Abstract
Bacterial anaerobic ammonium oxidation (anammox) is an important process in the marine nitrogen cycle. Because ongoing eutrophication of coastal bays contributes significantly to the formation of low-oxygen zones, monitoring of the anammox bacterial community offers a unique opportunity for assessment of anthropogenic perturbations in these environments. The current study used targeting of 16S rRNA and hzo genes to characterize the composition and structure of the anammox bacterial community in the sediments of the eutrophic Jiaozhou Bay, thereby unraveling their diversity, abundance, and distribution. Abundance and distribution of hzo genes revealed a greater taxonomic diversity in Jiaozhou Bay, including several novel clades of anammox bacteria. In contrast, the targeting of 16S rRNA genes verified the presence of only “Candidatus Scalindua,” albeit with a high microdiversity. The genus “Ca. Scalindua” comprised the apparent majority of active sediment anammox bacteria. Multivariate statistical analyses indicated a heterogeneous distribution of the anammox bacterial assemblages in Jiaozhou Bay. Of all environmental parameters investigated, sediment organic C/organic N (OrgC/OrgN), nitrite concentration, and sediment median grain size were found to impact the composition, structure, and distribution of the sediment anammox bacterial community. Analysis of Pearson correlations between environmental factors and abundance of 16S rRNA and hzo genes as determined by fluorescent real-time PCR suggests that the local nitrite concentration is the key regulator of the abundance of anammox bacteria in Jiaozhou Bay sediments.
Anaerobic ammonium oxidation (anammox, NH4+ + NO2− → N2 + 2H2O) was proposed as a missing N transformation pathway decades ago. It was found 20 years later to be mediated by bacteria in artificial environments, such as anaerobic wastewater processing systems (see reference 32 and references therein). Anammox in natural environments was found even more recently, mainly in O2-limited environments such as marine sediments (28, 51, 54, 67, 69) and hypoxic or anoxic waters (10, 25, 39-42). Because anammox may remove as much as 30 to 70% of fixed N from the oceans (3, 9, 64), this process is potentially as important as denitrification for N loss and bioremediation (41, 42, 73). These findings have significantly changed our understanding of the budget of the marine and global N cycles as well as involved pathways and their evolution (24, 32, 35, 72). Studies indicate variable anammox contributions to local or regional N loss (41, 42, 73), probably due to distinct environmental conditions that may influence the composition, abundance, and distribution of the anammox bacteria. However, the interactions of anammox bacteria with their environment are still poorly understood.
The chemolithoautotrophic anammox bacteria (64, 66) comprise the new Brocadiaceae family in the Planctomycetales, for which five Candidatus genera have been described (see references 32 and 37 and references therein): “Candidatus Kuenenia,” “Candidatus Brocadia,” “Candidatus Scalindua,” “Candidatus Anammoxoglobus,” and “Candidatus Jettenia.” Due to the difficulty of cultivation and isolation, anammox bacteria are not yet in pure culture. Molecular detection by using DNA probes or PCR primers targeting the anammox bacterial 16S rRNA genes has thus been the main approach for the detection of anammox bacteria and community analyses (58). However, these studies revealed unexpected target sequence diversity and led to the realization that due to biased coverage and specificity of most of the PCR primers (2, 8), the in situ diversity of anammox bacteria was likely missed. Thus, the use of additional marker genes for phylogenetic analysis was suggested in hopes of better capturing the diversity of this environmentally important group of bacteria. By analogy to molecular ecological studies of aerobic ammonia oxidizers, most recent studies have attempted to include anammox bacterium-specific functional genes. All anammox bacteria employ hydrazine oxidoreductase (HZO) (= [Hzo]3) to oxidize hydrazine to N2 as the main source for a useable reductant, which enables them to generate proton-motive force for energy production (32, 36, 65). Phylogenetic analyses of Hzo protein sequences revealed three sequence clusters, of which the cladistic structure of cluster 1 is in agreement with the anammox bacterial 16S rRNA gene phylogeny (57). The hzo genes have emerged as an alternative phylogenetic and functional marker for characterization of anammox bacterial communities (43, 44, 57), allowing the 16S rRNA gene-based investigation methods to be corroborated and improved.
The contribution of anammox to the removal of fixed N is highly variable in estuarine and coastal sediments (50). For instance, anammox may be an important pathway for the removal of excess N (23) or nearly negligible (48, 54, 67, 68). This difference may be attributable to a difference in the structure and composition of anammox bacterial communities, in particular how the abundance of individual cohorts depends on particular environmental conditions. Anthropogenic disturbance with variable source and intensity of eutrophication and pollution may further complicate the anammox bacterium-environment relationship.
Jiaozhou Bay is a large semienclosed water body of the temperate Yellow Sea in China. Eutrophication has become its most serious environmental problem, along with red tides (harmful algal blooms), species loss, and contamination with toxic chemicals and harmful microbes (14, 15, 21, 61, 71). Due to different sources of pollution and various levels of eutrophication across Jiaozhou Bay (mariculture, municipal and industrial wastewater, crude oil shipyard, etc.), a wide spectrum of environmental conditions may contribute to a widely varying community structure of anammox bacteria. This study used both 16S rRNA and hzo genes as targets to measure their abundance, diversity, and spatial distribution and assess the response of the resident anammox bacterial community to different environmental conditions. Environmental factors with potential for regulating the sediment anammox microbiota are discussed.
MATERIALS AND METHODS
Site description, sample collection, and environmental factor analyses.
The physical condition of the Jiaozhou Bay sediments and water column has been described previously in detail (12-14). In summary, limited water exchange and numerous sources of nutrient input from surrounding wastewater treatment plants (WWTPs), rivers, and intertidal and neritic mariculture fields made parts of the Jiaozhou Bay hypernutrified (see Fig. S1 in the supplemental material). Three WWTPs, Tuandao, Haibo, and Licun, are located on the eastern coast of Jiaozhou Bay. In addition, the rivers Haibo, Licun, and Lousan on the eastern coast serve as the conduits for industrial and domestic wastewater from Qingdao city, the rivers Baisha and Moshui carry mariculture wastewater from the north, and the rivers Dagu and Yang discharge agricultural fertilizer-rich freshwater from the west (76). A national crude oil reserve base with the capability of stockpiling more than 3 million tons of crude oil, a large oil refining project with a designed processing capacity of 10 million tons per year, and a sizeable shipping dock are located in the Huangdao area of the western coast of Jiaozhou Bay (see Fig. S1).
With its location in the temperate zone, Jiaozhou Bay sediment microbial processes may be the highest in summer. Using a stainless-steel 0.05-m2 Gray O'Hara box corer, sediment samples were collected from 8 stations in summer 2006 (see Fig. S1 in the supplemental material) as described in previous publications (12, 13), which also detailed the procedures of environmental analyses (see Table S1). Replicate surface sediment subcore samples up to a depth of 5 cm with an approximate 20-cm distance from each other inside the box core were taken (12). Black anoxic sediments were found at the deep layer in all the subcore samples, indicating a suitable environment for anammox in the sediments of our samples.
DNA extraction and anammox bacterial 16S rRNA and hzo gene analyses.
DNA was extracted from sediment samples as reported previously (12, 19). Replicate DNA extractions from three separate subcore samples were pooled for each sampling station. Anammox bacterial 16S rRNA and hzo gene fragments were amplified using published PCR protocols with the primer pairs Brod541F and Brod1260R (51) and hzoF1 and hzoR1 (43), derived from the primers Ana-hzo1F and Ana-hzo2R (53), respectively, with the latter specifically targeting cluster 1 hzo sequences (57). To test reproducibility of our experimental procedure and to identify any potential small-scale (∼20 cm) spatial variability of the sediment anammox bacterial community, two separate 16S rRNA gene clone libraries (D1-I and D1-II) were constructed for station D1 and two separate hzo clone libraries (A3-I and A3-II) for station A3, each from a distinct subcore DNA sample. PCR products from 5 or more reactions were pooled to minimize PCR bias, gel purified, ligated into pMD18-T or pMD19-T Simple vectors (Takara, Tokyo, Japan), and used to transform competent Escherichia coli TOP10 cells (16, 19). Plasmid insert-positive recombinants were selected using 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal)-isopropyl-β-d-thiogalactopyranoside (IPTG) LB indicator plates amended with 100 μg/ml ampicillin. A miniprep method was used to isolate insert-positive plasmids (11), and the cloning vector primers RV-M and M13-D were used to reamplify the cloned DNA fragments (16, 19). The resulting PCR products were screened for correct size and purity by electrophoresis using 1% agarose gels.
Amplicons of correct size obtained from 16S rRNA gene fragments were digested separately with the MspI, HhaI, and TaqI endonucleases (Fermentas, Glen Burnie, MD). Amplicons of the correct size obtained from hzo gene fragments were digested with the MspI and HhaI endonucleases. Restriction fragments were resolved by electrophoresis on 4% agarose gels in 0.5× Tris-borate-EDTA (TBE) buffer. Band patterns digitally photographed with an ImageMaster VDS (Pharmacia Biotech, Piscataway, NJ) or AlphaImager HP (Alpha Innotech, Santa Clara, CA) imaging system were compared for restriction fragment length polymorphisms (RFLP) to identify redundant clones.
