Abstract
Recent metagenomic sequencing studies of uncultured viral populations have provided novel insights into the ecology of environmental bacteriophage. At the same time, viral metagenomes could also represent a potential source of recombinant proteins with biotechnological value. In order to identify such proteins, a novel two-step screening technique was devised for cloning phage lytic enzymes from uncultured viral DNA. This plasmid-based approach first involves a primary screen in which transformed Escherichia coli clones that demonstrate colony lysis following exposure to inducing agent are identified. This effect, which can be due to the expression of membrane-permeabilizing phage holins, is discerned by the development a hemolytic effect in surrounding blood agar. In a secondary step, the clones identified in the primary screen are overlaid with autoclaved Gram-negative bacteria (specifically Pseudomonas aeruginosa) to assay directly for recombinant expression of lytic enzymes, which are often encoded proximally to holins in phage genomes. As proof-of-principle, the method was applied to a viral metagenomic library constructed from mixed animal feces, and 26 actively expressed lytic enzymes were cloned. These proteins include both Gram-positive-like and Gram-negative-like enzymes, as well as several atypical lysins whose predicted structures are less common among known phage. Overall, this study represents one of the first functional screens of a viral metagenomic population, and it provides a general approach for characterizing lysins from uncultured phage.
The field of metagenomics has expanded rapidly in recent years, providing access to environmental microorganisms that would remain unapproachable by standard, culture-based methods. The foundation of metagenomics lies in the direct extraction of DNA/RNA from environmental samples (e.g., soil, water, or feces) without prior isolation of individual microbial species (reviewed in references 18 and 32). It has been estimated that only a small proportion of naturally occurring microbes—approximately 1% of soil bacteria, for instance—are culturable under standard laboratory conditions (31). In this light, metagenomics has become an increasingly common tool for studying diverse ecosystems, from around the globe to within the human body.
Overall, metagenomics research can be divided into two general categories: sequence-based and functional. In the former, environmental DNA is sequenced in mass and compared with genetic databases to address broad questions of ecology, taxonomy, and diversity. Some of the most extensive metagenomic studies to date have been sequence based in nature, benefiting from the development of high-throughput sequencing technologies. Notable examples include a 76-megabase study of an acid mine biofilm (33), a 1-gigabase analysis of the Sargasso Sea (35), and a 6.3-gigabase sampling of global oceanic samples (25). In functional metagenomics, by contrast, environmental genes are recombinantly expressed within a host organism, which is monitored for the acquisition of a desired phenotype. Rather than providing insight into entire ecosystems, functional studies aim to identify individual molecules with biomedical or industrial value. Targeted compounds may be either proteins (usually enzymes) encoded directly by environmental genes or small molecules synthesized by several enzymes of a gene cluster. Numerous classes of molecules have been identified to date, with particular interest in the areas of biosynthesis, biomass degradation, and antibiotic discovery (reviewed in references 2, 34, and 36).
While bacteria provide the majority of DNA to most metagenomic pools, recent studies have begun focusing on subsets of total environmental populations. A prominent example is viral metagenomics, in which viral particles (predominately bacteriophage) are purified from cellular material prior to DNA extraction (reviewed in references 10 and 12). Although the yield of DNA from environmental phage isolates is generally low, PCR amplification techniques have been developed to overcome this issue (4, 26). Viral metagenomic analyses have been conducted on a growing number of samples, including ones purified from soil (15), seawater (4, 39), and human feces (3). These studies have revealed a remarkable abundance of novel sequences, supporting the notion that phage represent the largest source of untapped genetic diversity on the planet (19). Despite this wealth of information, viral metagenomic studies to date have remained predominantly sequence based in nature. In this regard, functional screens of viral metagenomes could provide a large source of recombinant molecules.
Recently one class of phage-encoded protein has received particular attention from the biotechnology field: phage lytic enzymes (also referred to as endolysins or lysins) (reviewed in references 16 and 17). These peptidoglycan hydrolases are expressed late in the infective cycle of double-stranded DNA phage, and—along with a membrane-permeabilizing protein known as a holin—they are responsible for disrupting the bacterial cell envelope and freeing progeny viral particles. Despite this conserved biological function, phage lysins (especially Gram-positive ones) are a tremendously diverse group of proteins whose enzymatic specificity includes various bonds within the peptidoglycan macromolecule. They include glycosyl hydrolases that target the polysaccharide backbone (muramidases/lysozymes and glucosaminidases), alanine amidases that target the initial l-alanine of the pentapeptide stem, and endopeptidases that target subsequent peptide bonds in the stem or cross bridge. While lysins of Gram-negative phage generally consist of an enzymatic domain alone, Gram-positive lysins are modular and combine an N-terminal lytic domain with a C-terminal binding domain that can recognize various epitopes within the target cell envelope.
Although researchers have known of lysins for decades, interest has increased markedly in recent years after it was proposed that they could act as novel anti-infective agents against Gram-positive pathogens, whose peptidoglycan is directly accessible from the extracellular space (8, 23, 28). A growing number of in vitro and in vivo studies have confirmed the ability of recombinantly expressed lysins to kill such organisms, and their appeal lies in both the potency and the specificity they demonstrate toward individual Gram-positive species. This enzybiotic value of phage lysins goes alongside additional proposed applications in the areas of food (11), agricultural (20), veterinary (7), and industrial science (21, 40).
