Abstract
A reporter assay-based screening method for enzymes, which we named product-induced gene expression (PIGEX), was developed and used to screen a metagenomic library for amidases. A benzoate-responsive transcriptional activator, BenR, was placed upstream of the gene encoding green fluorescent protein and used as a sensor. Escherichia coli sensor cells carrying the benR-gfp gene cassette fluoresced in response to benzoate concentrations as low as 10 μM but were completely unresponsive to the substrate benzamide. An E. coli metagenomic library consisting of 96,000 clones was grown in 96-well format in LB medium containing benzamide. The library cells were then cocultivated with sensor cells. Eleven amidase genes were recovered from 143 fluorescent wells; eight of these genes were homologous to known bacterial amidase genes while three were novel genes. In addition to their activity toward benzamide, the enzymes were active toward various substrates, including d- and l-amino acid amides, and displayed enantioselectivity. Thus, we demonstrated that PIGEX is an effective approach for screening novel enzymes based on product detection.
Environmental microorganisms carry great potential as sources of industrial enzymes (5, 21); however, the majority of these organisms are difficult to culture in the laboratory (1, 29). Metagenomics has emerged as a credible alternative to conventional, cultivation-based microbial screening. It allows exhaustive screening of microbial genomes in their natural environments by directly cloning environmental DNA in a surrogate host; however, functional screening of metagenomic libraries often suffers from low hit rates, largely as a result of insufficient expression of anonymous foreign genes in the host, which is commonly Escherichia coli (26).
One approach to overcoming this problem is to use a reporter assay (23), a strategy that is frequently employed in molecular biology (e.g., β-galactosidase assay and chloramphenicol acetyltransferase [CAT] assay) and biotechnology (e.g., to detect environmental pollutants [18] and food additives [17]). Previously, we developed a substrate-induced gene expression (SIGEX) method (25) to capture genetic elements that respond to exogenous compounds. By using green fluorescent protein (GFP) as a reporter and performing the screen using a fluorescence-activated cell sorter, we achieved high screening efficiency. The method was successfully used to retrieve genetic elements that sense exogenous compounds (e.g., benzoate and naphthalene).
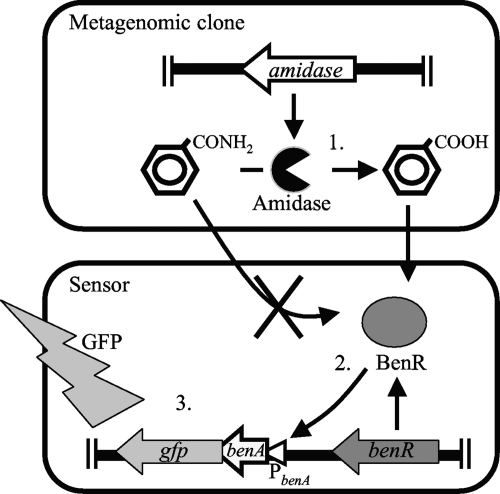
In the present study, we extended our reporter assay system to specifically retrieve enzymes. To this end, we utilized the previously identified transcriptional activator BenR, which was placed upstream of gfp. We assumed that E. coli cells harboring the benR-gfp cassette would fluoresce in the presence of a benzoate precursor compound (substrate) if they expressed an enzyme capable of actively transforming the precursor into benzoate (Fig. 1). This reporter assay system, which we named product-induced gene expression (PIGEX), should allow the identification of desired enzymatic activities by linking product formation to reporter gene expression. Using this system, we targeted amidases that convert benzamide to benzoate.
FIG. 1.
Schematic representation of the PIGEX (product-induced gene expression) method using amidase as the target enzyme. The amidase-positive clone catalyzes the conversion of benzamide to benzoate (step 1). Benzoate activates the transcriptional regulator BenR, which in turn activates the benA promoter (PbenA) (step 2) and induces the expression of a reporter gene (gfp; step 3). Enzymatic benzoate production is measured as GFP fluorescence.
Amidases (EC 3.5.1.4) catalyze the conversion of amides to their corresponding carboxylic acids (with ammonia being produced as a by-product) according to the following reaction: RCONH2 + H2O → RCOOH + NH3. Given that the function of amidases is to hydrolyze amide bonds, the biological consequences of this reaction are many and varied. In addition to their diverse biological roles, amidases have found a number of industrial applications, notably in the synthesis of chiral compounds (7, 20, 28). To date, numerous bacterial amidases have been purified and characterized, but no efforts have been made to recover these enzymes via culture-independent approaches. To assess the possibility of identifying novel amidases in metagenomes, we used PIGEX screening to search for amidases that use benzamide as a substrate. By coupling benzamidase activity to benzoate-responsive fluorescence, we successfully identified 11 amidase genes, including three genes with low sequence similarity to known amidases.
MATERIALS AND METHODS
Reagents.
Restriction and DNA modification enzymes and pHSG398 were purchased from Takara Bio (Shiga, Japan). Aromatic compounds, acetamide, acrylamide, and propionamide were purchased from Tokyo Chemical Industry (Tokyo, Japan), while cis,cis-muconic acid was purchased from Sigma-Aldrich (St. Louis, MO). Amino acid amides were purchased from Bachem (Bubendorf, Switzerland).
Bacterial strains, media, and growth conditions.
The E. coli strains used in this study were JM109 (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ[lac-proAB] [F′ tra36 proAB+ lacIq lacZΔM15]) and EPI300 (F− mcrA Δ[mrr-hsdRMS-mcrBC] φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL nupG trfA dhfr). Bacteria were cultured at 37°C in LB broth or on LB agar plates. Antibiotics (100 μg/ml ampicillin [Amp] and 34 μg/ml chloramphenicol [Cm]) were added to the culture medium as appropriate.