The primers RV-M and M13-D were used for sequencing plasmid inserts using an ABI 3770 automatic sequencer (Applied Biosystems, Foster City, CA). DNA sequences were checked for possible chimera with software programs CHIMERA_CHECK (7), Bellerophon (26), and Pintail (4). The BLAST program was used for retrieval of the top-hit 16S rRNA gene or Hzo protein sequences from GenBank (1). The 16S rRNA gene sequences were grouped into operational taxonomic units (OTUs) with 0.5% distance cutoff, and the Hzo sequences were grouped with 1% distance cutoff using the DOTUR software program (56). The small distance cutoffs were used because the potential for microdiversity among environmental anammox bacteria has been reported recently (75). Phylogenetic analyses followed previously reported procedures (16, 19).
Quantification of the sediment 16S rRNA and hzo genes.
Plasmids carrying a bacterial 16S rRNA gene fragment constructed previously (16) and a “Ca. Scalindua” 16S rRNA or hzo gene fragment constructed in this study were extracted from E. coli hosts using a plasmid minikit (Qiagen, Valencia, CA). Concentrations of plasmid DNAs and sediment genomic DNAs were measured using PicoGreen (Molecular Probes, Eugene, OR) and a Modulus single-tube multimode-reader fluorometer (Turner Biosystems, Sunnyvale, CA).
All real-time fluorescence quantitative PCR (qPCR) assays targeting the 16S rRNA or hzo genes were carried out in triplicate with an ABI Prism 7500 sequence detection system (Applied Biosystems, Foster City, CA) using the SYBR green qPCR method (13, 18, 52). The 25-μl qPCR mixture contained the following: 12.5 μl of 2× SYBR green Premix II (TaKaRa, Tokyo, Japan), 0.5 μM final concentration of each primer for the total bacterial 16S rRNA genes (0.2 μM final concentration of each primer for the “Ca. Scalindua” 16S rRNA or hzo genes), 0.5 μl Rox reference dye II, and 20 ng template environmental DNA. To increase specificity, 50 mM KCl (final concentration) and 0.8 μg/μl bovine serum albumin (BSA) (final concentration) were added to the qPCR mixture for quantification of the sediment total bacterial 16S rRNA genes, and 0.8 μg/μl BSA (final concentration) was added to the qPCR mixture for quantification of the sediment anammox bacterial hzo genes. All reactions were carried out in 8-strip thin-well PCR tubes with ultraclear cap strips (ABgene, Fpsom, United Kingdom). Agarose gel electrophoresis and melting-curve analysis were routinely employed to confirm specificities of the qPCRs. Standard curves were obtained with serial dilution of standard plasmids containing target 16S rRNA or hzo gene fragments as the insert. The abundance of standard plasmid inserts ranged from 1.02 × 106 to 1.02 × 1012 (bacterial 16S rRNA gene), 3.89 × 10 to 3.89 × 106 (“Ca. Scalindua” 16S rRNA gene), or 2.70 × 102 to 2.70 × 107 (hzo gene). In all experiments, negative controls containing no template DNA were subjected to the same qPCR procedure to exclude or detect any possible contamination or carryover.
Sediment bacterial 16S rRNA genes were quantified using the primers 341F and 518R (63), “Ca. Scalindua” 16S rRNA genes were quantified using the primers Brod541F and Brod1260R (28, 51), and anammox bacterial hzo genes were quantified using the primers hzoF1 and hzoR1 (43). The qPCR thermocycling parameters were as follows: an initial denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 94°C for 5 s, primer annealing at 60°C for 20 s for bacterial 16S rRNA genes (61°C for “Ca. Scalindua” 16S rRNA genes and 56°C for hzo genes), and elongation at 72°C for 45 s for bacterial or “Ca. Scalindua” 16S rRNA genes (60 s for hzo genes). Melting curves were obtained at 60°C to 95°C, with a read every 1°C held for 1 s between reads. Fluorescence reads for bacterial or “Ca. Scalindua” 16S rRNA genes were carried out at 72°C and those for hzo genes at 80°C to avoid detection of nonspecific PCR products (6). Data were analyzed with the second-derivative maximum method using the ABI PRISM 7500 SDS software (version 1.4; Applied Biosystems).
Statistical analyses.
Environmental classification was performed previously (13) via hierarchical clustering using normalized environmental data (i.e., adjusted to 0 mean and 1 standard deviation [SD] via Z transformation). Library coverage was calculated as C = [1 − (n1/N)] × 100, where n1 is the number of unique OTUs and N the total number of clones in a library (16). Indices of diversity (Shannon-Weiner H and Simpson D) and evenness (J) were calculated for each library. Rarefaction analysis and two nonparametric richness estimators, the abundance-based coverage estimator (SACE) and the bias-corrected Chao1 (SChao1), were calculated using DOTUR (56).
Community classification of the sediment anammox bacterial assemblages was performed using the Jackknife environment clustering and principal coordinates analysis (PCoA) of the UniFrac software program (17, 47). Correlations of the sediment anammox bacterial assemblages with environmental factors were explored using canonical correspondence analysis (CCA) as reported elsewhere (12, 13, 18-20).
Data normal distribution (normality) was examined using the statistics software program MINITAB (release 13.32; Minitab Inc., State College, PA). Pearson correlation analyses (significance level α = 0.05) of the abundance of sediment total bacterial 16S rRNA, “Ca. Scalindua” 16S rRNA, or anammox bacterial hzo genes with environmental factors were also performed using MINITAB (18).
Nucleotide sequence accession numbers.
The determined partial 16S rRNA gene sequences have been deposited in GenBank under accession numbers FJ668672 to FJ668707 and GU433661 to GU433695 and the partial hzo gene sequences under accession numbers GU433696 to GU433880.
RESULTS
Jiaozhou Bay environmental conditions.
Hypernutrification is one of the most notorious environmental problems of Jiaozhou Bay. This situation is aggravated by limited water exchange with the adjacent Yellow Sea, especially at the innermost stations A3, A5, and Y1 (see Fig. S1 in the supplemental material). High concentrations of dissolved inorganic nitrogen, especially NH4+-N, were evident at all the stations (see Table S1). Based on multivariate clustering of physicochemical factors, a previous study (13) identified two types of environmental conditions in Jiaozhou Bay: stations A5, C4, and Y1 of the eastern area established type 1, and the remaining stations belonged to type 2 (see Table S1). This classification was consistent with findings of other studies (12-15, 21, 45, 61, 71) that identified the eastern part of the bay as the most hypernutrified and polluted area, which is highly likely to be caused by river runoff and WWTP discharges from nearby Qingdao city (see Fig. S1). This uneven distribution of the various sources and intensities of eutrophication and pollution was suspected of also having an impact on the anammox microbiota in Jiaozhou Bay.
Diversity of anammox bacterial 16S rRNA and hzo gene sequences.
The two “Ca. Scalindua” 16S rRNA gene clone libraries (D1-I and D1-II) constructed from separate sediment subcore samples of station D1 had similar OTU diversities based on rarefaction analysis (see Fig. S2a in the supplemental material). Community classification using UniFrac environmental clustering (see Fig. S3a) and PCoA (see Fig. S4a) also shows that these two clone libraries are highly similar, indicating the reproducibility of our experimental procedures and the negligible within-site variability of the sediment anammox bacterial community probed by using the “Ca. Scalindua” 16S rRNA gene sequences. Thus, these two “Ca. Scalindua” 16S rRNA gene clone libraries were combined into a single D1 library.
Of the 8 “Ca. Scalindua” 16S rRNA gene clone libraries constructed for the Jiaozhou Bay sediment samples, each for a distinct sampling station (see Fig. S1), a total of 962 insert-positive clones were represented in 70 unique RFLP sequences and 41 OTUs. The values of library coverage (C) ranged from 93.8% to 98.9% (see Table S2 in the supplemental material), which together with rarefaction analysis (see Fig. S2a) indicated that the “Ca. Scalindua” anammox bacteria were sufficiently represented in these clone libraries. Station D1 had the highest OTU diversity and station D5 had the lowest diversity based on the majority of the diversity indices (H, 1/D, and J) and the SACE and SChao1 estimators (see Table S2).
The two anammox bacterial hzo clone libraries (A3-I and A3-II) constructed from separate sediment subcore samples of station A3 appeared to be slightly different based on rarefaction analysis (see Fig. S2b in the supplemental material). However, community classification using UniFrac environmental clustering (see Fig. S3b) and PCoA (see Fig. S4b) shows that these two libraries are highly similar. Neither the UniFrac significance (P = 1.000) nor the P-test significance (P = 1.000) statistic showed a significant difference, indicating a negligible within-site variability of the sediment anammox bacterial community probed by using the hzo sequences. Thus, these two hzo libraries were pooled into a single A3 clone library.
