. A., A. Rajić, J. Sargeant, J. Harris, R. Amezcua, L. Downey, S. Read, and S. A. McEwen. 2008. The zoonotic potential of Mycobacterium avium spp. paratuberculosis: a systematic review. Can. J. Public Health 99:145-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whan, L., H. J. Ball, I. R. Grant, and M. T. Rowe. 2005. Occurrence of Mycobacterium avium subsp. paratuberculosis in untreated water in Northern Ireland. Appl. Environ. Microbiol. 71:7107-7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whan, L., I. R. Grant, and M. T. Rowe. 2006. Interaction between Mycobacterium avium subsp. paratuberculosis and environmental protozoa. BMC Microbiol. 6:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittington, R. J., I. B. Marsh, P. J. Taylor, D. J. Marshall, C. Taragel, and L. A. Reddacliff. 2003. Isolation of Mycobacterium avium subsp. paratuberculosis from environmental samples collected from farms before and after destocking sheep with paratuberculosis. Aust. Vet. J. 81:559-563. [DOI] [PubMed] [Google Scholar]
- 27.Williams, M. M., M. A. Yakrus, M. J. Arduino, R. C. Cooksey, C. B. Crane, S. N. Banerjee, E. D. Hilborn, and R. M. Donlan. 2009. Structural analysis of biofilm formation by rapidly and slowly growing nontuberculous mycobacteria. Appl. Environ. Microbiol. 75:2091-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolinsky, E. 1979. Nontuberculous mycobacteria and associated diseases. Am. Rev. Respir. Dis. 119:107-159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.