Construction of a benzoate-responsive sensor plasmid.
A DNA fragment containing the benzoate-responsive transcriptional regulator benR upstream of gfp was excised from pbzo28 using EcoRI and SphI (25). The same restriction sites were used to insert the fragment into pHSG398, yielding pCmGFPbenR. E. coli JM109 cells harboring pCmGFPbenR were used as sensor cells.
A single colony of sensor cells was grown overnight in LB medium supplemented with Cm (LB-Cm) and diluted 100-fold in fresh LB-Cm medium. Cells were grown with shaking at 30°C until they reached an optical density at 600 nm (OD600) of 0.3 (mid-log phase) or overnight (stationary phase). The culture was subsequently divided into 0.5-ml aliquots and transferred to 96-deep-well plates containing 0.5 ml of LB-Cm medium supplemented with various concentrations of benzamide (or its derivatives), thus yielding a total volume of 1 ml per well. The plates were shaken at 1,200 rpm at 30°C for 1 to 24 h and then centrifuged (1,500 × g for 5 min). The supernatants were removed, and the cell pellets were washed with distilled water and resuspended in 200 μl of distilled water. Cell suspension (100 μl) from each well was transferred to the wells of a black, clear-bottomed 96-well plate. GFP fluorescence (488-nm excitation, 520-nm emission, and 515-nm cutoff) was measured using a fluorescence microplate reader (SpectraMAX Gemini XS; Molecular Devices, Sunnyvale, CA). Cell density (OD600) was measured using a UV-visible light (Vis) microplate reader (VERSAMax; Molecular Devices). Fluorescence was normalized to the cell density (OD600 of 1.0).
Screening of a metagenomic fosmid library.
The metagenomic fosmid library used in this study was previously constructed from DNA extracted from activated sludge used to treat industrial wastewater at a coke plant (22). The library consisted of 96,000 fosmid clones, which were divided among 10 96-well plates. We performed the screening process using the following equipment: a Biomek NX liquid handling system (Beckman Coulter, Brea, CA), a Maximizer MBR-420FL incubation shaker (Taitec, Tokyo, Japan), and a microplate reader (Molecular Devices).
To screen the library for amidase genes, small volumes of cells were taken from frozen glycerol stocks and grown in LB-Cm medium overnight at 37°C. An aliquot of the cells was then transferred to 500 μl of fresh LB-Cm containing medium 0.5 μl of CopyControl Induction Solution (Epicentre, Madison, WI) and grown with vigorous shaking at 1,200 rpm at 37°C for 18 h. Sensor cells carrying pCmGFPbenR were grown overnight at 37°C in LB-Cm medium. The cells were then diluted 100-fold in fresh LB-Cm medium and cultivated with shaking at 37°C. When the OD600 reached 0.6, benzamide was added to the growth medium to a final concentration of 10 mM. The cells were then divided into 0.5-ml aliquots in 96-deep-well plates containing 0.5 ml of metagenomic library cells, thus yielding a total volume of 1 ml per well. The 96-deep-well plates were shaken at 1,200 rpm at 37°C for 18 h and then centrifuged (1,500 × g for 5 min). The supernatants were discarded, and the cell pellets were washed in distilled water and resuspended in 200 μl of distilled water. Cell suspension (100 μl) from each well was transferred to the wells of a black 96-well microplate. GFP fluorescence was measured as described above.
Isolation of individual positive fosmid clones from hit wells.
Because each well contained ideally 100 fosmid clones, a second screen was performed to isolate individual positive clones from the hit wells. Cells were removed from the hit wells, diluted, and grown on LB-Cm agar plates. In total, 192 colonies from each well were screened as described above. When several colonies were retrieved from the same well, their individual identities were determined by restriction fragment length polymorphism (RFLP) analysis using EcoRI and/or PstI.
Subcloning, DNA sequencing, and gene annotation.
Fosmid DNA from individual positive clones was purified using a FosmidMAX DNA Purification Kit (Epicentre) and partially digested with Sau3AI. The resulting DNA fragments were separated by agarose gel electrophoresis, and bands ranging from 2 to 5 kb in size were excised, purified, and ligated into the BamHI site of pUC18. The resulting reaction mixture was used to transform E. coli JM109 by electroporation. Cells were grown on LB-Amp plates containing 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactoside and 0.1 mM isopropyl β-d-thiogalactoside. In total, 192 white colonies were picked, resuspended in separate wells of 96-deep-well plates containing 250 μl of LB-Amp medium, and screened for benzamidase activity as described above. When several positive clones were identified in the same library, their individual identities were determined by RFLP analysis using EcoRI. Plasmids were purified from the positive clones using a QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany), and the nucleotide sequences of the inserts were determined using an a BigDye Terminator, version 3.1, Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) in conjunction with an ABI 3730XL sequencer (Applied Biosystems). Nucleotide and amino acid sequences were analyzed using Genetyx-Mac, version 15.0.0, software (Software Development Co., Tokyo, Japan). BLASTX and BLASTP searches were performed using the NCBI database (with default parameters). A multiple amino acid sequence alignment was carried out using ClustalW software, version 1.83 (24), and refined by visual inspection. We subjected the entire amino acid sequences of our metagenomic amidases and 16 amidases that had previously been functionally characterized to phylogenetic analyses. Neighbor-joining (NJ) trees (19) were constructed using NJplot software, version 2.3 (16). The topology of the trees was determined using 1,000 bootstrap replicates, and the distances were corrected by the Kimura method (9).