Of the 8 anammox bacterial hzo clone libraries constructed for the Jiaozhou Bay sediment samples, each for a distinct sampling station (see Fig. S1 in the supplemental material), a total of 999 insert-positive clones represented 185 unique RFLP sequences and 56 OTUs. The values of library coverage (C) ranged from 90.9% to 99.2% (see Table S2), which, together with rarefaction analysis (see Fig. S2b), indicated that the anammox bacteria were sufficiently represented in these clone libraries. Station B2 had the highest diversity and station Y1 had the lowest diversity of OTUs based on the majority of diversity indices (H, 1/D, and J). However, stations C4 and D1 had the highest richness and station A5 had the lowest richness of OTUs based on the SACE and SChao1 estimators (see Table S2).
Phylogeny of “Ca. Scalindua” 16S rRNA gene sequences.
The obtained 70 distinct “Ca. Scalindua” 16S rRNA gene sequences were 93.9% to 99.9% identical with one other and 98.5% to 100.0% identical to the closest sequence matches in GenBank. All (100.0%) of the top-hit sequences in GenBank were originally retrieved from marine sediments or anoxic seawater (22, 25, 49, 51, 59, 70, 74, 75). Six of our sequences (11 clones) were also closely related to GenBank sequences originally retrieved from freshwater environments (51).
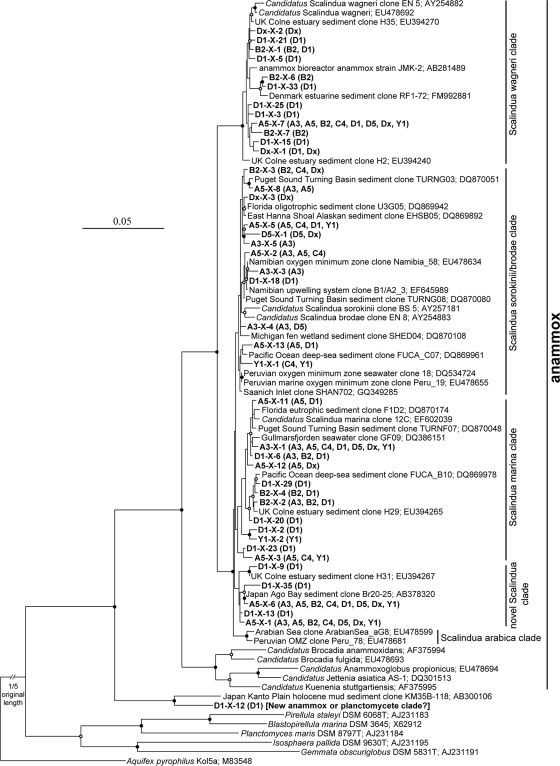
The phylogenetic tree based on the alignment of 16S rRNA gene sequences revealed that Jiaozhou Bay surface sediments harbored a diverse complement of anammox bacteria affiliated with the “Ca. Scalindua” lineage (Fig. 1). Our analysis identified four putative “Ca. Scalindua” clades, including the “Candidatus Scalindua wagneri” (60), “Candidatus Scalindua marina,” and “Candidatus Scalindua sorokinii/brodae” clades (60) and a novel clade of sequences of less than 98% identity with the sequences in other clades (Fig. 1).
FIG. 1.
Phylogenetic analysis of the “Ca. Scalindua” 16S rRNA gene sequences obtained from Jiaozhou Bay sediments. A Clustal_X alignment of environmental and publicly available 16S rRNA gene sequences was subjected to a distance neighbor-joining inference of phylogeny (PHYLIP package, version 3.67). The 16S rRNA gene sequence from Aquifex pyrophilus Kol5a was used as an outgroup. The tree branch distances represent nucleotide substitution rates, and the scale bar represents the expected number of changes per homologous position. Bootstrap values (100 resamplings) higher than 70% are shown with solid circle symbols, and those less than 70% but greater than or equal to 50% are shown with open circle symbols on the corresponding nodes. The “Ca. Scalindua” 16S rRNA gene sequences obtained in this study are shown in bold, along with their distribution in each clone library as depicted in parentheses. The sequence D1-X-12 is clustered outside all known anammox bacterial species, and thus, it might stand for a new cluster of anammox bacteria or just a new cluster of planctomycetes. Due to the uncertainty of its phylogenetic position, D1-X-12 is not included as a valid sequence for further analyses.
Two OTUs, A5-X-6 and A5-X-7, occurred in all 8 “Ca. Scalindua” 16S rRNA gene clone libraries (Fig. 1). Two other OTUs, A3-X-1 and A5-X-1, occurred in 7 of the 8 “Ca. Scalindua” 16S rRNA gene clone libraries (Fig. 1). Although these 4 OTUs accounted for only 9.8% of all “Ca. Scalindua” 16S rRNA gene OTUs, they accounted for 61.5% of all the clones in our libraries, potentially representing the most abundant and prevalent “Ca. Scalindua” anammox bacteria in Jiaozhou Bay sediments.
Phylogeny of anammox bacterial Hzo protein sequences.
The obtained 185 unique hzo sequences were 70.4% to 99.9% identical with one other and 75.4% to 95.2% identical to the closest-match GenBank sequences. The deduced partial Hzo protein sequences (331 amino acid residues) were 76.1% to 100.0% identical with one other and 80.7% to 99.1% identical to the closest-match GenBank sequences obtained from several natural and engineered environments, including Hong Kong coastal wetland sediments (43), anammox reactors, and enrichment cultures (53, 62). Some of our Hzo sequences are also closely related to GenBank sequences retrieved from South China Sea deep-sea sediments (43), northeastern Mexico wall biomats in cenote Zacaton, and other environments (57).
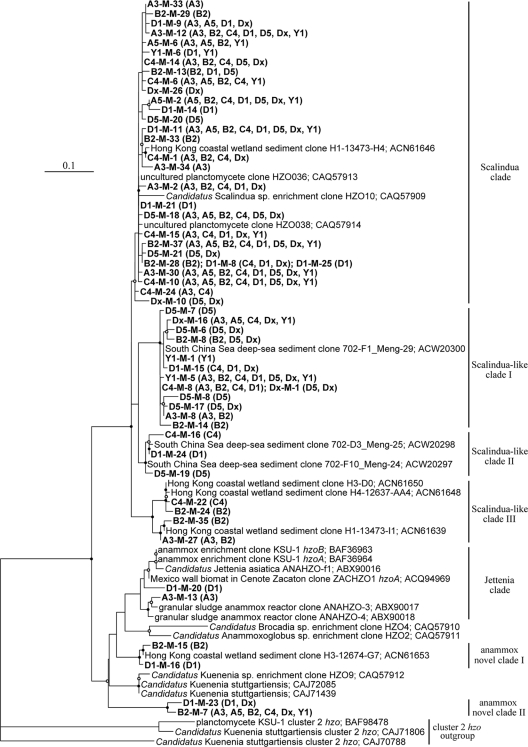
The phylogenetic tree constructed from an alignment of experimental and archived deduced Hzo protein sequences revealed diverse anammox bacteria in the sediments of Jiaozhou Bay (Fig. 2). In addition to the dominant “Ca. Scalindua” lineage, anammox bacteria related to “Ca. Jettenia,” “Ca. Anammoxoglobus,” “Ca. Brocadia,” and “Ca. Kuenenia” lineages were also found to occur in this coastal environment. Overall, this study identified 7 distinct Hzo sequence clades, including the clade of previously reported “Ca. Scalindua” sequences, “Ca. Scalindua”-like clade I, “Ca. Scalindua”-like clade II, “Ca. Scalindua”-like clade III, “Ca. Jettenia” clade, novel anammox clade I, and novel anammox clade II (Fig. 2). Based on DOTUR analysis, the sequences of any two of these clades were less than 90% identical. The known GenBank sequences affiliated within the “Ca. Scalindua” clade were obtained mainly from Hong Kong coastal wetland sediments and other environments (43, 57). The known GenBank sequences affiliated within the “Ca. Scalindua”-like clades I and II were obtained mainly from South China Sea deep-sea sediments (43), and those affiliated within the “Ca. Scalindua”-like clade III were obtained mainly from Hong Kong coastal wetland sediments (43). The known GenBank sequences affiliated within the “Ca. Jettenia” clade were obtained mainly from anammox reactors or enrichment cultures and from northeastern Mexico wall biomats in cenote Zacaton (53, 62). The known GenBank sequences affiliated with the novel anammox clade I were obtained mainly from Hong Kong coastal wetland sediments (43).
FIG. 2.
Phylogenetic consensus tree constructed after neighbor-joining distance analysis (PHYLIP package, version 3.67) of a Clustal_X alignment of publicly available and experimental anammox bacterial Hzo protein sequences deduced from hzo genes recovered from the Jiaozhou Bay sediments. The tree branch distances represent amino acid substitution rates, and the scale bar represents the expected number of changes per homologous position. The three cluster 2 Hzo sequences (BAF98478, CAJ71806, and CAJ70788) were used as an outgroup. Bootstrap values higher than 70% of 100 resamplings in support of the presented tree are shown with solid circle symbols, and those less than 70% but greater or equal to 50% are shown with open circle symbols on the corresponding nodes. The anammox bacterial Hzo sequences obtained in this study are shown in bold, along with their distribution in each clone library as depicted in parentheses.