Considering this potential, lytic enzymes represent an intriguing functional target for viral metagenomic screens. At the same time, identifying lysins in this manner would present several distinct challenges. Aside from general concerns common to all functional screens (e.g., protein expression and solubility), metagenomic lysin identification would face the following particular issues. (i) Clonal toxicity: recombinant lysin expression is typically well tolerated by host bacteria, since the enzymes are sequestered in the cytoplasm away from the peptidoglycan layer. Holins, on the other hand, interact nonspecifically with plasma membranes and are generally toxic to an Escherichia coli host, inducing bacteriolysis from within (9). Since holins are short (∼100 residues) and are often encoded adjacent to lysins, they can lead to selective toxicity of many of the clones one hopes to identify. In a metagenomic screen, where numerous lysins are present within a single library, this effect could lead to a significant loss of positive hits. (ii) Target bacterial species: in standard phage genomic screens, lysin-encoding clones are selected by their ability to kill the host bacterium of the encoding phage, which generally demonstrates the highest sensitivity (27). In a metagenomic screen, however, numerous host species of unknown origin could be present within a sample, confounding this choice of screening agent.
To address these issues, we have devised a novel functional strategy for the general cloning of lytic enzymes from uncultured phage DNA. It utilizes a plasmid-based E. coli expression system and consists of a two-step process. Following induction by arabinose, clones are first screened for holin-mediated lysis by a hemolytic effect they create in the surrounding blood agar. These initial hits are then restreaked as patches and overlaid with Gram-negative cells whose outer membranes have been permeabilized by autoclaving, serving as a general source of peptidoglycan. The clones are observed for surrounding Gram-negative clearing zones to assay directly for the recombinant production of lytic enzymes encoded adjacent to the holins. As proof-of-principle, we applied our methodology to a viral metagenomic library constructed from mixed animal feces, identifying 26 actively expressed lysins of diverse molecular architectures. The first of its kind, this study presents a general model for lysin identification through viral metagenomics, highlighting the potential of this field for cloning of proteins of biotechnological or academic value.
MATERIALS AND METHODS
DNA library construction.
Fecal specimens were collected at the Long Island Game Farm (Manorville, NY) from the following species: giraffe, zebra, donkey, domestic goat, llama, lion, and bison. Additionally, two dried fecal specimens were obtained from commercial sources: bat guano (Fox Farm Soil and Fertilizer Company) and cricket droppings (Ghann's Cricket Farm, Augusta, GA). Viral fractions were purified by an adaptation of the procedure of Casas and Rohwer (6). In summary, fecal samples (∼100 g each) were suspended in an equal volume of phosphate-buffered saline (PBS) (pH 7.4) and agitated overnight at 4°C. Particulate/cellular material was removed by centrifugation, followed by two passages through a 0.22-μm filter. Phage were precipitated by addition of polyethylene glycol (molecular weight, 10,000) (10% [wt/vol]. Centrifuged precipitates were pooled to form a collective phage library, which was subjected to phenol-chloroform extraction and ethanol precipitation. Phage DNA was separated from coprecipitated compounds by agarose gel electrophoresis and extraction of high-molecular-weight DNA. From this material, an expressible linker amplified shotgun library (E-LASL) was constructed, as previously described (26).
Lysin screening methodology.
Amplified metagenomic inserts were ligated into the arabinose-inducible pBAD plasmid using the TOPO-TA expression kit (Invitrogen). Transformed clones were initially plated on LB agar supplemented with 100 μg/ml ampicillin and 5% defibrinated sheep's blood. Following overnight growth at 37°C, the plates were placed in a sealed container that was attached to the outlet of a commercial nebulizer (Spider-Neb II). Nebulized arabinose (derived from an initial 20% [wt/vol] aqueous solution) was continuously pumped into this container for 1 h. The plates were returned to 37°C, and colonies were identified that developed a zone of hemolysis in the surrounding blood agar. Hits were identified over the subsequent 6- to 8-h period, since nonspecific blood-agar oxidation (i.e., alpha-hemolysis) would often appear around colonies at longer times (∼16 h). Chosen clones were streaked on separate LB-ampicillin plates (lacking arabinose) and allowed to repropagate without induced expression.
For the secondary screen, the above hits were streaked as ∼1-cm by 2-cm patches onto LB-ampicillin plates supplemented with 0.2% arabinose. Following overnight incubation at 37°C, the plates were exposed to chloroform vapor (15 min) to kill and permeabilize any still-viable E. coli. The patches were then overlaid with molten soft agar containing autoclaved Pseudomonas aeruginosa and observed for clearing zones for up to 24 h. This particular Gram-negative species was chosen as the screening agent based on the visual clarity of the clearing zones it produced in preliminary experiments. For all lytic clones, the encoded metagenomic insert was sequenced with plasmid-targeted primers (Genewiz; South Plainfield, NJ). When primer walking was required to sequence an insert in entirety, appropriate oligonucleotides were designed and ordered (Operon, Huntsville, AL).
Preparation of Gram-negative overlay.
Gram-negative soft agar was prepared as follows: P. aeruginosa strain PAO1 was grown to stationary phase (OD ≈ 2.5) in brain heart infusion medium. Cells were pelleted and resuspended in PBS (pH 7.4) to half the volume of the original liquid culture. Agar was added directly to this suspension (7.5 g/liter), which was autoclaved for 15 min at 122°C at 15 lb/in2. Solidified aliquots were stored at 4°C until the time of use, at which point they were melted and equilibrated at 55°C. For a single 150-mm petri dish, 15 ml of soft agar was overlaid.
Computational analysis.
