Abstract
The 37-kDa oncofetal antigen (OFA), a tumor immunogen expressed on all mammalian cancers examined to date, was secreted and anchored to the cell wall of Lactobacillus plantarum using homologous signal peptides and LPxTG anchors. Orally administered L. plantarum expressing anchored OFA induced a specific immune response against OFA in mice.
There is increasing interest in using lactic acid bacteria (LAB) as mucosal delivery vehicles, since these bacteria are normal inhabitants of the human intestine (23) and may exert probiotic effects (15, 16). The recent focus on LAB has resulted in several promising delivery strategies for therapeutics, microbicides, and vaccine antigens (5, 6, 11, 24). As delivery vectors, lactobacilli constitute an attractive alternative to the commonly used species Lactococcus lactis because of their immunostimulatory properties and ability to persist longer at the mucosal layer (10). The use of antigen-producing lactobacilli for cancer vaccine purposes has so far been limited to E7 and L1 (1, 9, 21), both derived from human papillomavirus type 16.
The 37-kDa oncofetal antigen (OFA) is a promising cancer vaccine candidate because it is a universal tumor immunogen expressed in all mammalian tumors tested so far (8), including human colon carcinomas (17). In this study, we express, secrete, and anchor OFA in the probiotic human saliva isolate Lactobacillus plantarum WCFS1 (14), using the pSIP system (22) for protein expression. All constructed OFA plasmids used in this study are based on pSIP derivatives previously constructed for secretion of staphylococcal nuclease (Nuc) and lactobacillal amylase (Amy) by using a variety of signal peptides (SPs) from L. plantarum WCFS1, which are designated by their gene codes (18, 19). Constructs were first established in Escherichia coli TOP10 cells and then transformed into L. plantarum by electroporation, as described previously (3) using the appropriate antibiotics as selection markers. Fragments obtained by PCR were first cloned in appropriate TOPO vectors before further handling.
For secretion only, the Nuc- or Amy-encoding genes were simply exchanged by a fragment encoding the complete OFA coding sequence with a SalI site and an Acc65I site (Fig. 1). The OFA sequence (accession no. AAD26866.1) was amplified from cDNA with primers OFASal (GTCGACTCCGGAGCCCTTGAC) (SalI site italic) and OFAAcc (GGTACCTCAGGACCACTCAGTGGT) (Acc65I site italic). The 0.89-kb OFA fragment was ligated into the SalI-Acc65I-digested vectors pLp_0297sAmyA, pLp_0373sNuc, pLp_0600AmyA, pLp_1447sAmyA, and pLp_2940sAmyA (19), yielding the plasmids pLp_0297sOFA, pLp_0373sOFA, pLp_0600sOFA, pLp_1447sOFA, and pLp_2940sOFA.
FIG. 1.
Schematic overview of the expression cassette for secretion and cell wall anchoring of OFA in L. plantarum. The vectors are based on previously described secretion vectors (19) in which a secretion cassette is translationally fused to the inducible PsppA promoter. All parts of the cassette are easily exchangeable using the introduced linker (L) restriction sites (SalI and MluI), the NdeI site at the translational fusion point, and the downstream multiple cloning site (MCS) containing the Acc65I and HindIII sites. The construction of the MluI linker site and the addition of a cell wall anchor (CWA) sequence are new in this study (see text). The primary sequence of Lp_2578 shows a signal peptide cleavage site (arrow), an LPxTG motif (gray box; the actual consensus sequence in L. plantarum is LPQTxE) (9, 14), and a proline-rich motif (underlined as predicted by MotifScan; http://myhits.isb-sib.ch/cgi-bin/motif_scan) running from amino acids (aa) 51 to 194 counted in the upstream direction from the LPxTG motif that may be adapted to a location inside the peptidoglycan layer (13). pLp_0373sOFAcwa1 encodes the longest anchor (644 aa), in which almost the entire mature Lp_2578 protein was fused to the C terminus of OFA using a serine (boldface S) close to the N terminus of mature Lp_2578). pLp_0373sOFAcwa2 encodes the medium-length anchor (194 aa), the fusion point being at a proline (boldface, underlined P). pLp_0373sOFAcwa3 encodes the shortest anchor (128 aa), the fusion point being a serine (boldface, underlined S).