Four deduced Hzo protein OTUs (A3-M-30, B2-M-37, C4-M-10, and D1-M-11) occurred in all 8 hzo clone libraries, and 5 OTUs (A3-M-12, A5-M-2, B2-M-7, D5-M-18, and Y1-M-5) occurred in 7 or 6 of the 8 hzo clone libraries. Although these 9 sequences accounted for only 16.1% of all the OTUs, they accounted for 65.1% of all the clones in our hzo clone libraries, potentially representing the most abundant and prevalent anammox bacteria in the Jiaozhou Bay sediments.
Anammox bacterial community classification and spatial distribution.
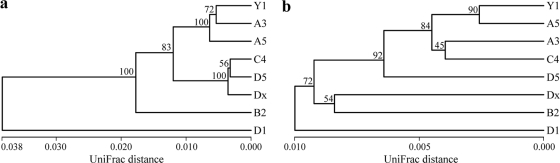
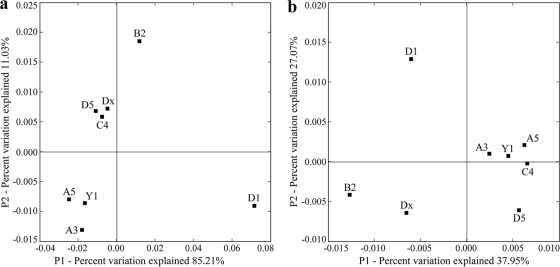
Weighted UniFrac environmental clustering analysis using the obtained 16S rRNA gene sequences indicated that several clusters of “Ca. Scalindua” anammox bacterial assemblages could be identified (Fig. 3a). The station D1 “Ca. Scalindua” assemblage appears to represent a distinct cluster, and the remaining stations seem to represent another cluster with 3 subclusters. Weighted UniFrac PCoA analysis confirmed this classification (Fig. 4a ). The “Ca. Scalindua” assemblage of station D1 could be separated from those of the other stations along the first PCoA principal coordinate (P1), which explained 80.8% of the total “Ca. Scalindua” anammox bacterial community variability among all the sampling stations (Fig. 4a). P1 and P2 (the second PCoA principal coordinate) together classified the “Ca. Scalindua” assemblages of the remaining 7 stations into 3 subclusters (Fig. 4a).
FIG. 3.
Dendrogram of the hierarchical clustering analysis of the Jiaozhou Bay sediment anammox bacterial assemblages as revealed by the “Ca. Scalindua” 16S rRNA gene sequences (a) or the Hzo protein sequences (b), constructed by using the UniFrac normalized and weighted jackknife environment clusters statistical method. The percentage supports of the classification tested with sequence jackknifing resamplings are shown near the corresponding nodes.
FIG. 4.
Ordination diagrams of the UniFrac weighted and normalized PCoA analyses of the Jiaozhou Bay sediment anammox bacterial assemblages as revealed by the “Ca. Scalindua” 16S rRNA gene sequences (a) or the Hzo protein sequences (b). Shown are the plots of the first two principal coordinate axes (P1 and P2) for PCoA and the distributions of the “Ca. Scalindua” 16S rRNA gene-typic assemblages (a) or the anammox bacterial Hzo protein-typic assemblages (designated with the sampling station names) (b) in response to these axes.
Weighted UniFrac environmental clustering analysis using the obtained Hzo protein sequences indicated the existence of several clusters of the total anammox bacterial assemblages (Fig. 3b). The station D1 anammox bacterial assemblage represented a distinct cluster, and the remaining stations comprised another cluster with several subclusters. Weighted UniFrac PCoA analysis further confirmed this classification (Fig. 4b). The anammox bacterial assemblage of station D1 could be separated from those of the other stations along the first and second PCoA principal coordinates (P1 and P2), which together explained 65.02% of the total anammox bacterial community variability among all the sampling stations (Fig. 4b).
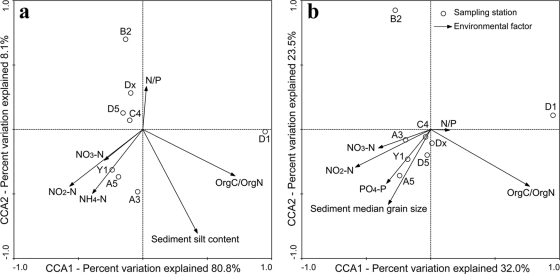
For the weighted CCA analysis using the “Ca. Scalindua” 16S rRNA gene sequences (Fig. 5a), the first two CCA axes (CCA1 and CCA2) explained 84.8% of the total variance in the anammox bacterial composition and 88.9% of the cumulative variance of the anammox bacterium-environment relationship. CCA1, which explained 80.8% of the cumulative variance of the anammox bacterium-environment relationship, clearly distinguished the assemblage of station D1 from those of the other stations (Fig. 5a). Of all the environmental factors analyzed, organic C/organic N (OrgC/OrgN) contributed significantly (P = 0.0260; 1,000 Monte Carlo permutations) to the anammox bacterium-environment relationship, which distinguished the anammox bacterial assemblage of station D1 from those of the other stations (Fig. 5a). The environmental factor NO2−-N also contributed to the anammox bacterium-environment relationship in Jiaozhou Bay, though with a lesser significance (P = 0.0929). These two environmental factors together provided 61.47% of the total CCA explanatory power. Although none of the other environmental factors shown in the CCA graph (Fig. 5a) contributed significantly (P > 0.200) to the anammox bacterium-environment relationship, the combination of these environmental factors provided additionally 33.94% of the total CCA explanatory power.
FIG. 5.
CCA ordination plots for the first two principal dimensions of the relationship between the sediment and porewater environmental parameters in Jiaozhou Bay and the distribution of the sediment anammox bacterial assemblages as reviewed by assessing the “Ca. Scalindua” 16S rRNA gene OTUs (a) or the anammox bacterial Hzo OTUs (b). Correlations between environmental variables and CCA axes are represented by the length and angle of arrows (environmental factor vectors). Due to the possible diel variation of seawater temperature, DO, and pH during our sampling period, these parameters were not included in the analyses. Covarying variables, such as NH4+ and dissolved inorganic nitrogen (DIN) (r = 0.9969) and OrgC and OrgN (r = 0.9826), were checked to minimize collinearity in the CCA analyses.
For the weighted CCA analysis using the Hzo protein sequences (Fig. 5b), the first two CCA axes (CCA1 and CCA2) explained 52.4% of the total variance in the anammox bacterial composition and 55.5% of the cumulative variance of the anammox bacterium-environment relationship. CCA1, which explained 32.0% of the cumulative variance of the anammox bacterium-environment relationship, clearly distinguished the anammox bacterial assemblage of station D1 from those of the other stations (Fig. 5b). CCA2, which explained 23.5% of the cumulative variance of the anammox bacterium-environment relationship, distinguished the anammox bacterial assemblage of station B2 from those of the other stations (Fig. 5b). Of all the environmental factors analyzed, OrgC/OrgN contributed significantly (P = 0.0310) to the anammox bacterium-environment relationship, which distinguished the anammox bacterial assemblage of station D1 and that of station B2 from those of the other stations (Fig. 5b). The environmental factor, sediment median grain size, also contributed to the anammox bacterium-environment relationship in Jiaozhou Bay, although with a lesser significance (P = 0.0739). These two environmental factors together provided 44.63% of the total CCA explanatory power. While none of the other environmental factors shown in the CCA graph (Fig. 5b) contributed significantly (P > 0.100) to the anammox bacterium-environment relationship, the combination of these environmental factors provided additionally 49.59% of the total CCA explanatory power.
Quantification of sediment anammox bacterial assemblages.
Melting-curve analyses of the total bacterial 16S rRNA, “Ca. Scalindua” 16S rRNA, and anammox bacterial hzo genes confirmed that the fluorescent signals were obtained from specific PCR products by our qPCR quantifications. Standard curves generated using plasmids containing cloned 16S rRNA or hzo gene fragments to relate the threshold cycle (Ct) to gene copy number revealed linearity (r2 ≥ 0.98) over 6 orders of magnitude of the standard plasmid DNA concentration (see Table S3 in the supplemental material). The obtained high correlation coefficients and similar slopes indicated high primer hybridization and extension efficiencies (see Table S3), making comparison of gene abundances reasonable (13, 18). The absence or undetectable influence of sediment PCR-inhibitory substances was confirmed by obtaining similar amplification efficiencies with 10-fold-diluted environmental DNAs extracted from the sediments of station A5 (data not show).
The qPCR results showed a heterogeneous distribution of the sediment total bacterial 16S rRNA gene abundance among the sampling sites of Jiaozhou Bay, where station Y1 had the highest gene abundance (2.8 × 1011 copies g−1 sediments) and station D1 had the lowest gene abundance (1.4 × 1010 copies g−1 sediments) (Table 1). The abundance of the sediment “Ca. Scalindua” 16S rRNA genes also showed a distributional heterogeneity, where station C4 had the highest abundance (8.7 × 105 copies g−1 sediments) and station D1 had the lowest abundance (2.0 × 104 copies g−1 sediments) (Table 1). The abundance of the sediment anammox bacterial hzo genes also showed a distributional heterogeneity, where station Dx had the highest abundance (5.9 × 106 copies g−1 sediments) and station D1 had the lowest abundance (3.5 × 105 copies g−1 sediments) (Table 1).