Protein sequences of the cloned lysins were subjected to BLASTP analysis to identify known homologues among defined organisms (blast.ncbi.nlm.nih.gov). The NCBI nonredundant sequence collection (nr) was utilized as the reference database, with the cutoff E value set at 10−3. Putative catalytic and binding domains were assigned via the Pfam (v24.0) software program (pfam.sanger.ac.uk). Multiple sequence alignment (MSA) of PlyM1-PlyM20 (amino acid sequences) was performed using the ClustalX algorithm (30), followed by 100 rounds of bootstrapping. Phylogenetic analysis was conducted with the PHYLIP v3.67 software package (13) using the protdist (Jones-Taylor-Thornton matrix) and kitch (Fitch-Margoliash method) algorithms. A consensus tree was generated from the individual bootstrap data sets with the consense program; a nonbootstrapped tree was also derived from the original MSA.
Nucleotide sequence accession numbers.
Nucleotide sequences of lysins determined in this work have been deposited in GenBank under accession no. HM011589 to HM011614 (for lysins PlyM1 to PlyM26; see Table 1).
TABLE 1.
Cloned metagenomic lysinsa
| Name | GenBank accession no. | Protein length (aa)b | Predicted enzymatic domain(s)c | Predicted cell wall binding domain(s)d | Database homologue(s)e |
|---|---|---|---|---|---|
| PlyM1 | HM011589 | 197+ (trunc.) | Amidase 2 | Numerous hits against genus Bacillus (10−65-10−20) | |
| PlyM2 | HM011590 | 249 | Amidase 2 | Amidase 2 associated | Same as above |
| PlyM3 | HM011591 | 307+ (trunc.) | Amidase 2 | PG-1 | Same as above |
| PlyM4 | HM011592 | 329 | Amidase 2 | PG-1 | Same as above |
| PlyM5 | HM011593 | 284+ (trunc.) | Amidase 3 | PG-1 | Two hits against Desulfotomaculum reducens and Alkaliphilus metalliredigens (∼10−16) |
| PlyM6 | HM011594 | 357 | Amidase 3 | PG-1 | Same as above |
| PlyM7 | HM011595 | 356 | Amidase 3 | PG-1 | Same as above |
| PlyM8 | HM011596 | 357 | Amidase 3 | PG-1 | Same as above |
| PlyM9 | HM011597 | 343+ (trunc.) | Amidase 3 | PG-1 | Same as above |
| PlyM10 | HM011598 | 282+ (trunc.) | Amidase 3 | PG-1 | Same as above |
| PlyM11 | HM011599 | 356 | Amidase 3 | PG-1 | Same as above |
| PlyM12 | HM011600 | 338 | Amidase 2 | PG-1 and PG-3 | Numerous hits against broad variety of firmicutes (all >10−42) |
| PlyM13 | HM011601 | 232 | Amidase 3 | SPOR | Numerous hits against genera Bacillus and Geobacillus (all >10−60) |
| PlyM14 | HM011602 | 263 | Amidase 3 | SPOR | Same genera as above (all >10−49) |
| PlyM15 | HM011603 | 230 | Amidase 3 | SPOR | Same genera as above (all >10−26) |
| PlyM16 | HM011604 | 233 | Amidase 3 | SPOR | Same genera as above (all >10−28) |
| PlyM17 | HM011605 | 256 | Amidase 2 | Not predicted | Numerous hits against genus Bacillus (all >10−65); homologies do not extend into C-term binding region |
| PlyM18 | HM011606 | 253 | Amidase 2 | LysM | Three hits against Renibacterium salmoninarum, Actinomyces urogenitalis (10−30-10−26) |
| PlyM19 | HM011607 | 272 | Amidase 3 | SH3-5 | Numerous hits against genus Bacillus (all >10−77) |
| PlyM20 | HM011608 | 302 | Amidase 2 | SPOR | ∼10 hits against clostridium-like genera (10−67-10−51) |
| PlyM21 | HM011609 | 363 | Endopeptidase, muramidase | Not predicted | Numerous hits against diverse G+ and G− bacteria (all >10−25) |
| PlyM22 | HM011610 | 159 | Muramidase | Numerous hits against diverse G− bacteria (all >10−38) | |
| PlyM23 | HM011611 | 149 | Muramidase | Numerous hits against diverse gammaproteobacteria (all >10−41) | |
| PlyM24 | HM011612 | 181 | Muramidase | Thirteen hits against Acinetobacter (10−64-10−45) | |
| PlyM25 | HM011613 | 256 | PG-1 (N-terminal) | Numerous hits against Burkholderia, other G- bacteria (all >10−40) | |
| PlyM26 | HM011614 | 188 | Numerous hits against various G− bacterial species (all >10−49) |
The 26 actively-expressed lysins cloned in this study are summarized here.
aa, amino acid residues. Five lysins (indicated with “trunc.”g) were cloned as enzymatically active C-terminal truncations; for these proteins, the indicated length is that which was included on the plasmid insert.
See the text or Document S1 in the supplemental material for corresponding Pfam accession numbers. Enzymatic domains of PlyM25 and PlyM26 are not fully defined from sequence information (see the text).
For PlyM1, the protein was truncated within the C-terminal region, preventing an accurate prediction of a binding domain. Pfam analysis did not recognize a conserved binding domain for the Gram-positive lysins PlyM17 and PlyM21, although a distinct C-terminal region exists that presumably serves this purpose. Binding domains were not predicted for 4/5 Gram-negative lysins (PlyM22 to PlyM24 and PlyM26).