For construction of the anchoring vectors, the ofa gene was amplified from pLp_0373sOFA with primers OFASal and OFAMlu (GGTACCTACGCGTGGACCACTCAGTGGT, containing both MluI and Acc65I sites [italic]) to remove the stop codon in OFA and to introduce a C-terminal MluI site (Fig. 1). The resulting fragment was ligated back into SalI-Acc65I-digested pLp_0373sOFA, yielding the intermediate plasmid pLp_0373sOFA-I. Three versions of the anchoring sequences were PCR amplified from the lp_2578 gene on the L. plantarum chromosome by using the same reverse primer, 2578Hind, with a HindIII restriction site (italic) (AAGCTTTCAAGCACGACGGCGAT) and three different forward primers with an MluI restriction site (italic) for amplification of the long anchor (2578MluI; ACGCGTAGTGTTACGGGTTTAACGGC), medium anchor (2578Mlu2; ACGCGTGTCACTGAACCAGGAAC), and short anchor (2578Mlu3; ACGCGTAGCCAACCAGGCAAAC) (see Fig. 1 for details). The anchor-encoding DNA fragments were inserted into the pLp_0373sOFA-I vector, using the MluI and HindIII sites, yielding the plasmids pLp_0373sOFAcwa1, pLp_0373sOFAcwa2, and pLp_0373sOFAcwa3, respectively. A plasmid encoding intracellular OFA (with no signal peptide or anchor) was constructed by amplifying the OFA fragment from pLp_0373sOFA with the primers OFANde (CATATGTCCGGAGCCCTTGAC) (NdeI site italic) and OFAAcc and by religating the resulting fragment into the NdeI-Acc65I-digested Lp_0373sOFA plasmid, yielding pCytOFA.
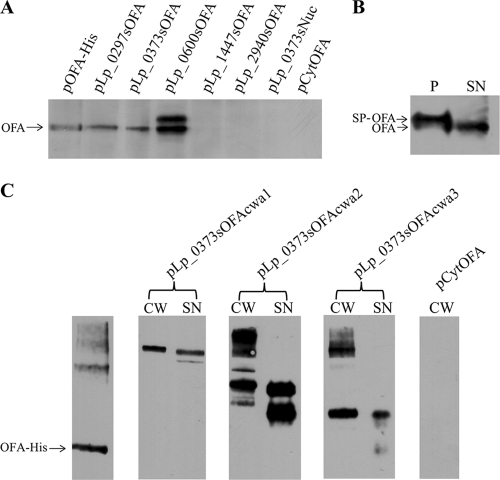
The production and localization of proteins in the recombinant L. plantarum strains were analyzed by studying protein extracts from the supernatant (SN), the cell wall fraction (CW), and protoplasts (P). Cells were grown until they reached an optical density at 600 nm (OD600) of ∼0.3 and induced by adding 25 ng ml−1 pheromone peptide as previously described (22). The cells were harvested 4 h after induction and washed twice with cold Tris-buffered sucrose (pH 7.0, 10 mM MgCl2, 250 mM sucrose). The supernatants were filtered through 0.22-μm-pore Millex GP filter units (Millipore, Carrigtwohill, Co. Cork, Ireland), and 1 mM phenylmethylsulfonyl fluoride (PMSF) was added. The pH of the supernatants was adjusted to ∼7 by addition of NaOH prior to addition of 0.2 mg ml−1 (final concentration) sodium deoxycholate. After incubation for 30 min on ice, the supernatant proteins were precipitated by adding ice-cold 100% trichloroacetic acid (TCA) to 16% (vol/vol) final concentration. After incubation on ice for 20 min, proteins were collected by centrifugation at 16,100 × g for 15 min and washed once with chilled acetone. The protoplasts and cell wall fractions were prepared from the cell pellets essentially as described previously (20). Sodium deoxycholate was added to the cell wall fraction to a final concentration of 0.2 mg ml−1, and the cell wall proteins were precipitated and washed as described for the supernatant fraction above. For analysis by Western blotting, precipitated proteins were dried in a vacuum centrifuge and solubilized in a buffer containing 50 mM dithiothreitol (DTT), 4 M urea, 1.1 M thiourea, 1% ASB-14, and 1× NuPage loading buffer (Invitrogen, Carlsbad, CA). The buffer volumes were adjusted such as to have a fixed ratio between the volume and the OD600 value of the cultures from which the proteins were extracted. All samples were vortexed and boiled for 10 min prior to electrophoresis on 10% NuPage Novex Bis-Tris gels