TABLE 1.
Abundance of total bacterial and “Ca. Scalindua” 16S rRNA genes and anammox bacterial hzo genes in sediments of the eight sampling stations of the Jiaozhou Bay
| Sampling station | No. of target gene copies g−1 sedimentsa |
Ratio |
||||
|---|---|---|---|---|---|---|
| Total bacterial 16S rRNA | “Ca. Scalindua” 16S rRNA | Anammox bacterial hzo | “Ca. Scalindua” 16S rRNA/bacterial 16S rRNA (×10−6) | Anammox bacterial hzo/bacterial 16S rRNA (×10−6) | Anammox bacterial hzo/“Ca. Scalindua” 16S rRNA | |
| A3 | 1.4 × 1011 (2.5 × 1010) | 2.8 × 105 (2.0 × 104) | 1.4 × 106 (8.7 × 104) | 2.0 | 10.0 | 5.0 |
| A5 | 1.0 × 1011 (1.3 × 1010) | 4.1 × 105 (4.5 × 104) | 1.9 × 106 (1.0 × 105) | 4.1 | 19.0 | 4.6 |
| B2 | 4.2 × 1010 (1.7 × 109) | 1.4 × 105 (8.0 × 103) | 1.8 × 106 (1.5 × 105) | 3.3 | 42.9 | 12.9 |
| C4 | 1.0 × 1011 (1.3 × 1010) | 8.7 × 105 (2.3 × 104) | 2.1 × 106 (1.9 × 105) | 8.7 | 21.0 | 2.4 |
| D1 | 1.4 × 1010 (2.0 × 109) | 2.0 × 104 (8.7 × 102) | 3.5 × 105 (3.9 × 104) | 1.4 | 25.0 | 17.5 |
| D5 | 5.8 × 1010 (1.5 × 109) | 1.8 × 105 (5.1 × 103) | 6.7 × 105 (4.0 × 104) | 3.1 | 11.6 | 3.7 |
| Dx | 2.0 × 1011 (5.4 × 1010) | 1.3 × 105 (1.3 × 104) | 5.9 × 106 (5.9 × 105) | 0.7 | 29.5 | 45.4 |
| Y1 | 2.8 × 1011 (2.6 × 1010) | 3.8 × 105 (2.1 × 104) | 2.7 × 106 (2.7 × 105) | 1.4 | 9.4 | 7.1 |
Standard errors are indicated in parentheses.
The ratio of sediment anammox bacteria hzo genes to “Ca. Scalindua” 16S rRNA genes ranged from 2.4:1 to 45.4:1 in Jiaozhou Bay, where station Dx had the highest ratio (45.4:1) and station C4 had the lowest ratio (2.4:1).
Pearson correlation analyses (see Table S4 in the supplemental material) indicated that the abundance of the sediment total bacterial 16S rRNA genes was strongly (P < 0.01) correlated with sediment porewater PO43−-P and moderately (P < 0.05) correlated positively with sediment OrgN and porewater NH4+-N and negatively with sediment porewater dissolved oxygen (DO). The abundance of the sediment “Ca. Scalindua” 16S rRNA genes was strongly correlated with sediment porewater NO2−-N. The abundance of the sediment anammox bacterial hzo genes did not show any significant correlation with any environmental factor measured. The relative abundance of sediment anammox bacteria, revealed as the ratio of “Ca. Scalindua” 16S rRNA gene abundance (or anammox bacterial hzo gene abundance) and total bacterial 16S rRNA gene abundance, did not show any significant correlation with any environmental factor measured. The ratio of sediment anammox bacterial hzo gene abundance to “Ca. Scalindua” 16S rRNA gene abundance was moderately negatively correlated with sediment porewater NO2−-N (see Table S4).
DISCUSSION
Anammox bacterial diversity in coastal marine environments.
Anammox bacteria are ubiquitous in O2-limited environments (see references 5, 29, 32, and 50 and references therein). Most of the 16S rRNA gene-based studies indicate that the diversity of anammox bacteria in marine environments is quite low, with domination by strains in the “Ca. Scalindua” lineage (22, 25, 30, 34, 40, 51, 54, 59, 75). There is speculation that the detected low diversity may be an experimental artifact due to low or biased PCR primer coverage and specificity (2, 8, 43). In contrast, two more-recent studies suggested that anammox bacteria from other lineages in addition to “Ca. Scalindua” can also be detected in marine environments by using novel PCR strategies or complex PCR primer sets (2, 8). A systematic primer evaluation study indicates that most of the PCR primers targeting the 16S rRNA genes do not cover all the known anammox bacterial lineages and that hzo might provide a better target to improve coverage (43). The hzo gene-based approach indeed detected diverse anammox bacterial lineages from Jiaozhou Bay, while the “Ca. Scalindua” 16S rRNA gene primers confirm the previously found high microdiversity of the “Ca. Scalindua” anammox bacteria in marine environments (75). Our hzo gene primers provide a simpler PCR method to target the major anammox bacterial lineages than the 16S rRNA gene primers used in previous studies (2, 8).
Diverse anammox bacterial phylotypes were found in the sediments of Jiaozhou Bay based on both the 16S rRNA and hzo gene sequences (Fig. 1 and 2). In addition to the common “Ca. Scalindua” guild, anammox bacteria affiliated more or less closely to the “Ca. Jettenia,” “Ca. Brocadia,” “Ca. Anammoxoglobus,” and “Ca. Kuenenia” guilds were also detected from the sediments of Jiaozhou Bay (Fig. 2). Similarly, non-“Ca. Scalindua” anammox bacteria were detected in coastal marine sediments of Japan; however, no anammox activities were detected (2), suggesting that the detected DNA might have originated from dormant or dead anammox bacteria. Anammox bacteria related to the “Ca. Brocadia” and “Ca. Kuenenia” lineages were also found at low-salinity sites in the Cape Fear River estuary, where a nearby WWTP might provide the actual source of these bacteria found in estuarine sediments (8).
Anammox bacteria in the “Ca. Jettenia,” “Ca. Brocadia,” “Ca. Anammoxoglobus,” and “Ca. Kuenenia” lineages are usually associated with wastewater processing systems, freshwater, or diverse terrestrial environments (27, 31-33, 77). It is possible that some anammox bacteria are more resistant to stressful and adverse environmental changes than others (5), which may explain that anammox bacteria belonging to predominantly nonmarine cohorts can survive in marine environments at low numbers without detectable anammox activity (2). Our findings confirm those of other studies (2, 8) that detected anammox bacteria in the “Ca. Jettenia,” “Ca. Brocadia,” “Ca. Anammoxoglobus,” and “Ca. Kuenenia” lineages in coastal marine sediments, where they can exist either autochthonously (residential to the coastal environments) or allochthonously (transported to the coastal environments through river runoffs and wastewater effluents) (9). “Ca. Scalindua wagneri” was originally discovered in a WWTP (60); however, coastal sediment bacteria affiliated with this clade appear to be the main cohort of active anammox bacteria in the Japan Yodo River estuary (2), indicating that “Ca. Scalindua wagneri”-like bacteria may have a broad niche adaptation in both manmade and natural environments.
In addition to several novel subclusters in the “Ca. Scalindua” lineage (Fig. 1), our current study also identified a few novel clusters related to the rarely detected non-“Ca. Scalindua” lineages in marine environments (Fig. 2). One of these clusters, the anammox bacteria novel clade II, appears to be unique to the marine sediments of Jiaozhou Bay, or it was not yet detected anywhere else. This finding suggests that the diversity of environmental anammox bacteria may have been underestimated in previous studies.
Anammox bacterial ecophysiology in response to changes in coastal marine environments.
Quantification of the “Ca. Scalindua” 16S rRNA and anammox bacterial hzo genes indicates that the hzo gene abundance was higher than the abundance of “Ca. Scalindua” anammox bacterial 16S rRNA genes (Table 1). This is reasonable, since (i) the hzo gene primers have a broader coverage for the anammox bacteria than the 16S rRNA gene primers, which target only the “Ca. Scalindua” group, and (ii) there may be multiple hzo gene copies but only one 16S rRNA gene copy in anammox bacteria, as shown by the sequenced genome of “Candidatus Kuenenia stuttgartiensis” (65). In light of these considerations, our data suggest that the “Ca. Scalindua” lineage bacteria may account for the majority of the sediment anammox bacteria at some of the Jiaozhou Bay sampling sites (Table 1).