Cloned lysins were subjected to BLASTP analysis to identify homologues among sequenced bacteria/phage. In many instances, a lysin demonstrated highest homology to proteins encoded by a particular bacterial genus/species (or its phage). The identities of these taxa are identified here, and the degree of homology is indicated numerically with E values (or a minimum E value if numerous proteins demonstrated a continuous range of homology). In other cases, as noted, no genus/species was preferentially represented among the closest BLASTP hits. A complete list of homologues for each lysin is included in the online supplemental material. G+ and G−, Gram-positive and Gram-negative.
RESULTS AND DISCUSSION
A plasmid-based library was constructed with pooled metagenomic DNA extracted from the phage fraction of animal fecal samples. To clone lytic enzymes from this pool, we first addressed the issue of holin-based clonal toxicity. Rather than mitigate the effect, we instead exploited it by specifically identifying the toxic clones. There exists one published example where a lysin was cloned through the toxicity of its adjacent holin. When screening a library from Actinomyces naeslundii phage AV-1, Delisle et al. utilized a “plasmid release” protocol in which mixed E. coli transformants were grown in a single liquid culture (see reference 9 for protocol details). Holin-encoding cells would undergo lysis following induced expression, releasing their plasmids into the medium, from which they could be purified and characterized. That approach was adapted here for metagenomic libraries, in which numerous targeted clones are present.
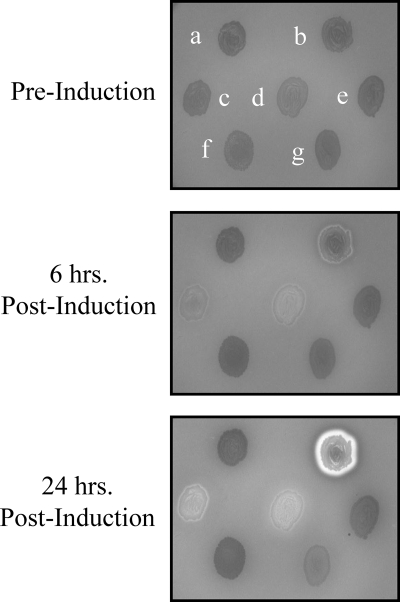
Plated E. coli transformants, already visible as individual colonies, were supplied a nebulized mist of arabinose-inducing agent. Colonies undergoing lysis could subsequently be visualized by the appearance of subtle yet definitive hemolytic zones in the surrounding blood agar (Fig. 1). This effect was often accompanied by the development of colony viscosity: when a pipette tip was touched to a hemolytic colony and lifted up, the bacterial mass would adhere in a string-like manner between the tip and the agar surface. Presumably, both the hemolysis and the viscosity were due to the release of intracellular contents from the lysing colonies. The hemolytic effect could likewise be due to holin-induced lysis of blood agar erythrocytes, since holins have been shown to be capable of permeabilizing eukaryotic membranes (1).
FIG. 1.
Depicted here are seven E. coli Top-10 clones on LB-ampicillin supplemented with 5% sheep's blood. Each clone is transformed with a pBAD plasmid containing a different protein-encoding insert. a, top left: empty plasmid; b, top right: aerolysin, a hemolytic bacterial exotoxin encoded by the genus Aeromonas; c, center left: Bacillus anthracis phage alanine amidase with adjacent holin; d, center middle: lysin PlyM14 from this study with adjacent holin; e, center right: arbitrary metagenomic insert from this study; f, bottom left: Bacillus anthracis phage alanine amidase without adjacent holin; g, bottom right: Bacillus anthracis phage muramidase without adjacent holin. Prior to induction with nebulized arabinose, the clones are indistinguishable against the blood agar background. Following induction (apparent at 6 h and more prominent at 24 h), a zone of hemolysis is present around the clones encoding holin/lysin combinations (c and d). No such zones are present surrounding the clones encoding empty plasmid (a), random sequence (e), or lysin only (f and g). The aerolysin-encoding clone (b) is included as a positive control of a dedicated hemolytic transformant.
Approximately two hundred thousand clones were screened in this manner, and 502 preliminary hits were identified. To gauge their identity, 52 of these hits were subjected to DNA sequencing. Forty-one unique clones were observed here—of these, 17 contained open reading frames (ORFs) that encoded both a complete holin and a complete lysin, 4 encoded a complete holin with only a partial (i.e., truncated) lysin, 2 encoded complete holins without any recognizable lysin, and 18 demonstrated homology to neither holins nor lysins. The latter 18 inserts encoded ORFs with a variety of predicted functions, including phage structural proteins and DNA-interacting proteins. For 5 inserts, BLAST analysis revealed homology only to phage ORFs of unknown function; for 1 insert, no significant BLAST hits were returned. This indicates that the hemolysis screen was not absolutely specific for holin/lysin cassettes, although the finding is not unexpected. This technique could select for any toxic proteins that compromise the viability (and, resultantly, the envelope integrity) of the host. Moreover, even among lysin-encoding clones, the screen does not reveal which ones express soluble, active enzymes. A secondary screen was thus necessary to specify these clones.