PCR genotyping.
Cells were removed from the positive wells and cultured in 1 ml of LB-Cm medium at 37°C for 18 h. They were then centrifuged, washed twice with water, and suspended in 200 μl of water. Oligonucleotide primers were designed to amplify part of the gene encoding 8D amidase (5′-GTACCCACCACGGCATCG-3′ and 5′-CACTTCAGGCAGACGCAG-3′). One microliter of cells was added to a PCR mixture containing a 0.4 μM concentration of each primer and 1 U of EX Taq polymerase (Takara Bio). The cycling conditions were as follows: denaturation for 1 min at 94°C followed by 32 cycles of 20 s at 98°C, 20 s at 53°C, and 1 min at 72°C, with a final extension at 72°C 10 min. The products were analyzed by agarose gel electrophoresis.
Substrate specificities of the retrieved amidases.
E. coli JM109 cells carrying the metagenomic amidase genes were cultivated at 37°C for 18 h in 250 ml of LB-Amp medium. Cells were then harvested and washed twice with 10 mM sodium phosphate buffer (pH 7.0), resuspended in 5 ml of 10 mM sodium phosphate buffer (pH 7.0), and disrupted using a cell disrupter (One Shot Model; Constant Systems, Warwick, United Kingdom). Cell debris was removed by centrifugation twice (36,000 × g for 5 min at 4°C). The protein concentration in the cell extracts was determined using a Quick Start Bradford Dye Reagent Kit (Bio-Rad, Richmond, CA) with bovine gamma globulin as the standard. The amidase reaction mixture consisted of 10 mM sodium phosphate buffer (pH 7.0), 1 mM amide substrate, and 100 μg/ml crude protein. The reaction mixture was incubated at 37°C for 1 h, after which the reactions were stopped by the addition of 10% (vol/vol) trichloroacetate. Ammonium concentrations were determined spectrophotometrically according to the phenol-hypochlorite method (4) using a Spectroquant Ammonium Test Kit (Merck, Darmstadt, Germany). Ammonium chloride was used as the standard.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this article have been submitted to GenBank/EMBL/DDBJ under accession numbers AB500111 to AB500114 and AB545788 to AB545794.
RESULTS AND DISCUSSION
PIGEX.
The PIGEX method is based on the reporter assay illustrated in Fig. 1. This system consists of a sensor gene paired with a reporter gene. To screen for amidase genes, we used as the sensor a benzoate-responsive BenR transcriptional regulator that we ourselves had previously retrieved from a metagenome (25). As the reporter, we used GFP. Cells carrying this genetic unit fluoresced upon the addition of benzoate to the culture medium while those lacking benR did not. An important factor in the choice of sensor is the ability to distinguish between substrate and product. To confirm the suitability of BenR for use as a sensor, we tested its substrate specificity. A range of benzoate analogs were tested for their ability to activate BenR (Table 1) by separately adding them to LB medium at a concentration of 1 mM. Significant fluorescence was observed for benzoate (3,682 ± 160 arbitrary fluorescence units [AU]), 2-hydroxybenzoate (2,215 ± 177 AU), 3-hydroxybenzoate (631 ± 103 AU), and benzaldehyde (840 ± 64 AU). In contrast, benzamide did not yield significant fluorescence (39 ± 1 AU), making it a suitable sensor for PIGEX screening. The second factor that needs to be considered is the absence of endogenous inducers. The E. coli strains used in this study did not produce detectable levels of benzoate when grown in LB medium in the presence or absence of benzamide, allowing us to isolate clones capable of catalyzing the conversion of benzamide to benzoate. Third, products generated by amidase reactions must be inert in vivo to ensure constant induction of the sensor. From this point of view, substrates such as amino acid amides are unsuitable because the products of their metabolism, amino acids, are readily metabolized in E. coli cells. The use of benzamide as the sole substrate may potentially have limited our ability to retrieve functionally diverse amidases although it should be noted that amidases are known to be catalytically promiscuous.
TABLE 1.
Induction of green fluorescence in E. coli JM109/pCmGFPbenR by various compounds
| Inducing compound | Fluorescence (AU)a |
|---|---|
| None | 44 ± 2 |
| Benzoate and derivatives | |
| Benzoate | 3,682 ± 160 |
| 2-Hydroxybenzoate | 2,215 ± 177 |
| 3-Hydroxybenzoate | 631 ± 103 |
| 4-Hydroxybenzoate | 44 ± 3 |
| 3,4-Hydroxybenzoic acid | 47 ± 1 |
| 2,5-Hydroxybenzoic acid | 67 ± 8 |
| Benzene and derivatives | |
| Benzene | 44 ± 3 |
| Toluene | 38 ± 2 |
| Ethylbenzene | 45 ± 3 |
| Benzyl alcohol | 61 ± 5 |
| Benzaldehyde | 840 ± 64 |
| Aniline | 40 ± 1 |
| Benzylamine | 45 ± 2 |
| Benzonitrile | 40 ± 0 |
| Benzamide | 39 ± 1 |
| Acetophenone | 43 ± 1 |
| Benzoic acid methyl ester | 41 ± 2 |
| Phenol and derivatives | |
| Phenol | 47 ± 2 |
| Catechol | 40 ± 2 |
| o-Cresol | 42 ± 2 |
| m-Cresol | 41 ± 2 |
| p-Cresol | 39 ± 1 |
| Polyaromatics | |
| Naphthalene | 47 ± 3 |
| 1,2-Dihydroxynaphthalene | 41 ± 2 |
| Biphenyl | 47 ± 1 |
| 2,3-Dihydroxybiphenyl | 60 ± 6 |
| Other | |
| cis,cis-Muconic acid | 43 ± 1 |
Fluorescence intensity values were normalized to cell densities. All values are the means of five measurements. Arbitrary fluorescence units (AU) represent the fluorescence due to GFP production.