and subsequent immunoblotting using the iblot system (Invitrogen). Protein detection was performed with the SNAP i.d. System (Millipore, Billerica, MA), using anti-laminin R polyclonal antibodies (Santa Cruz Biotechnology, Inc., CA) or OFA monoclonal antibodies combined with horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (Bio-Rad) (Fig. 2 A and B) or rabbit anti-mouse antibody (Dako, Denmark) (Fig. 2C), respectively. Proteins were visualized with the SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL). Western analysis of the supernatant fractions of the various L. plantarum transformants (Fig. 2A) showed correctly processed OFA in two strains (harboring pLp_0297sOFA or pLp0373sOFA). The supernatant of cells harboring pLp_0600sOFA showed two bands, most likely representing processed and unprocessed OFA. The other two tested strains (L. plantarum harboring pLp_1447sOFA and pLp_2940sOFA) did not secrete detectable amounts of OFA, nor did cells harboring pCytOFA (with no signal peptide). Western blot analysis of protoplast fractions confirmed that OFA was produced by all five SP constructs and the pCytOFA construct (data not shown). Analysis of the secretion efficiency of the L. plantarum strain harboring pLp_0373sOFA (Fig. 2B) showed that even in this well-secreting strain, the majority of the produced protein is not processed and secreted. Figure 2C shows that all three anchors led to retention of OFA in the cell wall fraction. OFA was also observed in the supernatant fractions from L. plantarum strains harboring the constructs for OFA anchoring. Finding heterologously expressed proteins destined for the cell wall in the supernatant fraction is not unusual (7, 12). The samples with purified OFA and the cell wall fractions for the two strains with the shorter anchors show high-molecular-weight bands, dominated by a band of approximately twice the expected size of OFA. This suggests some degree of di- and oligomerization, which would be in accordance with the OFA protein's known natural tendency to dimerize (4). The supernatant fractions show some lower-molecular-weight bands, in particular for pLp_0373sOFAcwa2, indicating the occurrence of proteolytic degradation.
FIG. 2.
Western analysis of secretion and anchoring of OFA in L plantarum. (A) Supernatant fractions from L. plantarum harboring various secretion vectors. The plasmid present in each L. plantarum strain is indicated above the wells; the lane marked “OFA-His” contains 240 ng purified hexahistidine-tagged OFA. Negative controls were supernatants from L. plantarum harboring pLp_0373sNuc (19) and pCytOFA (OFA without signal peptide). (B) Analysis of secretion efficiency in L. plantarum harboring pLp_0373sOFA. (P and SN indicate the protoplast and the supernatant fraction, respectively.) All samples in panels A and B represent the same amount of culture, except for sample P in panel B, which was diluted 20-fold relative to the other samples. (C) The blot shows purified His-tagged OFA (60 ng) and the cell wall (CW) and supernatant (SN) fractions from L. plantarum harboring the three OFA anchoring vectors, as indicated above the wells. The cell wall fraction from L. plantarum harboring pCytOFA (OFA with no signal peptide) was used as a negative control. The arrows indicate the expected sizes of the cell-wall-anchored OFA. All samples in panel C came from the same blot, and all samples represent equivalent amounts of cells.
