Abstract
We report the selection and amplification of the broad-host-range Salmonella phage phi PVP-SE1 in an alternative nonpathogenic host. The lytic spectrum and the phage DNA restriction profile were not modified upon replication in Escherichia coli Bl21, suggesting the possibility of producing this phage in a nonpathogenic host, contributing to the safety and easier approval of a product based on this Salmonella biocontrol agent.
Salmonella enterica has long been recognized as an important zoonotic pathogen of economic significance in animals and humans and remains the primary cause of reported food poisonings worldwide, with massive outbreaks occurring in recent years (9, 14, 33). The serovars most commonly reported in human infections in the European Union (EU) and the United States have been Enteritidis and Typhimurium, comprising, respectively, 76% and 14% of the cases in the EU (14, 15, 27). Therefore, controlling Salmonella infections has become an important goal for the poultry industry, the most common source of Salmonella Enteritidis infections (3, 7, 14, 33).
The use and misuse of antimicrobials in both humans and animals have given rise to the emergence of infectious bacteria displaying resistance toward many, and in some cases all, effective antimicrobials (2, 14, 33). Thus, the development of alternatives to chemotherapy is imperative and a critical priority. The use of (bacterio)phages, viruses that specifically infect and lyse bacteria, as a therapeutic agent (phage therapy) is one possible option for controlling pathogenic bacteria. Phages have already been tested as biocontrol agents for salmonellae and other pathogens in humans and animals, with success showing advantages over antibiotics (4, 23, 25, 29-31). However, due to the high specificity of phages, they often can only be produced in their natural hosts (7, 35), which, in the case of pathogenic hosts, turns out to be problematic due to the release of cell debris and large quantities of both endotoxins and exotoxins and the presence of live cells that are then found in the crude phage lysate (10, 24, 29, 32).
The use of a nonpathogenic host in the production process would eliminate the risk of accidentally administering a pathogen (6, 7, 24). Consequently, it would greatly simplify the purification process, with a consequent reduction in cost and would furthermore increase the safety of phage preparations, leading to easy and faster approval of phage products. Nevertheless, this tends to be a difficult approach since multivalent phages are rare (6, 7, 19, 29).
The aims of this work were to select and characterize a Salmonella phage with a broad lytic spectrum and study the possibility of producing it in a nonpathogenic Escherichia coli strain.
Phage lytic spectrum.
Thirteen Salmonella phages isolated in the scope of the European project Phagevet-P were used, and the three with the broadest host ranges are presented in Table 1 along with their hosts. Also, broad-host-range Salmonella phage Felix-O1, acquired from the Profos AG collection (Regensburg, Germany), was used. To determine the phages' lytic spectra, drops (10 μl) of serial dilutions of the different phage suspensions were added to bacterial lawns (Tables 1 and 2). LB broth, Miller (Sigma-Aldrich Inc., St. Louis, MO), was the medium used, and it was prepared according to the manufacturer's instructions. Agar plates and soft agar were prepared by adding, respectively, 1.2% and 0.6% agar (Applichem, Darmstadt, Germany) to the LB broth. The plates were incubated overnight at 37°C, and the lytic activity was checked for the formation of clear areas and phage plaque formation on the bacterial lawns. Thirty-eight bacterial strains were used (Tables 1 and 2). The strains in Table 1 belong to serovar Enteritidis (detailed information about these strains can be found in reference 28), and two E. coli strains (N5-bovine and N9-porcine) were also isolated (Table 2). The remaining strains were from the NCTC (National Collection of Type Cultures), ATCC (American Type Culture Collection), SGSC (Salmonella Genetic Stock Centre), and CECT (Colección Española de Cultivos Tipo) type culture collections. At least two salmonellae from each subspecies were used. The two well-known E. coli strains BL21 [BL21-Gold(DE3), purchased from Stratagene] and K-12 were also used to test the ability of the phage to be amplified in an alternative host.
TABLE 1.
Comparison of the lytic spectra of isolated Salmonella phages and phage Felix-O1 against poultry Salmonella isolatesa
| Salmonella Enteritidis strain | Lysis by phage (host): |
|||
|---|---|---|---|---|
| phi PVP-SE1 (S1400/94) | phi PVP-SE2 (821) | phi PVP-SE3 (869) | Felix-O1 | |
| AL855 | + | + | + | NT |
| EX2 | + | L | + | − |
| S1400/94 | + | + | + | + |
| 269 | + | + | + | + |
| 546 | + | + | + | + |
| 629B | + | + | + | + |
| 657 | + | L | L | + |
| 821 | + | + | + | + |
| 855 | + | L | + | + |
| 869 | + | + | + | + |
| 905 | + | + | + | + |
| 932 | + | + | + | − |
| 9510.85 | + | L | L | − |
−, absence of a phage halo and phage plaques; +, presence of a phage halo and phage plaques; L (lysis from without), presence of a phage halo and absence of phage plaques; NT, not tested.