Growth phase dependence and benzoate dose dependence of GFP fluorescence.
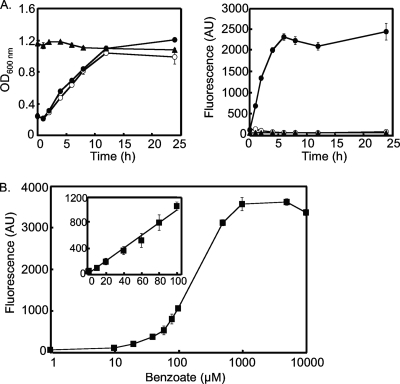
We first examined the growth phase dependence and benzoate dose dependence of GFP fluorescence in response to benzoate. As shown in Fig. 2 A, in the absence of benzoate, log phase (OD600 of 0.6) pCmGFPbenR-carrying E. coli JM109 cells did not fluoresce. However, upon the addition of 0.5 mM benzoate to the culture medium, the cells began to fluoresce within 1 h. Fluorescence peaked at 6 h and persisted for at least 24 h. In contrast, when cells were grown to the stationary phase before induction, they did not fluoresce, even after the addition of 0.5 mM benzoate, suggesting that gfp expression is growth phase dependent in our system. Thus, our subsequent experiments employed mid-log-phase cells as a sensor. We also examined the benzoate dose dependence of GFP fluorescence (Fig. 2B). The fluorescence intensity of E. coli JM109 cells carrying pCmGFPbenR was 44 ± 2 AU in the absence of benzoate and increased concomitantly with an increase in benzoate concentration (52 ± 2 AU at 1 μM benzoate and 100 ± 10 AU at 10 μM benzoate). The cells exhibited a linear response up to 1 mM, and no stronger signal was measured thereafter. Taking into account the detection limit of the fluorescence plate reader, we judged 10 μM to be the lowest concentration of benzoate that could be used to identify positives.
FIG. 2.
Growth- and dose-dependent induction of GFP by benzoate. Error bars represent 1 standard deviation from the mean (n = 4). (A) Growth phase dependence of GFP fluorescence. E. coli JM109/pCmGFPbenR sensor cells were grown to the log or stationary phase, and then benzoate was added to the culture medium (final concentration of 0.5 mM). Cell density (left panel) and fluorescence intensity (right panel) were monitored after induction. Open circle, log-phase cells (no benzoate induction); filled circle, log-phase cells induced with 0.5 mM benzoate; closed triangle, stationary-phase cells induced with 0.5 mM benzoate. (B) GFP fluorescence in sensor cells (E. coli JM109/pCmGFPbenR) induced with different concentrations of benzoate. The inset panel shows a close-up of the graph in the 0 to 100 μM benzoate region.
PIGEX screening of a metagenomic library.
Metagenomic library cells (96,000 clones divided among 960 wells) were grown overnight and combined with an equal volume of sensor cells grown separately to mid-log phase with benzamide. After the mixed culture was grown overnight, we identified 143 fluorescent wells (Table 2) (maximum fluorescence intensity, 2,625 AU [in well 10D9]; minimum, 152 AU [well 3E2]). Of these, four wells (2B1, 3E2, 9C7, and 10D9) were selected, and their contents were subjected to a second round of screening to isolate individual clones. Fosmid DNA (insert length, 30 to 40 kb) was purified from the amidase-positive clones obtained from each well. We then created fosmid shotgun libraries (insert length, 2 to 5 kb) and functionally screened them by PIGEX. Short DNA fragments responsible for the observed benzamidase activity were subjected to DNA sequencing.
TABLE 2.
Summary of hit wells identified in the first screen
| Amidase name or type | Fluorescent well(s) (identification no.)a |
|---|---|
| 8D-type | 1A5, 1D7, 1G1, 1G8, 1G12, 1H5, 1H7, 1H9, 1H12, 2B1, 2D3, 2E4, 2F1, 2F4, 2F11, 2H6, 3A10, 3B1, 3B8, 3C12, 3D1, 3D4, 3D7, 3E2, 3E3, 3E4, 3F3, 3F7, 3F10, 3G10, 3H1, 4A1, 4A4, 4A8, 4B4, 4B6, 4B8, 4F8, 4G11, 4H6, 4H9, 4H11, 5A9, 5B1, 5B4, 5C5, 5D3, 5D7, 5E6, 5F6, 5G8, 5G10, 6A2, 6A11, 6B1, 6B10, 6B12, 6C4, 6E3, 6F2, 6H4, 7A10, 7B1, 7B2, 7B4, 7B10, 7C8, 7D1, 7F3, 7F4, 7F9, 7G1, 7G2, 7G10, 7H1, 7H9, 8B10, 8C2, 8D11, 8E7, 8F5, 8F12, 8G3, 8H5, 9A4, 9A5, 9A8, 9A11, 9C7, 9D10, 9E11, 9F1, 9F3, 9F4, 9G5, 9G6, 9G7, 9H9, 10A7, 10C6, 10C12, 10D2, 10D5, 10D9, 10E3, 10F6, 10F7, 10F11, 10G3, 10G9, 10H6 |
| 3D8-type | 1D11, 3D8, 4A10, 5A1, 5B9, 6D9, 6E7, 7F1, 7G5, 8B11, 8G12, 9D12, 9E6, 10A11 |
| 6H10-type | 1H4, 3H2, 5F2, 5G9, 6H10, 7F7 |
| 10A5-type | 8C3, 8D3, 10A5 |
| 3B10-type | 1F5, 3B10, 6E2 |
| 1B9 | 1B9 |
| 3B3 | 3B3 |
| 4E7 | 4E7 |
| 5C8 | 5C8 |
| 9D11 | 9D11 |
| 10H1 | 10H1 |
A metagenomic library (96,000 clones divided in 10 96-well plates) was used for screening. Wells in boldface were used for single clone isolation.