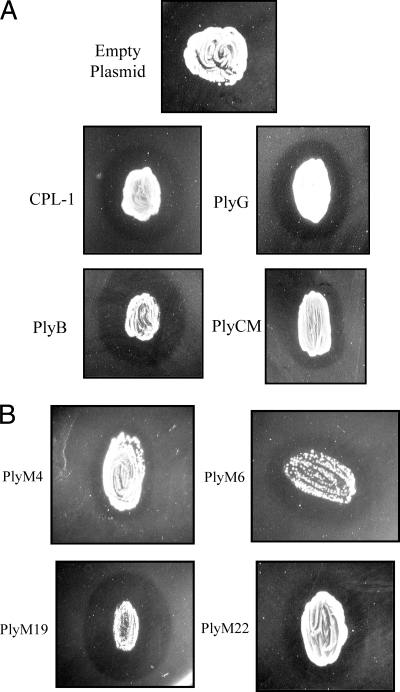
For this step, we exploited the cell envelope properties of Gram-negative bacteria. By way of explanation, viable Gram-negative cells are generally resistant to lysin treatment due to their protective outer membrane. Once this membrane is compromised, however, they become highly sensitized to enzyme action (much more so than Gram-positive bacteria) due to the thinness of their peptidoglycan. This includes lysis by both nonspecific eukaryotic lysozymes (forming the basis of commercially available extraction kits) and phage lysins from viruses that infect other bacterial species (5). Consistent with these properties, we have observed that clearing zones appear in soft-agar experiments in which autoclaved Gram-negative cells are exposed to diverse Gram-positive lysins (Fig. 2 A). While this phenomenon was of little significance when cloning lysins by standard genomic techniques—here, the encoding phage and target species are known a priori—it could be useful for identifying unknown lysins from a metagenomic pool.
FIG. 2.
Various lysin-encoding clones overlaid with autoclaved P. aeruginosa PAO1. (A) Previously characterized phage lysins. As opposed to the negative control (top), Gram-negative clearing zones are apparent around E. coli clones encoding CPL-1 (Streptococcus pneumoniae muramidase), PlyG (Bacillus anthracis alanine-amidase), PlyB (Bacillus anthracis muramidase), and PlyCM (Clostridium perfringens muramidase). In each case, the cells were streaked onto LB agar containing arabinose inducing agent, allowed to proliferate overnight, and chloroform permeabilized prior to Gram-negative overlay. (B) Metagenomic lysin hits. Four representative examples of the 26 enzymes cloned during this study are depicted here (PlyM4, PlyM6, PlyM19, and PlyM22). As mentioned in the text, these clones (which encode holin/lysin combinations) often demonstrated toxicity when streaked on arabinose-containing agar. This is particularly evident for the clones encoding PlyM4, PlyM6, and PlyM19, in which punctate colonies (presumably downregulated mutants) have proliferated upon a lysed E. coli patch.
To these ends, the above 502 hits were restreaked as patches onto arabinose-containing agar and tested for their ability to lyse a soft-agar overlay of autoclaved P. aeruginosa. Since these strains were selected for their clonal toxicity, the streaked patches obviously did not proliferate well on the arabinose plates; following overnight incubation, they were often comprised of lysed cells with occasional punctate colonies (presumably recombinant mutants with downregulated insert expression; Fig. 2B). Nevertheless, even with little growth, enough lysin was synthesized to produce distinct overlying clearing zones. Sixty-five hits were identified here (Fig. 2B), which were shown to encode 26 unique lysins, summarized in Table 1 (PlyM1 to PlyM26). Of these enzymes, 15 were observed for the first time during the secondary screen, while 11 had been identified previously during the sequencing of 52 hemolytic clones. Conversely, 6 of the putative lysins identified during the initial sequencing (PlyM27 to PlyM32 [GenBank accession no. HM011615 to HM011620]) were not detected by the secondary screen. This could be attributable to insolubility or insufficient expression under the secondary screen's induction conditions (i.e., a biomass effect).
The 26 actively expressed enzymes contain a variety of enzymatic and binding domains (the sequences themselves and the exact positions of the predicted domains are detailed in Document S1 in the supplemental material). The majority of these (PlyM1 to PlyM20) encode typical Gram-positive lysins, with both an N-terminal catalytic region and a C-terminal binding region. Of these 20 genes, 15 encode full-length lysins and 5 encode truncated proteins (an artifact of library construction) that lack the final portion of the C terminus but retain catalytic activity. Of the 6 remaining lysins, 3 represent typical Gram-negative lysins (comprising only a catalytic domain, PlyM22 to PlyM24), while 3 possess atypical lysin architectures (PlyM21 and PlyM25 to PlyM26). As expected, a majority of the hits (24/26) also encode a short, adjacent ORF that can be assigned putative holin functionality.
Among the Gram-positive lysins, Pfam domain analysis suggests that all 20 possess N-acetylmuramoyl-l-alanine-amidase activity: 8 are predicted to be type 2 amidases (Pfam family PF01510), and 12 are predicted to be type 3 amidases (PF01520). At the C-terminal ends of the Gram-positive lysins, a variety of binding domains are likewise predicted. These include 10 PG-1 motifs (PF01471), 1 PG-3 motif (PF09374), 5 SPOR motifs (PF05036), 1 SH3 type 5 motif (PF08460), 1 LysM motif (PF01476), and 1 amidase-2-associated domain (PF12123). For two of the Gram-positive enzymes (PlyM17 and PlyM21), a clear C-terminal region is present, even though Pfam analysis fails to predict a binding motif. These regions likely possess binding functions, albeit ones that have not yet been categorized as conserved families.