For detection of an OFA immune response, inbred female 7- to 8-week-old BALB/c mice were purchased from Taconic (Bomholt, Denmark). Animal care was in accordance with national legislation and institutional guidelines, and the experimental protocol was accepted by the local ethical committee. L. plantarum was grown and induced as described above. Four hours after induction, the cells were harvested by centrifugation at 3,500 × g for 10 min at 4°C and washed with cold phosphate-buffered saline (PBS) buffer. Cell suspensions of L. plantarum or L. plantarum harboring pLp_0373sOFAcwa2 were placed in ball-tipped syringes for oral gavage inoculation (1 × 109 bacteria in 500 μl PBS buffer, three times, with 1-week intervals). Lp_0373sOFAcwa2 was chosen because an anchor length corresponding to the predicted proline-rich region a priori seems the most optimal anchor length. Thirty days after the first immunization, mice were euthanized and sera were tested for the presence of antibodies against the OFA-expressing 4T1 breast cancer cell line (2). In order to visualize a specific OFA immune response, 4T1 protein extracts (40 μg per lane) were separated by SDS-PAGE and subsequently analyzed by Western blotting (Fig. 3). A specific OFA immune response was demonstrated in mice immunized with L. plantarum expressing the cell-wall-anchored OFA, whereas such a response did not occur in mice immunized with wild-type L. plantarum. The character of the IgG response against OFA was determined by coating a 96-well microtiter plate with hexahistidine-tagged OFA (0.5 μg in 100 μl PBS per well). After being blocked with 5% fetal calf serum in PBS for 1 h at room temperature and subsequent washing, the plate was incubated with serum samples (1/100) from immunized mice. Subsequent to washing with PBS, either anti-mouse IgG1 or anti-mouse IgG2a conjugated to alkaline phosphatase (1/1,000; Serotec, Oxford, United Kingdom) was used to determine the type of IgG response. After 1 h of incubation at room temperature, the plate was washed and the immune complexes were detected by adding p-nitrophenyl phosphate as substrate and reading the OD at 405 nm. Samples were tested in triplicate. The OD values measured at 405 nm were 2.1 (±0.3) and 0.6 (±0.2) for the IgG1 and IgG2a antibodies, respectively (standard deviations are shown in parentheses). The generated antibodies were dominated by isotype IgG1, associated with a humoral immune response.
FIG. 3.
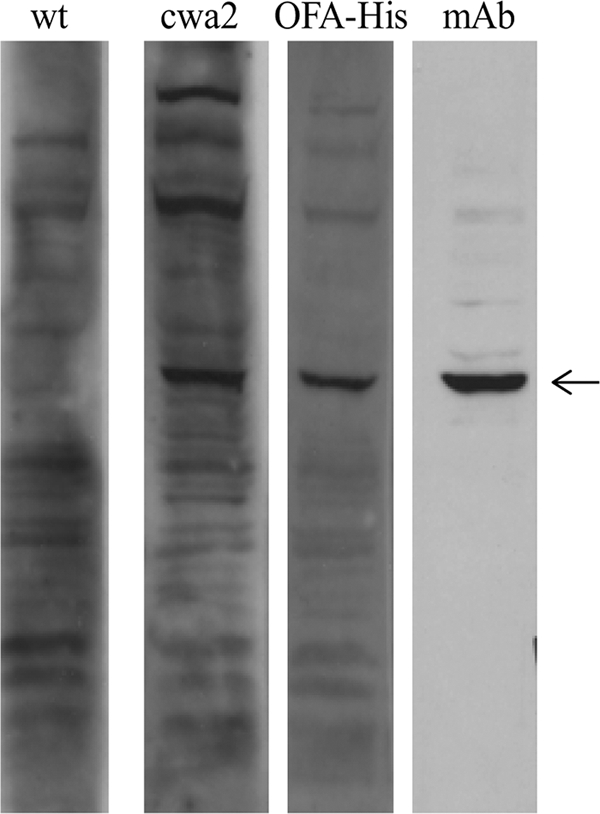
Representative example of OFA immune response in vivo. The lanes contain protein extracts (40 μg per lane) from OFA-expressing 4T1 cells, incubated with sera from mice orally immunized with wild-type (wt) L. plantarum or L. plantarum harboring pLp_0373sOFAcwa2 (cwa2), or from mice subcutaneously immunized with His-tagged OFA in Freund's complete adjuvant (OFA-His). Anti-OFA monoclonal mouse IgG antibody (mAb) was used as a positive control. Antibody binding was visualized using HRP conjugated to anti-mouse IgG and the ECL system (Amersham Life Science, Buckinghamshire, United Kingdom). The arrow indicates OFA.