Unexpectedly, all four clones shared a nearly identical (99%) DNA sequence, containing a putative amidase gene (p8D; open reading frame 3 [ORF3]) (see Fig. S1 and Table S1 in the supplemental material). This bias could be due to the use of activated sludge (which has the specific biological function of degrading compounds in wastewater) as a source of DNA. A similar bias was noted in a screen for extradiol dioxygenase genes using the same library. Of 43 extradiol dioxygenase genes identified, 20 shared nearly identical sequences (22). Alternatively, the bias may be a consequence of using E. coli as the host. It has been suggested that host genetic systems (i.e., transcription and translation systems) can affect the expression of foreign genes and may limit the diversity of retrieved genes (26).
To explore the types of amidase genes in the 143 fluorescent wells we initially identified, we performed PCR-based genotyping. PCR primers were designed to target genes homologous to 8D amidase. A set of primers specific for the internal region of the amidase gene (nucleotide positions 262 to 279) and its 64-bp downstream region was designed. As a result, 8D-type PCR products were obtained from 111 wells, indicating a predominance of this type of gene in our metagenomic library. The remaining 32 wells yielded no product, suggesting the presence of different types of amidase genes. We applied PIGEX screening to these 32 wells to recover individual fosmid clones; RFLP analysis confirmed their uniqueness. We produced pUC-based shotgun libraries for these fosmids, screened the libraries by the PIGEX procedure, and then determined the nucleotide sequences of the inserts of the positive plasmids by DNA sequencing. Genes showing high similarity were identified in a number of clones (3D8-type, 14 clones; 6H10-type, 6 clones; 10A5-type, 3 clones; and 3B10-type, 3 clones); unique genes were found in 6 clones (1B9, 3B3, 4E7, 5C8, 9D11, and 10H1) (Table 2). In total, we recovered 11 different benzamidase genes. We confirmed the activity of these amidases (1B9, 3B3, 3B10, 3D8, 4E7, 5C8, 6H10, 8D, 9D11, 10A5, and 10H1) by the appearance of benzoate and disappearance of benzamide in HPLC analysis.
In this study, our screening process consisted of three stages: (i) well screening, (ii) individual isolation from hit wells, and (iii) gene identification through functional screening of shotgun libraries. This apparently complex procedure was necessitated by the use of a fosmid-based library with approximately 100 clones per well (22). One could screen a plasmid-based library with one clone per well in a single step.
Comparison of PIGEX and other reporter assay methods.
Reporter assay systems similar to PIGEX have been described by Williamson et al. (30), Mohn et al. (14), and van Sint Fiet et al. (27). Williamson et al. (30) used a recombinant E. coli carrying luxR and gfp genes on a single reporter plasmid, which fluoresced in the presence of bacterial quorum-sensing compounds. They screened a metagenomic library for quorum-sensing compounds using an intracellular system in which both sensor and reporter vectors were included in a single cell. They identified 11 quorum-sensing genes, only one of which was identified as positive in their extracellular system. Based on these results, they concluded that their intracellular system was superior to their extracellular one in terms of sensitivity.
We also performed—in parallel—amidase screening using an intracellular system and observed cross talk between positive and negative clones (data not shown). When we cocultivated E. coli cells carrying amidase-positive fosmids and the benzoate sensor on agar plates, we found that negative colonies in close proximity with positive colonies themselves fluoresced. This phenomenon most likely resulted from the diffusion of benzoate produced by amidase-expressing clones. We believe that such cross talk may be a common problem in intracellular screens performed using compounds that freely permeate cell membranes. Thus, determining which system works better is not straightforward.
Sequence analysis of metagenomic amidases.
Plasmids were purified from the 11 positive clones, and their nucleotide sequences were determined. The genetic organization of the plasmid inserts is shown in Fig. S1 in the supplemental material. The amidase genes carried by eight of the clones (1B9, 3B3, 3D8, 4E7, 6H10, 8D, 9D11, and 10A5) were readily identified on the basis of amino acid sequence identity (see Table S1). In contrast, 3B10, 5C8, and 10H1 had no obvious identity to known amidase-coding genes, indicating that PIGEX was effective for screening our metagenome for novel enzymes.
The last three clones contained a single ORF in their inserts. We assumed that these ORFs were responsible for their amidase activity. The ORF of 3B10 shared 54% amino acid sequence identity with a putative isochorismatase hydrolase from Streptomyces sp. strain AA4. Although this enzyme bore no apparent relationship to benzamidase, a conserved domain database search (13) revealed homology between the ORF and the cysteine hydrolase superfamily, which includes nicotinamidase. Thus, it is likely that this ORF is responsible for the observed benzamidase activity. The ORF included in the 5C8 clone shared 45% primary amino acid sequence identity with a hypothetical protein. The ORF included in 10H1 shared 57% primary amino acid sequence identity with a transmembrane ABC transporter signature motif-containing protein, a component of the periplasmic binding protein-dependent ABC transporter involved in the uptake of branched-chain amino acids. Despite its apparent independence from amidases, one well-characterized d-stereospecific alanine amidase from Brevibacillus borstelensis exhibits 44% amino acid sequence identity to an ABC transporter (2). Therefore, it is possible that the ORF encodes a polypeptide with amidase activity.