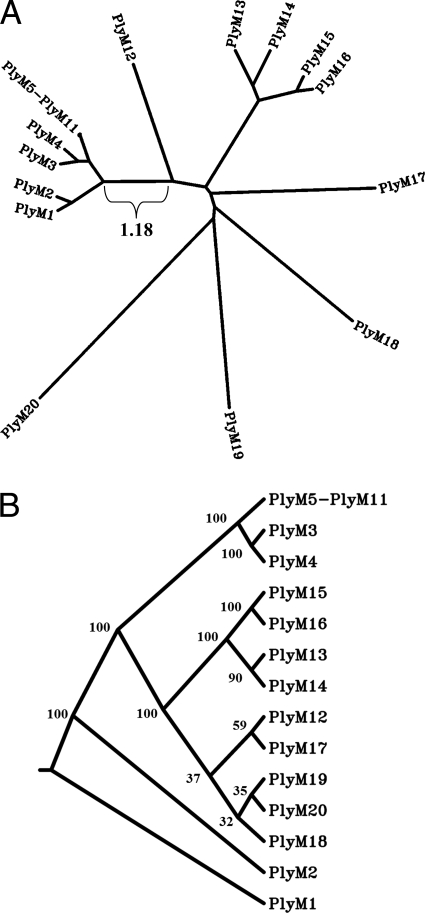
Multiple sequence alignment of PlyM1 to PlyM20 reveals strong similarities among some of the cloned proteins, summarized in Fig. 3 as a distance tree. In particular, PlyM5 to PlyM11 demonstrate high sequence homology with one another, with pairwise sequence identities of 91 to 95% on the nucleotide level and 95 to 98% on the amino acid level (all E values < 10−159). PlyM3 and PlyM4 share this homology with PlyM5 to PlyM11 at the C terminus but diverge in their enzymatic regions. These lysins were presumably derived from a group of similar phage infecting one of the component bacterial species of the fecal sample. When subject to BLAST analysis, PlyM5 to PlyM11 demonstrate closest homology (E values ≈ 10−16) to two putative prophage lysins of Delsulfotomaculum reducens MI-1 and Alkaliphilus metalliredigens QYMF, both spore-forming organisms of the class Clostridia. The other Gram-positive lysins were likewise analyzed via BLAST; for many (PlyM1 to PlyM4, PlyM13 to PlyM17, and PlyM19), the closest homologues are encoded by phage/prophage infecting Bacillus and related genera. (For a complete list of BLAST homologues for all cloned lysins, the reader is referred to Document S2 in the supplemental material). These homology findings are consistent with the origin of the library, although one should not draw conclusions on the ecology of the sample based on this information alone. Overall, if one is interested in viral metagenomics as a tool for studying microbial ecology, sequenced-based approaches are superior to functional screens.
FIG. 3.
Similarities among Gram-positive hits. (A) The sequence homology among the typical Gram-positive lysins (PlyM1 to PlyM20) is organized here as a contemporaneous-tip distance tree (one branch length is included as a reference). Several clusterings are apparent, especially PlyM5 to PlyM11; these enzymes are depicted as a single node in the image, since their degree of homology (90+ pairwise nucleotide/amino acid identity) prevents individual visual differentiation. The reader should note that this tree was generated from a single multiple sequence alignment (MSA) so that the distance values between proteins would be most visually apparent as the branch lengths. (B) Bootstrap analysis (100 rounds) was also conducted on the original MSA of PlyM1 to PlyM20, and a consensus cladogram was generated. As in panel A, PlyM5 to PlyM11 have been collapsed here to a single node. The bootstrap consensus values are indicated at their respective nodes.
Among the lysins that do not demonstrate typical Gram-positive architectures, PlyM21 possesses two distinct catalytic regions: an N-terminal M23 endopeptidase domain (Pfam family PF01551) and a central lambda phage-like muramidase domain (PF00959). While less common, several Gram-positive lysins have been characterized with multiple catalytic domains (8, 38). It is currently unclear what, if any, advantage is offered by this extended architecture. In contrast, the lysins PlyM22 to PlyM24 are relatively short in length and consist of a single muramidase domain (PF00959) without any C-terminal binding region, typical for Gram-negative phage.
PlyM25 and PlyM26 differ significantly from the other enzymes cloned here and, in general, represent poorly characterized varieties of lytic enzymes. Domain analysis of PlyM25 predicts an N-terminal PG-1 binding motif but fails to recognize a definitive catalytic domain. Rather, the central/C-terminal region corresponds to a domain of unknown function (DUF 3380, PF11860), from which the enzymatic activity presumably originates. BLAST analysis of PlyM25 reveals several dozen ORFs of moderate homology (E values, 10−20 to 10−40) among Gram-negative phage/prophage. Virtually none of these homologues, however, are currently annotated as lysins, exceptions being ORF12 of P. aeruginosa phage phiCTX (22) and ORF27 of Burkholderia cepacia phage Bcep781 (29). These last two enzymes share the PG-1/DUF, although only the Burkholderia protein has been assigned lytic function experimentally (its targeted bond remains undetermined).
For PlyM26, Pfam predicts neither a binding nor a peptidoglycan hydrolase motif but rather a chitinase enzymatic domain (Pfam family PF00182). It is conceivable that PlyM26 represents a dedicated phage-encoded chitinase or a nonviral chitinase derived from free genomic DNA in the original fecal sample. Nevertheless, these seem unlikely scenarios; the first four nucleotides of PlyM26 overlap with an upstream 101-amino-acid ORF containing three predicted transmembrane domains, making it a strong candidate for an adjacent holin. Moreover, given the similarity of chitin to peptidoglycan and the fact that chitinases belong to the same protein clan (Pfam CL0037) as muramidases and glucosaminidases, it is more probable that PlyM26 represents one of the latter functionalities. Like PlyM25, PlyM26 does possess a number of homologues among Gram-negative phage/prophage. Again, however, few are annotated as phage lysins and none (to our knowledge) have been characterized biochemically.