This study presents, for the first time, bacterial surface display of OFA and illustrates the potential of using L. plantarum for mucosal delivery of cancer vaccines. It was shown that L. plantarum cells expressing cell-wall-anchored OFA were capable of inducing specific antibodies in mice. Whether the surface-expressed OFA antigen will be sufficient to induce tumor regression is the subject of ongoing studies in a mouse model.
Acknowledgments
This work was supported by Norwegian Research Council grants 159058 and 183637.
We thank Adel Barsoum for providing the anti-OFA monoclonal antibody and Dag Sørensen for helping with the animal experiments.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Aires, K. A., A. M. Cianciarullo, S. M. Carneiro, L. L. Villa, E. Boccardo, G. Perez-Martinez, I. Perez-Arellano, M. L. Oliveira, and P. L. Ho. 2006. Production of human papillomavirus type 16 L1 virus-like particles by recombinant Lactobacillus casei cells. Appl. Environ. Microbiol. 72:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslakson, C. J., and F. R. Miller. 1992. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 52:1399-1405. [PubMed] [Google Scholar]
- 3.Aukrust, T. W., M. B. Brurberg, and I. F. Nes. 1995. Transformation of Lactobacillus by electroporation. Methods Mol. Biol. 47:201-208. [DOI] [PubMed] [Google Scholar]
- 4.Barsoum, A. L., J. W. Rohrer, and J. H. Coggin. 2000. 37kDa oncofetal antigen is an autoimmunogenic homologue of the 37kDa laminin receptor precursor. Cell. Mol. Biol. Lett. 5:207-230. [Google Scholar]
- 5.Bermudez-Humaran, L. G., N. G. Cortes-Perez, F. Lefevre, V. Guimaraes, S. Rabot, J. M. Alcocer-Gonzalez, J. J. Gratadoux, C. Rodriguez-Padilla, R. S. Tamez-Guerra, G. Corthier, A. Gruss, and P. Langella. 2005. A novel mucosal vaccine based on live lactococci expressing E7 antigen and IL-12 induces systemic and mucosal immune responses and protects mice against human papillomavirus type 16-induced tumors. J. Immunol. 175:7297-7302. [DOI] [PubMed] [Google Scholar]
- 6.Braat, H., P. Rottiers, D. W. Hommes, N. Huyghebaert, E. Remaut, J. P. Remon, S. J. van Deventer, S. Neirynck, M. P. Peppelenbosch, and L. Steidler. 2006. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn's disease. Clin. Gastroenterol. Hepatol. 4:754-759. [DOI] [PubMed] [Google Scholar]
- 7.Brinster, S., S. Furlan, and P. Serror. 2007. C-terminal WxL domain mediates cell wall binding in Enterococcus faecalis and other gram-positive bacteria. J. Bacteriol. 189:1244-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coggin, J. H., Jr., A. L. Barsoum, J. W. Rohrer, M. Thurnher, M. Zeis, and P. O. Schwarzenberger. 2005. Cancer vaccine technology update: the immunobiology of 37 kDa oncofetal antigen/immature laminin receptor protein (OFA/iLRP) and RNA, a universal tumor immunogen. Mod. Aspects Immunobiol. 16:27-34. [Google Scholar]
- 9.Cortes-Perez, N. G., V. Azevedo, J. M. Alcocer-Gonzalez, C. Rodriguez-Padilla, R. S. Tamez-Guerra, G. Corthier, A. Gruss, P. Langella, and L. G. Bermudez-Humaran. 2005. Cell-surface display of E7 antigen from human papillomavirus type-16 in Lactococcus lactis and in Lactobacillus plantarum using a new cell-wall anchor from lactobacilli. J. Drug Target. 13:89-98. [DOI] [PubMed] [Google Scholar]