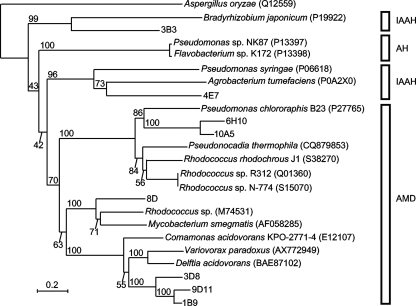
A primary sequence alignment of eight canonical metagenomic benzamidases and functionally characterized bacterial amidases is shown in Fig. S2 in the supplemental material, and their phylogenetic relationships are shown in Fig. 3. Our metagenomic amidases are sparsely distributed in the amidase family. Chebrou et al. (3) compared the amino acid sequences of 22 amidases from both prokaryotes and eukaryotes and identified a highly conserved GGSS(G/S/A)G sequence. This motif is conserved in canonical metagenomic benzamidases though the second Gly is replaced with Val in compounds 1B9 (residue 154), 3D8 (residue 154), and 9D11 (residue 154). The active site (residues Asp191 and Ser195 of Rhodococcus rhodochrous J1 [10]) containing the motif G/A-XDX-G/A-G/A-S-I/V/L-RXP-A/S (residues 173 to 184 of 9D11) is also conserved in our metagenomic amidases.
FIG. 3.
Phylogenetic (neighbor-joining) tree of the metagenomic and representative amidases. The topology of the tree was determined by 1,000 bootstrap replicates, and the distances were corrected by the Kimura method (9). Amidases from isolated bacteria are represented with their organismal origin, and accession numbers are in parentheses. The numbers above the nodes represent the bootstrap values. The scale bar indicates the expected number of substitutions per residue. The acetoamidase of Aspergillus oryzae (EA2 subfamily) was used as the outgroup (3). Abbreviations for the distinct enzyme subfamilies are as follows: IAAH, indole-acetamide hydrolases; AH, 6-aminohexanoate-cyclic-dimer hydrolases; AMD, bacterial amidases.
Substrate specificities of metagenomic amidases.
The substrate specificities of our metagenomic amidases were determined using E. coli cell extracts (Table 3). All of the clones, including those that lacked sequence homology to known amidases (3B10, 5C8, and 10H1), showed benzamidase activity, but clone-to-clone variations were observed for other substrates. Recently, the crystal structure of an amidase from Rhodococcus sp. strain N-771 (identical to Rhodococcus sp. strain N-774 amidase) was determined (15). This amidase was found to prefer benzamide over all other substrates tested (acetamide, acrylamide, and propionamide). Hydrophobic residues in the active site (Phe146, Ile227, Trp328, Leu447, and Ile450) were proposed to be the basis for the specificity of the enzyme for the hydrophobic substrate benzamide. Among the metagenomic amidases in the current study, the 6H10 and 10A5 amidases had hydrophobic residues at positions corresponding to the active site in Rhodococcus sp. N-771 amidase (Phe, Leu, Trp, Trp, and Ile for 6H10; Phe, Phe, Trp, Trp, and Leu for 10A5) and showed similar activity profiles using acetamide, acrylamide, propionamide, and benzamide. However, for the other metagenomic amidases, hydrophilic residues were found in the active site. Notably, the residue corresponding to Leu447 in Rhodococcus sp. N-771 amidase was changed to Asp (1B9, 3D8, 4E7, and 9D11) or Met (8D), and those metagenomic amidases with Asp in place of Leu tended to have higher activity against substrates other than benzamide.
TABLE 3.
Relative activities of the 11 clone types on different amide substrates
| Substrate | Relative activity of the indicated clone carrying a recombinant plasmid (nmol/min/mg of protein)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pHSG398b | 1B9 | 3B3 | 3B10 | 3D8 | 4E7 | 5C8 | 6H10 | 8D | 9D11 | 10A5 | 10H1 | |
| Benzamide | <12 | 160.9 ± 3.2 | 233.0 ± 7.7 | 263.5 ± 14.1 | 36.4 ± 3.2 | 149.3 ± 5.6 | 108.6 ± 2.3 | 277.8 ± 6.0 | 176.0 ± 9.3 | 72.2 ± 7.2 | 266.6 ± 10.2 | 115.7 ± 10.3 |
| Acetamide | <12 | <12 | <12 | <12 | <12 | 22.3 ± 0.5 | 13.3 ± 0.4 | 13.0 ± 0.4 | 13.7 ± 1.7 | <12 | <12 | 13.6 ± 0.6 |
| Acrylamide | <12 | 16.4 ± 0.3 | <12 | <12 | <12 | 15.9 ± 0.7 | <12 | <12 | 21.2 ± 1.2 | <12 | <12 | <12 |