The question naturally arises at this point: what applied or basic purposes could metagenomic lysin screens serve? One possibility involves the identification of new enzymes with antibiotic activity against medically relevant bacteria. The lysins cloned here were, in fact, tested against a panel of Gram-positive pathogens. Although none of them demonstrated activity comparable to those of previously characterized lysins (and hence they were not pursued further), metagenomics could in principle be another avenue for enzybiotic discovery. Metagenomic lysin screens could also be useful for identifying enzymes that are active under a desired set of biochemical conditions (temperature, pH, salts, etc.). For instance, several lysins from thermophilic species have been characterized with the motivation that temperature resistance is an industrially attractive feature (21, 40). In general, a strength of metagenomics is its ability to study extreme environments whose bacteria/phage are difficult to propagate in the laboratory (14). One could easily envision isolating the phage from an extreme environment and screening them for lysins with the above technique, circumventing the need for laboratory culture or prophage induction.
That said, one must note that sequence-based metagenomics is also capable of identifying lysins through homology analysis. Nevertheless, if one's specific goal is to identify these enzymes, clonal screening represents a far more efficient approach than bulk sequencing, and sequencing data alone do not predict what lysins are actively expressed in a recombinant system. All of this, moreover, presupposes that a lysin is sufficiently homologous to other lysins annotated in the database. In fact, metagenomic lysin screening is perhaps most useful in an academic sense—the identification of new classes of catalytic and binding domains. Although already quite diverse, lysins are still being characterized with previously unrecognizable lytic sequences (21, 24). Even the simple library employed here yielded several clones whose identity as phage lysins would not have been obvious from their sequences alone.
Finally, it is important to note a limitation of this protocol and to mention how it could be adapted to address the issue. While it is clearly capable of identifying glycosyl-hydrolases and alanine-amidases, only one endopeptidase was cloned here (PlyM21), and this lysin possessed a secondary muramidase domain. In fact, endopeptidases present an obstacle for the technique, since it would be impossible to identify all endopeptidases using a single bacterial species in the soft agar overlay. While the polysaccharide backbone of peptidoglycan and the initial l-alanine of the pentapeptide stem are conserved among bacteria, considerable variability exists at the other stem positions and within the cross-bridge (37). Without the targeted bond, Gram-negative peptidoglycan would not be susceptible to Gram-positive endopeptidases. To ensure endopeptidase coverage against a particular Gram-positive species, therefore, one could conduct the secondary screen in duplicate, including that species along with the Gram-negative bacteria.
In conclusion, this study represents one of the first functional screens of a viral metagenomic sample. The technique is straightforward and generalizable, and it allows one to mine for biotechnologically relevant enzymes from the largest genetic pool on the planet.
Supplementary Material
Acknowledgments
This research was funded by NIH/NIAID grants AI057472 and AI11822 to V.A.F. J.E.S. acknowledges the kind support of the Pharmaceutical Research and Manufacturers of America Foundation and the NIH MSTP program (Weill Cornell/Rockefeller/Sloan-Kettering grant GM 07739).
We thank the Long Island Game Farm for providing fecal specimens.
Footnotes
Published ahead of print on 17 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Agu, C. A., R. Klein, J. Lengler, F. Schilcher, W. Gregor, T. Peterbauer, U. Bläsi, B. Salmons, W. H. Günzburg, and C. Hohenadl. 2007. Bacteriophage-encoded toxins: the lambda-holin protein causes caspase-independent non-apoptotic death of eukaryotic cells. Cell. Microbiol. 9:1753-1765. [DOI] [PubMed] [Google Scholar]
- 2.Brady, S. F., L. Simmons, J. H. Kim, and E. W. Schmidt. 2009. Metagenomic approaches to natural products from free-living and symbiotic organisms. Nat. Prod. Rep. 26:1488-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitbart, M. I., I. Hewson, B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2003. Metagenomic analysis of an uncultured viral community from human feces. J. Bacteriol. 185:6220-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitbart, M. I., P. Salamon, B. Anderson, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rowher. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briers, Y., G. Volckaert, A. Cornelissen, S. Lagaert, C. W. Michiels, K. Hertveldt, and R. Lavigne. 2007. Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages φKZ and EL. Mol. Microbiol. 65:1334-1344. [DOI] [PubMed] [Google Scholar]
- 6.Casas, V., and F. Rohwer. 2007. Phage metagenomics. Method. Enzymol. 421:259-268. [DOI] [PubMed] [Google Scholar]