- 10.Cortes-Perez, N. G., F. Lefevre, G. Corthier, K. Adel-Patient, P. Langella, and L. G. Bermudez-Humaran. 2007. Influence of the route of immunization and the nature of the bacterial vector on immunogenicity of mucosal vaccines based on lactic acid bacteria. Vaccine 25:6581-6588. [DOI] [PubMed] [Google Scholar]
- 11.Diep, D. B., G. Mathiesen, V. G. H. Eijsink, and I. F. Nes. 2009. Use of lactobacilli and their pheromone-based regulatory mechanism in gene expression and drug delivery. Curr. Pharm. Biotechnol. 10:62-73. [DOI] [PubMed] [Google Scholar]
- 12.Dieye, Y., S. Usai, F. Clier, A. Gruss, and J. C. Piard. 2001. Design of a protein-targeting system for lactic acid bacteria. J. Bacteriol. 183:4157-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 14.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U. S. A. 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebeer, S., J. Vanderleyden, and S. C. De Keersmaecker. 2008. Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 72:728-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ljungh, A., and T. Wadström. 2006. Lactic acid bacteria as probiotics. Curr. Issues Intest. Microbiol. 7:73-89. [PubMed] [Google Scholar]
- 17.Mafune, K., T. S. Ravikumar, J. M. Wong, H. Yow, L. B. Chen, and G. D. Steele, Jr. 1990. Expression of a Mr 32,000 laminin-binding protein messenger RNA in human colon carcinoma correlates with disease progression. Cancer Res. 50:3888-3891. [PubMed] [Google Scholar]
- 18.Mathiesen, G., A. Sveen, M. B. Brurberg, L. Fredriksen, L. Axelsson, and V. G. Eijsink. 2009. Genome-wide analysis of signal peptide functionality in Lactobacillus plantarum WCFS1. BMC Genomics 10:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathiesen, G., A. Sveen, J. C. Piard, L. Axelsson, and V. G. Eijsink. 2008. Heterologous protein secretion by Lactobacillus plantarum using homologous signal peptides. J. Appl. Microbiol. 105:215-226. [DOI] [PubMed] [Google Scholar]
- 20.Mujahid, S., T. Pechan, and C. Wang. 2007. Improved solubilization of surface proteins from Listeria monocytogenes for 2-DE. Electrophoresis 28:3998-4007. [DOI] [PubMed] [Google Scholar]
- 21.Poo, H., H. M. Pyo, T. Y. Lee, S. W. Yoon, J. S. Lee, C. J. Kim, M. H. Sung, and S. H. Lee. 2006. Oral administration of human papillomavirus type 16 E7 displayed on Lactobacillus casei induces E7-specific antitumor effects in C57/BL6 mice. Int. J. Cancer 119:1702-1709. [DOI] [PubMed] [Google Scholar]
- 22.Sørvig, E., G. Mathiesen, K. Naterstad, V. G. Eijsink, and L. Axelsson. 2005. High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors. Microbiology 151:2439-2449. [DOI] [PubMed] [Google Scholar]
- 23.Vaughan, E. E., M. C. de Vries, E. G. Zoetendal, K. Ben-Amor, A. D. Akkermans, and W. M. de Vos. 2002. The intestinal LABs. Antonie Van Leeuwenhoek 82:341-352. [PubMed] [Google Scholar]
- 24.Yuvaraj, S., S. Lahham, R. K. Marreddy, G. Dijkstra, W. A. Wolken, J. S. Lolkema, W. Helfrich, F. E. Johansen, M. P. Peppelenbosch, and N. A. Bos. 2008. Human scFv SIgA expressed on Lactococcus lactis as a vector for the treatment of mucosal disease. Mol. Nutr. Food Res. 52:913-920. [DOI] [PubMed] [Google Scholar]