| Indole actamide | <12 | 174.0 ± 3.7 | 27.0 ± 0.9 | 15.2 ± 0.2 | 73.5 ± 4.4 | 217.1 ± 2.4 | 24.0 ± 0.3 | 30.9 ± 0.7 | 181.8 ± 5.6 | 60.3 ± 5.3 | 20.7 ± 0.1 | 84.2 ± 0.2 |
| Nicotine amide | 17.5 ± 2.4 | 153.6 ± 4.5 | 83.6 ± 1.3 | 56.8 ± 0.4 | 31.8 ± 1.5 | 68.8 ± 1.0 | 52.8 ± 1.1 | 265.1 ± 2.2 | 175.6 ± 3.7 | 76.9 ± 3.8 | 287.8 ± 3.5 | 77.9 ± 1.0 |
| Propionamide | <12 | 149.4 ± 5.1 | 35.9 ± 0.9 | <12 | 17.5 ± 4.2 | 141.2 ± 5.1 | <12 | 22.7 ± 0.9 | 177.2 ± 12.1 | 55.8 ± 5.1 | 15.9 ± 0.4 | <12 |
| Glycine amide | <12 | 61.9 ± 4.8 | 137.3 ± 4.8 | 127.0 ± 6.2 | 46.5 ± 6.2 | 122.5 ± 5.4 | 136.4 ± 3.3 | 128.3 ± 3.7 | 52.9 ± 13.0 | 55.5 ± 8.5 | 127.3 ± 9.0 | 123.7 ± 5.7 |
| l-Alanine amide | <12 | 92.2 ± 1.8 | 118.1 ± 1.8 | 108.4 ± 3.4 | 37.6 ± 1.5 | 107.6 ± 3.4 | 124.3 ± 7.2 | 117.6 ± 2.6 | 61.9 ± 3.4 | 42.8 ± 10.7 | 114.2 ± 5.5 | 112.0 ± 4.2 |
| d-Alanine amide | <12 | 124.8 ± 15.1 | 121.7 ± 8.8 | 108.5 ± 0.8 | 36.2 ± 3.2 | 105.0 ± 2.3 | 118.3 ± 2.8 | 114.1 ± 2.4 | 92.4 ± 5.9 | 57.4 ± 2.3 | 116.4 ± 9.9 | 108.5 ± 3.8 |
| l-Leucine amide | <12 | 120.8 ± 5.4 | <12 | <12 | 27.5 ± 2.5 | <12 | <12 | <12 | 100.1 ± 2.7 | 51.6 ± 5.4 | <12 | <12 |
| d-Leucine amide | 20.2 ± 2.2 | 116.3 ± 6.4 | <12 | <12 | 104.3 ± 4.7 | <12 | <12 | <12 | <12 | 103.7 ± 2.4 | <12 | <12 |
| l-Proline amide | <12 | 103.9 ± 11.3 | <12 | <12 | <12 | <12 | <12 | <12 | 39.6 ± 2.2 | 60.7 ± 3.3 | <12 | <12 |
| d-Proline amide | <12 | 101.6 ± 8.4 | <12 | <12 | <12 | <12 | <12 | <12 | 54.8 ± 3.8 | 67.4 ± 1.7 | <12 | <12 |
| l-Phenylalanine amide | 18.9 ± 1.4 | 112.9 ± 10.9 | <12 | <12 | 101.7 ± 4.9 | 180.6 ± 4.8 | <12 | <12 | 99.3 ± 3.2 | 107.3 ± 3.5 | <12 | <12 |
| d-Phenylalanine amide | 30.9 ± 1.9 | 111.5 ± 10.7 | <12 | <12 | 98.3 ± 3.9 | 206.2 ± 7.2 | <12 | 15.9 ± 0.5 | 57.4 ± 2.6 | 102.0 ± 4.1 | <12 | <12 |
Reactions were done under the standard conditions described in Materials and Methods. All measurements were done in triplicate.
E. coli JM109 was transformed by pHSG398.
From a biotechnological point of view, the stereospecific hydrolysis of racemic amino acid amides is particularly useful as a cost-effective approach for the production of optically pure l- and d-amino acids (2, 6, 8, 11, 12). Of our metagenomic amidases, 1B9, 3D8, and 9D11 share ∼68% amino acid sequence identity with each other and ∼53% with known d-amino acid amidases from Delftia acidovorans (8) and Variovorax paradoxus (12). The amidase from D. acidovorans is highly selective for the d-enantiomers of proline amide, leucine amide, and phenylalanine amide, whereas the amidase from V. paradoxus prefers the d-enantiomers of alanine amide, leucine amide, and phenylalanine amide and the l-enantiomer of proline amide. In our study, the 3D8 and 9D11 amidases preferred the d-enantiomer of leucine amide, while the 1B9 amidase, the closest homolog of the 9D11 enzyme (84% primary sequence identity) lacked this selectivity (Table 3). All three of these enzymes lacked selectivity for phenylalanine and proline amides, and the 3D8 amidase would not metabolize proline amide. Interestingly, the 8D amidase had the opposite selectivity, favoring the l-enantiomers of leucine and phenylalanine amides. The 6H10 and 10A5 amidases share high amino acid sequence identity (76%), and their substrate specificities were similar. The 5C8 and 10H1 amidases also showed similar activity profiles despite a lack of sequence similarity. Therefore, the relationship between sequence and selectivity is not straightforward.
We used benzamide as the sole substrate for screening. However, owing to the intrinsic catalytic promiscuity of amidases, our metagenomic amidases displayed various substrate specificities and stereospecificities. They may form a unique starting point for further protein engineering.
Supplementary Material
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science (grants 19880040 and 22780081).