- 7.Celia, L. K., D. Nelson, and D. E. Kerr. 2008. Characterization of a bacteriophage lysin (Ply700) from Streptococcus uberis. Vet. Microbiol. 130:107-117. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, Q., D. Nelson, S. Zhu, and V. A. Fischetti. 2005. Removal of group B streptococci colonizing the vagina and oropharynx of mice with a bacteriophage lytic enzyme. Antimicrob. Agents Chemother. 49:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delisle, A. L., G. J. Barcak, and M. Guo. 2006. Isolation and expression of the lysin genes of Actinomyces naeslundii phage Av-1. Appl. Environ. Microbiol. 72:110-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delwart, E. L. 2007. Viral metagenomics. Rev. Med. Virol. 17:115-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutsch, S., S. Guezenec, M. Piot, S. Foster, and S. Lortal. 2004. Mur-LH, the broad-spectrum endolysin of Lactobacillus helveticus temperate bacteriophage phi-0303. Appl. Environ. Microbiol. 70:96-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, R. A., and F. Rohwer. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3:504-510. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 14.Ferrer, M., O. Golyshina, A. Beloqui, and P. N. Golyshin. 2007. Mining enzymes from extreme environments. Curr. Opin. Microbiol. 10:207-214. [DOI] [PubMed] [Google Scholar]
- 15.Fierer, N., M. Breitbart, J. Nulton, P. Salamon, C. Lozupone, R. Jones, M. Robeson, R. A. Edwards, B. Felts, S. Rayhawk, R. Knight, F. Rohwer, and R. B. Jackson. 2007. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fishetti, V. A. 2005. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 13:491-496. [DOI] [PubMed] [Google Scholar]
- 17.Fishetti, V. A., D. Nelson, and R. Schuch. 2006. Reinventing phage therapy: are the parts greater than the sum? Nat. Biotechnol. 24:1508-1511. [DOI] [PubMed] [Google Scholar]
- 18.Green, B. D., and M. Keller. 2006. Capturing the uncultivated majority. Curr. Opin. Biotechnol. 17:236-240. [DOI] [PubMed] [Google Scholar]
- 19.Hatfull, G. F. 2008. Bacteriophage genomics. Curr. Opin. Microbiol. 11:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, W., H. Salm, and K. Geider. 2004. Expression of bacteriophage phiEa1h lysozyme in Escherichia coli and its activity in growth inhibition of Erwinia amylovora. Microbiology 150:2702-2714. [DOI] [PubMed] [Google Scholar]
- 21.Matsushita, I., and H. Yanase. 2008. A novel thermophilic lysozyme from bacteriophage phiIN93. Biochem. Biophys. Res. Commun. 377:89-92. [DOI] [PubMed] [Google Scholar]
- 22.Nakayama, K., S. Kanaya, M. Ohnishi, Y. Terawaki, and T. Hayashi. 1999. The complete nucleotide sequence of phiCTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: implications for phage evolution and horizontal gene transfer via bacteriophages. Mol. Microbiol. 31:399-419. [DOI] [PubMed] [Google Scholar]
- 23.Nelson, D., L. Loomis, and V. A. Fischetti. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. U. S. A. 98:4107-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson, D., R. Schuch, P. Chahales, S. Zhu, and V. A. Fischetti. 2006. PlyC: a multimeric bacteriophage lysin. Proc. Natl. Acad. Sci. U. S. A. 103:10765-10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Wu, J. A. Eisen, J. M. Hoffman, K. Remington, K. Beeson, B. Tran, H. Smith, H. Baden-Tillson, C. Stewart, J. Thorpe, J. Freeman, C. Andrews-Pfannkoch, J. E. Venter, K. Li, S. Kravitz, J. F. Heidelberg, T. Utterback, Y. H. Rogers, L. I. Falcón, V. Souza, G. Bonilla-Rosso, L. E. Eguiarte, D. M. Karl, S. Sathyendranath, T. Platt, E. Bermingha, V. Gallardo, G. Tamayo-Castillo, M. R. Ferrari, R. L. Strausberg, K. Nealson, R. Friedman, M. Frazier, and J. C. Venter. 2007. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern tropical Pacific. PLoS Biol. 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitz, J. E., A. Daniel, M. Collin, R. Schuch, and V. A. Fischetti. 2008. Rapid DNA library construction for functional genomic and metagenomic screening. Appl. Environ. Microbiol. 74:1649-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuch, R., V. A. Fischetti, and D. Nelson. 2008. A genetic screen to identify bacteriophage lysins. In R. J. Clokie and A. M. Kropinski (ed.), Bacteriophages: methods and protocols, vol. 2. Molecular and applied aspects. Humana Press, Totowa, NJ. [DOI] [PMC free article] [PubMed]
- 28.Schuch, R., D. Nelson, and V. A. Fischetti. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884-889. [DOI] [PubMed] [Google Scholar]
- 29.Summer, E. J., C. F. Gonzalez, M. Bomer, T. Carlile, A. Embry, A. M. Kucherka, J. Lee, L. Mebane, W. C. Morrison, L. Mark, M. D. King, J. J. LiPuma, A. K. Vidaver, and R. Young. 2006. Divergence and mosaicism among virulent soil phages of the Burkholderia cepacia complex. J. Bacteriol. 188:255-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torsvik, V., and L. Ovreas. 2002. Microbial diversity and function in soil. Curr. Opin. Microbiol. 5:240-245. [DOI] [PubMed] [Google Scholar]
- 32.Tringe, S. G., and E. M. Rubin. 2005. Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 6:805-814. [DOI] [PubMed] [Google Scholar]
- 33.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 34.Uchiyama, T., and K. Miyazaki. 2009. Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr. Opin. Biotechnol. 20:616-622. [DOI] [PubMed] [Google Scholar]
- 35.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 36.Voget, S., H. Steele, and W. R. Streit. 2005. Metagenomes—an unlimited resource for novel genes, biocatalysts and metabolites. Minerva Biotecnol. 17:47-53. [Google Scholar]
- 37.Vollmer, W., D. Blanot, and M. A. de Pedro. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149-167. [DOI] [PubMed] [Google Scholar]
- 38.Wang, Y., J. H. Sun, and C. P. Lu. 2009. Purified recombinant phage lysin LySMP: an extensive spectrum of lytic activity for swine streptococci. Curr. Microbiol. 58:609-615. [DOI] [PubMed] [Google Scholar]
- 39.Williamson, S. J., D. B. Rusch, S. Yooseph, A. L. Halpern, K. B. Heidelberg, et al. 2008. The Sorcerer II global ocean sampling expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS One 3:e1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye, T., and X. Zhang. 2008. Characterization of a lysin from deep-sea thermophilic bacteriophage GVE2. Appl. Microbiol. Biotechnol. 78:635-641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.