Anammox activity requires the simultaneous presence of ammonium and nitrite, which may be found at or near the aerobic-anaerobic interface of sediments (37). NO2−-N is often limiting in eutrophic coastal sediments with excess NH4+-N and organic matter (48). Our environmental geochemical analyses indicate that NH4+-N is rarely limiting in the Jiaozhou Bay sediments (see Table S1 in the supplemental material) and that availability of NO2−-N most likely is the limiting factor that controls the abundance and activity of the in situ anammox bacteria (48, 55). Indeed, a strong positive correlation of the sediment anammox bacterial abundance as revealed by the “Ca. Scalindua” 16S rRNA gene abundance with sediment porewater NO2−-N is found (see Table S4). A previous study found a strong correlation between “Ca. Scalindua” abundance and anammox activity in the African coast Benguela upwelling system (40). Thus, the anammox activity might also be controlled by sediment NO2−-N in the eutrophic sediments of Jiaozhou Bay, since the in situ “Ca. Scalindua” anammox bacteria might actively consume nitrite and their population size and anammox activity might be regulated by sediment NO2−-N. Strong correlations between anammox potential and in situ availability of NO2−-N were also found in other estuarine sediments (48, 68, 69), indicating NO2−-N to be a key regulator of anammox in these environments. The correlation of NO2−-N with the “Ca. Scalindua” anammox bacterial assemblages (Fig. 5a) indicates that the composition, community structure, and distribution of the sediment anammox bacterial assemblages in Jiaozhou Bay might also be regulated by sediment porewater NO2−-N. These data indicate that the “Ca. Scalindua” lineage bacteria may be not only the major component of the sediment anammox microbiota but also the one responsible for the in situ anammox activity, regulated by sediment porewater NO2−-N. These data also imply that the “Ca. Scalindua” lineage bacteria might control sediment porewater NO2−-N levels by actively consuming it.
Both the UniFrac environmental clustering and PCoA analyses revealed considerable heterogeneity of the sediment anammox bacterial assemblages in Jiaozhou Bay (Fig. 3 and 4). Analyses of both target genes (16S rRNA and hzo) indicate that the anammox bacterial assemblage of station D1 is distinct from those of the other stations. CCA analyses indicate that the environmental factor OrgC/OrgN might contribute significantly to the uniqueness of the anammox bacterial assemblage at station D1 (Fig. 5). An anticlockwise eddy was found in the north of Huangdao at station D1 (46), which might make the in situ hydrological conditions more stagnant than those of the other nearby stations in Jiaozhou Bay. This may reduce material transport and water exchange between station D1 with the Yellow Sea outside Jiaozhou Bay. Station D1 also had the highest sediment OrgC/OrgN in Jiaozhou Bay (see Table S1 in the supplemental material). High OrgC/OrgN is usually associated with old or recalcitrant organic matter and may serve as a signature of long organic matter detention or terrestrial input intensity (38). The high OrgC/OrgN might also be caused by oil contamination from the nearby crude oil refining plants on the western coast of Jiaozhou Bay (see Fig. S1). Thus, the unique sediment anammox bacterial assemblage of station D1 (Fig. 3, 4, and 5) detected in the current study might be a result of a unique in situ hydrological, geochemical, or pollution condition. This is consistent with a previous study that also identified station D1 as unique in regard to its nirS-encoding denitrifying bacterial assemblage in the sediments of Jiaozhou Bay (12).
One of the sedimentological variables with a significant impact on anammox bacterial community structure, sediment median grain size (Fig. 5b), may depend on in situ hydrological conditions, such as currents, tides, waves, upwelling, lateral transport, water mixing, and the intensity and dynamics of these activities. These hydrological activities influence sediment composition, organic matter content, O2 content, porewater redox state, and nutrient composition and concentration (19) and thus the sediment anammox microbiota. This is in agreement with recent reports on the influence of sedimentological parameters on the structure and distribution of bacterial and archaeal assemblages implicated in sediment N cycling (12, 13, 19).
Our current study identified NO2−-N and a few other geochemical and sedimentological factors, such as sediment OrgC/OrgN and median grain size, as potentially the key regulators of the sediment anammox bacterial composition, community structure, abundance, distribution, or potential activity in the eutrophic Jiaozhou Bay. Our previous studies also identified porewater NO2−-N as contributing significantly to the composition, structure, and distribution of the sediment ammonia-oxidizing Betaproteobacteria community (13) and sediment OrgC/OrgN as contributing significantly to the community structure and spatial distribution of the nirS-encoding denitrifying bacterial assemblages in Jiaozhou Bay (12). N cycling in eutrophic coastal sediments is very complicated. Nitrate reduction as part of denitrification and ammonification processes, as well as nitrification, may contribute nitrite at different levels for anammox, depending on the specific environmental conditions and the availability of N compounds (41, 42, 55). Competition or collaboration for nitrite may occur between anammox and denitrification (55). The in situ nitrite supply or consumption may also be influenced by hydrological and geochemical processes at the seawater-sediment interface and the sediment oxic-suboxic-anoxic interfaces. The relative contribution of the various nitrite production or consumption processes is still poorly understood and needs to be further investigated in this coastal bay. Our previous studies indicate that the Jiaozhou Bay innermost stations A5 and Y1 were severely eutrophic and polluted and that stations A3 and C4 were also eutrophic and polluted to a certain degree (12-15, 71). Our current study correlates these findings with the fact that these sites, more or less, have distinct anammox bacterial assemblages (Fig. 3, 4, and 5). Our results suggest that anthropogenic activities may have an impact on the coastal anammox microbiota and activity in Jiaozhou Bay.
Supplementary Material
Acknowledgments
We thank Linbao Zhang, Ting Zhu, Xiaomin Yin, Jun Zuo, Dong Jiang, Jin He, Xinwei Du, Daigang Yang, and Wei Jin for their assistance in the project and Ying Ye for valuable discussion.
This work was supported by China Ocean Mineral Resources R&D Association grants DYXM-115-02-2-20 and DYXM-115-02-2-6, Hi-Tech Research and Development Program of China grant 2007AA091903, China National Natural Science Foundation grants 40576069 and 41076091, National Basic Research Program of China grant 2009CB219506, Key Scientific and Technological Development Program of the National Qingdao Economic & Technical Development Zone (Huangdao Area) grant 2009-2-34, Fundamental Research Funds for the Central Universities of China grant 09CX05005A, and Foundation of the State Key Laboratory of Heavy Oil Processing, China University of Petroleum grant SKL2010-02. M.G.K. was funded by incentive funds provided by the UofL-EVPR office and the U.S. National Science Foundation (EF-0412129).
Footnotes
Published ahead of print on 10 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano, T., I. Yoshinaga, K. Okada, T. Yamagishi, and S. Ueda. 2007. Detection of anammox activity and diversity of anammox bacteria-related 16S rRNA genes in coastal marine sediments in Japan. Microbes Environ. 22:232-242. [Google Scholar]
- 3.Arrigo, K. R. 2005. Marine microorganisms and global nutrient cycles. Nature 437:349-355. [DOI] [PubMed] [Google Scholar]