Footnotes
Published ahead of print on 10 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baek, D. H., S.-J. Kwon, S.-P. Hong, M.-S. Kwak, M.-H. Lee, J. J. Song, S.-G. Lee, K.-H. Yoon, and M.-H. Sung. 2003. Characterization of a thermostable d-stereospecific alanine amidase from Brevibacillus borstelensis BCS-1. Appl. Environ. Microbiol. 69:980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chebrou, H., F. Bigey, A. Arnaud, and P. Galzy. 1996. Study of the amidase signature group. Biochim. Biophys. Acta 1298:285-293. [DOI] [PubMed] [Google Scholar]
- 4.Conway, E. J., and A. Byrne. 1933. An absorption apparatus for the microdetermination of certain volatile substances. I. The microdetermination of ammonia. Biochem. J. 27:419-429. [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrer, M., A. Beloqui, K. N. Timmis, and P. N. Golyshin. 2009. Metagenomics for mining new genetic resources of microbial communities. J. Mol. Microbiol. Biotechnol. 16:109-123. [DOI] [PubMed] [Google Scholar]
- 6.Hermes, H. F., R. F. Tandler, T. Sonke, L. Dijkhuizen, and E. M. Meijer. 1994. Purification and characterization of an l-amino amidase from Mycobacterium neoaurum ATCC 25795. Appl. Environ. Microbiol. 1:153-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirrlinger, B., A. Stolz, and H. J. Knackmuss. 1996. Purification and properties of an amidase from Rhodococcus erythropolis MP50 which enantioselectively hydrolyzes 2-arylpropionamides. J. Bacteriol. 178:3501-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hongpattarakere, T., H. Komeda, and Y. Asano. 2005. Purification, characterization, gene cloning and nucleotide sequencing of d-stereospecific amino acid amidase from soil bacterium: Delftia acidovorans. J. Ind. Microbiol. Biotechnol. 32:567-576. [DOI] [PubMed] [Google Scholar]
- 9.Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, United Kingdom.
- 10.Kobayashi, M., Y. Fujiwara, M. Goda, H. Komeda, and S. Shimizu. 1997. Identification of active sites in amidase: evolutionary relationship between amide bond- and peptide bond-cleaving enzymes. Proc. Natl. Acad. Sci. U. S. A. 94:11986-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komeda, H., and Y. Asano. 2000. Gene cloning, nucleotide sequencing, and purification and characterization of the d-stereospecific amino-acid amidase from Ochrobactrum anthropi SV3. Eur. J. Biochem. 267:2028-2035. [DOI] [PubMed] [Google Scholar]
- 12.Krieg, L., M. B. Ansorge-Schumacher, and M. R. Kula. 2002. Screening for amidases: isolation and characterization of a novel d-amidase from Variovorax paradoxus. Adv. Synth. Catal. 344:965-973. [Google Scholar]
- 13.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohn, W. W., J. Garmenda, T. C. Galvao, and V. de Lorenzo. 2006. Surveying biotransformations with à la carte genetic traps: translating dehydrochlorination of lindane (gamma-hexachlorocyclohexane) into lacZ-based phenotypes. Environ. Micobiol. 8:546-555. [DOI] [PubMed] [Google Scholar]
- 15.Ohtaki, A., K. Murata, Y. Sato, K. Noguchi, H. Miyatake, N. Dohmae, K. Yamada, M. Yohda, and M. Odaka. 2010. Structure and characterization of amidase from Rhodococcus sp. N-771: insight into the molecular mechanism of substrate recognition. Biochim. Biophys. Acta 1804:184-192. [DOI] [PubMed] [Google Scholar]
- 16.Perrière, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 17.Reunanen, J., and P. E. J. Saris. 2003. Microplate bioassay for nisin in foods, based on nisin-induced green fluorescent protein fluorescence. Appl. Environ. Microbiol. 69:4214-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ron, E. Z. 2007. Biosensing environmental pollution. Curr. Opin. Biotechnol. 18:252-256. [DOI] [PubMed] [Google Scholar]
- 19.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 20.Shaw, N. M., and A. B. Naughton. 2004. The substrate specificity of the heat-stable stereospecific amidase from Klebsiella oxytoca. Tetrahedron 60:747-752. [Google Scholar]
- 21.Steele, H. L., K. E. Jaeger, R. Daniel, and W. R. Streit. 2009. Advances in recovery of novel biocatalysts from metagenomes. J. Mol. Microbiol. Biotechnol. 16:25-37. [DOI] [PubMed] [Google Scholar]
- 22.Suenaga, H., T. Ohnuki, and K. Miyazaki. 2007. Functional screening of a metagenomic library for genes involved in microbial degradation of aromatic compounds. Environ. Microbiol. 9:2289-2297. [DOI] [PubMed] [Google Scholar]
- 23.Tecon, R., and J. R. van der Meer. 2006. Information from single-cell bacterial biosensors: what is it good for? Curr. Opin. Biotechnol. 17:4-10. [DOI] [PubMed] [Google Scholar]
- 24.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uchiyama, T., T. Abe, T. Ikemura, and K. Watanabe. 2005. Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat. Biotechnol. 23:88-93. [DOI] [PubMed] [Google Scholar]
- 26.Uchiyama, T., and K. Miyazaki. 2009. Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr. Opin. Biotechnol. 20:616-622. [DOI] [PubMed] [Google Scholar]
- 27.van Sint Fiet, S., J. B. van Beilen, and B. Witholt. 2006. Selection of biocatalysts for chemical synthesis. Proc. Natl. Acad. Sci. U. S. A. 103:1693-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei, Y., T. Kurihara, T. Suzuki, and N. Esaki. 2003. A novel esterase from a psychrotrophic bacterium, Acinetobacter sp. strain no. 6, that belongs to the amidase signature family. J. Mol. Catal. B Enzym. 23:357-365. [Google Scholar]
- 29.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williamson, L. L., B. R. Borlee, P. D. Schloss, C. Guan, H. K. Allen, and J. Handelsman. 2005. Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl. Environ. Microbiol. 71:6335-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.