- 4.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandes, J. A., A. H. Devol, and C. Deutsch. 2007. New developments in the marine nitrogen cycle. Chem. Rev. 107:577-589. [DOI] [PubMed] [Google Scholar]
- 6.Caffrey, J. M., N. Bano, K. Kalanetra, and J. T. Hollibaugh. 2007. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J. 1:660-662. [DOI] [PubMed] [Google Scholar]
- 7.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dale, O. R., C. R. Tobias, and B. Song. 2009. Biogeographical distribution of diverse anaerobic ammonium oxidizing (anammox) bacteria in Cape Fear River Estuary. Environ. Microbiol. 11:1194-1207. [DOI] [PubMed] [Google Scholar]
- 9.Dalsgaard, T., B. Thamdrup, and D. E. Canfield. 2005. Anaerobic ammonium oxidation (anammox) in the marine environment. Res. Microbiol. 156:457-464. [DOI] [PubMed] [Google Scholar]
- 10.Dalsgaard, T., D. Canfield, J. Petersen, B. Thamdrup, and J. Acuña-González. 2003. N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature 422:606-608. [DOI] [PubMed] [Google Scholar]
- 11.Dang, H. Y., and C. R. Lovell. 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang, H. Y., C. Y. Wang, J. Li, T. G. Li, F. Tian, W. Jin, Y. S. Ding, and Z. N. Zhang. 2009. Diversity and distribution of sediment nirS-encoding bacterial assemblages in response to environmental gradients in the eutrophied Jiaozhou Bay, China. Microb. Ecol. 58:161-169. [DOI] [PubMed] [Google Scholar]
- 13.Dang, H. Y., J. Li, R. P. Chen, L. Wang, L. Z. Guo, Z. N. Zhang, and M. G. Klotz. 2010. Diversity, abundance and spatial distribution of sediment ammonia-oxidizing Betaproteobacteria in response to environmental gradients and coastal eutrophication in Jiaozhou Bay, China. Appl. Environ. Microbiol. 76:4691-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang, H. Y., J. Ren, L. S. Song, S. Sun, and L. G. An. 2008. Diverse tetracycline resistant bacteria and resistance genes from coastal waters of Jiaozhou Bay. Microb. Ecol. 55:237-246. [DOI] [PubMed] [Google Scholar]
- 15.Dang, H. Y., J. Ren, L. S. Song, S. Sun, and L. G. An. 2008. Dominant chloramphenicol-resistant bacteria and resistance genes in coastal marine waters of Jiaozhou Bay, China. World J. Microbiol. Biotechnol. 24:209-217. [Google Scholar]
- 16.Dang, H. Y., T. G. Li, M. N. Chen, and G. Q. Huang. 2008. Cross-ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang, H. Y., X. W. Luan, J. Y. Zhao, and J. Li. 2009. Diverse and novel nifH and nifH-like gene sequences in the deep-sea methane seep sediments of the Okhotsk Sea. Appl. Environ. Microbiol. 75:2238-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang, H. Y., X. W. Luan, R. P. Chen, X. X. Zhang, L. Z. Guo, and M. G. Klotz. 2010. Diversity, abundance and distribution of amoA-encoding archaea in deep-sea methane seep sediments of the Okhotsk Sea. FEMS Microbiol. Ecol. 72:370-385. [DOI] [PubMed] [Google Scholar]
- 19.Dang, H. Y., X. X. Zhang, J. Sun, T. G. Li, Z. N. Zhang, and G. P. Yang. 2008. Diversity and spatial distribution of sediment ammonia-oxidizing Crenarchaeota in response to estuarine and environmental gradients in the Changjiang Estuary and East China Sea. Microbiology 154:2084-2095. [DOI] [PubMed] [Google Scholar]
- 20.Dang, H., J. Li, X. Zhang, T. Li, F. Tian, and W. Jin. 2009. Diversity and spatial distribution of amoA-encoding archaea in the deep-sea sediments of the tropical West Pacific Continental Margin. J. Appl. Microbiol. 106:1482-1493. [DOI] [PubMed] [Google Scholar]
- 21.Deng, B., J. Zhang, G. Zhang, and J. Zhou. 2010. Enhanced anthropogenic heavy metal dispersal from tidal disturbance in the Jiaozhou Bay, North China. Environ. Monit. Assess. 161:349-358. [DOI] [PubMed] [Google Scholar]
- 22.Dong, L. F., C. J. Smith, S. Papaspyrou, A. Stott, A. M. Osborn, and D. B. Nedwell. 2009. Changes in benthic denitrification, nitrate ammonification, and anammox process rates and nitrate and nitrite reductase gene abundances along an estuarine nutrient gradient (the Colne estuary, United Kingdom). Appl. Environ. Microbiol. 75:3171-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engström, P., T. Dalsgaard, S. Hulth, and R. C. Aller. 2005. Anaerobic ammonium oxidation by nitrite (anammox): implications for N2 production in coastal marine sediments. Geochim. Cosmochim. Acta 69:2057-2065. [Google Scholar]
- 24.Francis, C. A., J. M. Beman, and M. M. M. Kuypers. 2007. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1:19-27. [DOI] [PubMed] [Google Scholar]
- 25.Hamersley, M. R., G. Lavik, D. Woebken, J. E. Rattray, P. Lam, E. C. Hopmans, J. S. Sinninghe Damsté, S. Krüger, M. Graco, D. Gutierrez, and M. M. M. Kuypers. 2007. Anaerobic ammonium oxidation in the Peruvian oxygen minimum zone. Limnol. Oceanogr. 52:923-933. [Google Scholar]
- 26.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 27.Humbert, S., S. Tarnawski, N. Fromin, M. P. Mallet, M. Aragno, and J. Zopfi. 2010. Molecular detection of anammox bacteria in terrestrial ecosystems: distribution and diversity. ISME J. 4:450-454. [DOI] [PubMed] [Google Scholar]
- 28.Jaeschke, A., B. Abbas, M. Zabel, E. C. Hopmans, S. Schouten, and J. S. Sinninghe Damsté. 2010. Molecular evidence for anaerobic ammonium-oxidizing (anammox) bacteria in continental shelf and slope sediments off northwest Africa. Limnol. Oceanogr. 55:365-376. [Google Scholar]
- 29.Jaeschke, A., H. J. Op den Camp, H. Harhangi, A. Klimiuk, E. C. Hopmans, M. S. Jetten, S. Schouten, and J. S. Sinninghe Damsté. 2009. 16S rRNA gene and lipid biomarker evidence for anaerobic ammonium-oxidizing bacteria (anammox) in California and Nevada hot springs. FEMS Microbiol. Ecol. 67:343-350. [DOI] [PubMed] [Google Scholar]
- 30.Jayakumar, A., G. D. O'Mullan, S. W. Naqvi, and B. B. Ward. 2009. Denitrifying bacterial community composition changes associated with stages of denitrification in oxygen minimum zones. Microb. Ecol. 58:350-362. [DOI] [PubMed] [Google Scholar]
- 31.Jetten, M. S., I. Cirpus, B. Kartal, L. van Niftrik, K. T. van de Pas-Schoonen, O. Sliekers, S. Haaijer, W. van der Star, M. Schmid, J. van de Vossenberg, I. Schmidt, H. Harhangi, M. van Loosdrecht, J. G. Kuenen, H. Op den Camp, and M. Strous. 2005. 1994-2004: 10 years of research on the anaerobic oxidation of ammonium. Biochem. Soc. Trans. 33:119-123. [DOI] [PubMed] [Google Scholar]
- 32.Jetten, M. S., L. van Niftrik, M. Strous, B. Kartal, J. T. Keltjens, and H. J. Op den Camp. 2009. Biochemistry and molecular biology of anammox bacteria. Crit. Rev. Biochem. Mol. Biol. 44:65-84. [DOI] [PubMed] [Google Scholar]
- 33.Kindaichi, T., I. Tsushima, Y. Ogasawara, M. Shimokawa, N. Ozaki, H. Satoh, and S. Okabe. 2007. In situ activity and spatial organization of anaerobic ammonium-oxidizing (anammox) bacteria in biofilms. Appl. Environ. Microbiol. 73:4931-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirkpatrick, J., B. Oakley, C. Fuchsman, S. Srinivasan, J. T. Staley, and J. W. Murray. 2006. Diversity and distribution of Planctomycetes and related bacteria in the suboxic zone of the Black Sea. Appl. Environ. Microbiol. 72:3079-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klotz, M. G., and L. Y. Stein. 2008. Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol. Lett. 278:146-156. [DOI] [PubMed] [Google Scholar]
- 36.Klotz, M. G., M. C. Schmid, M. Strous, H. J. M. op den Camp, M. S. M. Jetten, and A. B. Hooper. 2008. Evolution of an octahaem cytochrome c protein family that is key to aerobic and anaerobic ammonia oxidation by bacteria. Environ. Microbiol. 10:3150-3163. [DOI] [PubMed] [Google Scholar]
- 37.Kuenen, J. G. 2008. Anammox bacteria: from discovery to application. Nat. Rev. Microbiol. 6:320-326. [DOI] [PubMed] [Google Scholar]
- 38.Kukal, Z. 1971. Geology of recent sediments, p. 490. Academic Press, New York, NY.
- 39.Kuypers, M. M. M., A. O. Sliekers, G. Lavik, M. Schmid, B. B. Jøgensen, J. G. Kuenen, J. S. Sinninghe Damsté, M. Strous, and M. S. M. Jetten. 2003. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422:608-611. [DOI] [PubMed] [Google Scholar]
- 40.Kuypers, M. M. M., G. Lavik, D. Woebken, M. Schmid, B. M. Fuchs, R. Amann, B. B. Jørgensen, and M. S. Jetten. 2005. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc. Natl. Acad. Sci. U. S. A. 102:6478-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam, P., G. Lavik, M. M. Jensen, J. van de Vossenberg, M. Schmid, D. Woebken, D. Gutiérrez, R. Amann, M. S. Jetten, and M. M. Kuypers. 2009. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc. Natl. Acad. Sci. U. S. A. 106:4752-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lam, P., M. M. Jensen, G. Lavik, D. F. McGinnis, B. Muller, C. J. Schubert, R. Amann, B. Thamdrup, and M. M. Kuypers. 2007. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc. Natl. Acad. Sci. U. S. A. 104:7104-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, M., Y. G. Hong, M. G. Klotz, and J. D. Gu. 2010. A comparison of primer sets for detecting 16S rRNA and hydrazine oxidoreductase genes of anaerobic ammonium-oxidizing bacteria in marine sediments. Appl. Microbiol. Biotechnol. 86:781-790. [DOI] [PubMed] [Google Scholar]
- 44.Li, X. R., B. Du, H. X. Fu, R. F. Wang, J. H. Shi, Y. Wang, M. S. Jetten, and Z. X. Quan. 2009. The bacterial diversity in an anaerobic ammonium-oxidizing (anammox) reactor community. Syst. Appl. Microbiol. 32:278-289. [DOI] [PubMed] [Google Scholar]
- 45.Liu, S. M., J. Zhang, H. T. Chen, and G. S. Zhang. 2005. Factors influencing nutrient dynamics in the eutrophic Jiaozhou Bay, North China. Prog. Oceanogr. 66:66-85. [Google Scholar]
- 46.Liu, Z., H. Wei, G. Liu, and J. Zhang. 2004. Simulation of water exchange in Jiaozhou Bay by average residence time approach. Estuar. Coast. Shelf Sci. 61:25-35. [Google Scholar]
- 47.Lozupone, C. A., M. Hamady, S. T. Kelley, and R. Knight. 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73:1576-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer, R. L., N. Risgaard-Petersen, and D. E. Allen. 2005. Correlation between anammox activity and microscale distribution of nitrite in a subtropical mangrove sediment. Appl. Environ. Microbiol. 71:6142-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakajima, J., M. Sakka, T. Kimura, and K. Sakka. 2008. Detection of anaerobic ammonium-oxidizing bacteria in Ago Bay sediments. Biosci. Biotechnol. Biochem. 72:2195-2198. [DOI] [PubMed] [Google Scholar]
- 50.Op den Camp, H. J., B. Kartal, D. Guven, L. A. van Niftrik, S. C. Haaijer, W. R. van der Star, K. T. van de Pas-Schoonen, A. Cabezas, Z. Ying, M. C. Schmid, M. M. Kuypers, J. van de Vossenberg, H. R. Harhangi, C. Picioreanu, M. C. van Loosdrecht, J. G. Kuenen, M. Strous, and M. S. Jetten. 2006. Global impact and application of the anaerobic ammonium-oxidizing (anammox) bacteria. Biochem. Soc. Trans. 34:174-178. [DOI] [PubMed] [Google Scholar]
- 51.Penton, C. R., A. H. Devol, and J. M. Tiedje. 2006. Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl. Environ. Microbiol. 72:6829-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poret-Peterson, A. T., J. E. Graham, J. Gulledge, and M. G. Klotz. 2008. Transcription of nitrification genes by the methane-oxidizing bacterium, Methylococcus capsulatus strain Bath. ISME J. 2:1213-1220. [DOI] [PubMed] [Google Scholar]
- 53.Quan, Z. X., S. K. Rhee, J. E. Zuo, Y. Yang, J. W. Bae, J. R. Park, S. T. Lee, and Y. H. Park. 2008. Diversity of ammonium-oxidizing bacteria in a granular sludge anaerobic ammonium-oxidizing (anammox) reactor. Environ. Microbiol. 10:3130-3139. [DOI] [PubMed] [Google Scholar]
- 54.Risgaard-Petersen, N., R. Meyer, M. Schmid, M. Jetten, A. Enrich-Prast, S. Rysgaard, and N. Revsbech. 2004. Anaerobic ammonium oxidation in an estuarine sediment. Aquat. Microb. Ecol. 36:293-304. [Google Scholar]
- 55.Rysgaard, S., R. N. Glud, N. Risgaard-Petersen, and T. Dalsgaard. 2004. Denitrification and anammox activity in Arctic marine sediments. Limnol. Oceanogr. 49:1493-1502. [Google Scholar]
- 56.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmid, M. C., A. B. Hooper, M. G. Klotz, D. Woebken, P. Lam, M. M. M. Kuypers, A. Pommerening-Roeser, H. J. M. op den Camp, and M. S. M. Jetten. 2008. Environmental detection of octahaem cytochrome c hydroxylamine/hydrazine oxidoreductase genes of aerobic and anaerobic ammonium-oxidizing bacteria. Environ. Microbiol. 10:3140-3149. [DOI] [PubMed] [Google Scholar]
- 58.Schmid, M. C., B. Maas, A. Dapena, K. van de Pas-Schoonen, J. van de Vossenberg, B. Kartal, L. van Niftrik, I. Schmidt, I. Cirpus, J. G. Kuenen, M. Wagner, J. S. Sinninghe Damsté, M. Kuypers, N. P. Revsbech, R. Mendez, M. S. Jetten, and M. Strous. 2005. Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl. Environ. Microbiol. 71:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmid, M. C., N. Risgaard-Petersen, J. van de Vossenberg, M. M. M. Kuypers, G. Lavik, J. Petersen, S. Hulth, B. Thamdrup, D. Canfield, T. Dalsgaard, S. Rysgaard, M. K. Sejr, M. Strous, H. O. den Camp, and M. S. Jetten. 2007. Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ. Microbiol. 9:1476-1484. [DOI] [PubMed] [Google Scholar]
- 60.Schmid, M., K. Walsh, R. Webb, W. I. Rijpstra, K. van de Pas-Schoonen, M. J. Verbruggen, T. Hill, B. Moffett, J. Fuerst, S. Schouten, J. S. Sinninghe Damsté, J. Harris, P. Shaw, M. Jetten, and M. Strous. 2003. Candidatus “Scalindua brodae,” sp. nov., Candidatus “Scalindua wagneri,” sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 26:529-538. [DOI] [PubMed] [Google Scholar]
- 61.Shen, Z. L. 2001. Historical changes in nutrient structure and its influences on phytoplankton composition in Jiaozhou Bay. Estuar. Coast. Shelf Sci. 52:211-224. [Google Scholar]
- 62.Shimamura, M., T. Nishiyama, H. Shigetomo, T. Toyomoto, Y. Kawahara, K. Furukawa, and T. Fujii. 2007. Isolation of a multiheme protein with features of a hydrazine-oxidizing enzyme from an anaerobic ammonium-oxidizing enrichment culture. Appl. Environ. Microbiol. 73:1065-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shinkai, T., and Y. Kobayashi. 2007. Localization of ruminal cellulolytic bacteria on plant fibrous materials as determined by fluorescence in situ hybridization and real-time PCR. Appl. Environ. Microbiol. 73:1646-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strous, M., and M. S. M. Jetten. 2004. Anaerobic oxidation of methane and ammonium. Annu. Rev. Microbiol. 58:99-117. [DOI] [PubMed] [Google Scholar]
- 65.Strous, M., E. Pelletier, S. Mangenot, T. Rattei, A. Lehner, M. W. Taylor, M. Horn, H. Daims, D. Bartol-Mavel, P. Wincker, V. Barbe, N. Fonknechten, D. Vallenet, B. Segurens, C. Schenowitz-Truong, C. Medigue, A. Collingro, B. Snel, B. E. Dutilh, H. J. Op den Camp, C. van der Drift, I. Cirpus, K. T. van de Pas-Schoonen, H. R. Harhangi, L. van Niftrik, M. Schmid, J. Keltjens, J. van de Vossenberg, B. Kartal, H. Meier, D. Frishman, M. A. Huynen, H. W. Mewes, J. Weissenbach, M. S. Jetten, M. Wagner, and D. Le Paslier. 2006. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440:790-794. [DOI] [PubMed] [Google Scholar]
- 66.Strous, M., J. A. Fuerst, E. H. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. Webb, J. G. Kuenen, and M. S. Jetten. 1999. Missing lithotroph identified as new planctomycete. Nature 400:446-449. [DOI] [PubMed] [Google Scholar]
- 67.Thamdrup, B., and T. Dalsgaard. 2002. Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl. Environ. Microbiol. 68:1312-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trimmer, M., J. C. Nicholls, and B. Deflandre. 2003. Anaerobic ammonium oxidation measured in sediments along the Thames estuary, United Kingdom. Appl. Environ. Microbiol. 69:6447-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trimmer, M., J. C. Nicholls, N. Morley, C. A. Davies, and J. Aldridge. 2005. Biphasic behavior of anammox regulated by nitrite and nitrate in an estuarine sediment. Appl. Environ. Microbiol. 71:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walsh, D. A., E. Zaikova, C. L. Howes, Y. C. Song, J. J. Wright, S. G. Tringe, P. D. Tortell, and S. J. Hallam. 2009. Metagenome of a versatile chemolithoautotroph from expanding oxygen minimum zones. Science 326:578-582. [DOI] [PubMed] [Google Scholar]
- 71.Wang, C. Y., H. Y. Dang, and Y. S. Ding. 2008. Incidence of diverse integrons and β-lactamase genes in environmental Enterobacteriaceae isolates from Jiaozhou Bay, China. World J. Microbiol. Biotechnol. 24:2889-2896. [Google Scholar]
- 72.Ward, B. B. 2003. Significance of anaerobic ammonium oxidation in the ocean. Trends Microbiol. 11:408-410. [DOI] [PubMed] [Google Scholar]
- 73.Ward, B. B., A. H. Devol, J. J. Rich, B. X. Chang, S. E. Bulow, H. Naik, A. Pratihary, and A. Jayakumar. 2009. Denitrification as the dominant nitrogen loss process in the Arabian Sea. Nature 461:78-81. [DOI] [PubMed] [Google Scholar]
- 74.Woebken, D., B. M. Fuchs, M. M. Kuypers, and R. Amann. 2007. Potential interactions of particle-associated anammox bacteria with bacterial and archaeal partners in the Namibian upwelling system. Appl. Environ. Microbiol. 73:4648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woebken, D., P. Lam, M. M. Kuypers, S. W. Naqvi, B. Kartal, M. Strous, M. S. Jetten, B. M. Fuchs, and R. Amann. 2008. A microdiversity study of anammox bacteria reveals a novel Candidatus Scalindua phylotype in marine oxygen minimum zones. Environ. Microbiol. 10:3106-3119. [DOI] [PubMed] [Google Scholar]
- 76.Zhang, G. L., J. Zhang, J. Xu, and F. Zhang. 2006. Distributions, sources and atmospheric fluxes of nitrous oxide in Jiaozhou Bay. Estuar. Coast. Shelf Sci. 68:557-566. [Google Scholar]
- 77.Zhang, Y., X. H. Ruan, H. J. Op den Camp, T. J. Smits, M. S. Jetten, and M. C. Schmid. 2007. Diversity and abundance of aerobic and anaerobic ammonium-oxidizing bacteria in freshwater sediments of the Xinyi River (China). Environ. Microbiol. 9:2375-2382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.