TABLE 2.
Lytic spectra of isolated Salmonella phages against different Salmonella subtypes and bacteria other than salmonellaea
| Strain | Lysis by phage: |
||
|---|---|---|---|
| phi PVP-SE1 | phi PVP-SE2 | phi PVP-SE3 | |
| Salmonella Typhimurium NCTC 12416 subsp. I | + | − | − |
| Salmonella NCTC 13349 subsp. I | + | + | + |
| Salmonella sp. strain SGSC 3047 subsp. II | + | − | − |
| Salmonella sp. strain SGSC 3039 subsp. II | + | − | − |
| Salmonella Arizonae SGSC 3063 subsp. IIIa | L | − | − |
| Salmonella Arizonae 83 (isolate) subsp. IIIa | − | − | − |
| Salmonella sp. strain SGSC 3069 subsp. IIIb | + | − | − |
| Salmonella sp. strain SGSC 3068 subsp. IIIb | + | − | − |
| Salmonella sp. strain SGSC 3086 subsp. IV | L | − | − |
| Salmonella sp. strain SGSC 3074 subsp. IV | + | − | − |
| Salmonella Bongori SGSC 3103 subsp. V | + | − | − |
| Salmonella Bongori SGSC 3100 subsp. V | + | − | − |
| Salmonella sp. strain SGSC 3118 subsp. VI | + | − | − |
| Salmonella sp. strain SGSC 3116 subsp. VI | + | − | − |
| Salmonella sp. strain SGSC 3121 subsp. VII | + | − | − |
| Salmonella sp. strain SGSC 3120 subsp. VII | + | − | − |
| Escherichia coli N9 | L | − | − |
| Escherichia coli N5 | L | − | − |
| Escherichia coli CECT 434 (ATCC 25922) | L | − | − |
| Escherichia coli BL21 | + | − | − |
| Escherichia coli K-12 | + | − | − |
| Enterobacter amnigenus CECT 4078 (ATCC 33072) | L | − | − |
| Enterobacter aerogenes CECT 684 (ATCC 13048) | − | − | − |
| Klebsiella pneumoniae 11296 | − | − | − |
| Shigella flexneri ATCC 12022 | − | − | − |
−, absence of a phage halo and phage plaques; +, presence of a phage halo and phage plaques; L (lysis from without), presence of a phage halo and absence of phage plaques; NT, not tested.
Phage phi PVP-SE1 showed the broadest lytic spectrum. Besides being able to lyse almost all Salmonella strains (of all Salmonella subtypes except IIIa), it was also able to infect E. coli BL21 and K-12, and thus, phi PVP-SE1 can be considered a multivalent phage. Moreover, this phage was able to infect more strains than Felix-O1 (Table 1), which is known for its wide lytic spectrum among salmonellae. This result supports the evidence that this newly isolated phage has a very broad lytic spectrum, and therefore it is an interesting candidate for use in phage therapy. The ability of phi PVP-SE1 to infect the two E. coli strains (BL21 and K-12) and the production of very clear lysis areas on lawns of BL21 encourage the selection of this nonpathogenic strain for production of the phage.
Phage morphology.
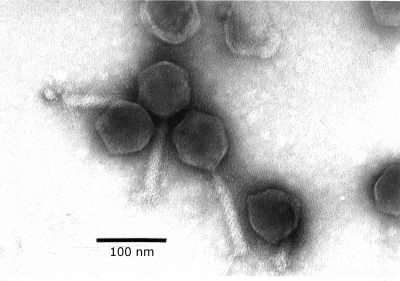
The morphology of phage phi PVP-SE1 was analyzed using transmission electron microscopy (TEM) (Fig. 1). Phage particles were sediment at 25,000 × g for 60 min using a Beckman (Palo Alto, CA) J2-21 centrifuge with a JA 18.1 fixed-angle rotor. Phages were washed twice in 0.1 M ammonium acetate (pH 7.0), deposited on copper grids provided with carbon-coated Formvar films, stained with 2% potassium phosphotungstate (pH 7.2), and examined in a Philips EM 300 electron microscope (by H.-W. Ackermann, Laval University, Quebec, Canada). Morphologically, this phage belongs to the Myoviridae family, is characterized by its contractile tail, and resembles typical O1-like phages (Fig. 1) (H.-W. Ackermann, Université Laval, Quebec, Canada, personal communication). However, this phage has a head size of 84 nm and a tail of 120 by 18 nm, which are considerably larger than those of typical O1-like phages (72 nm, 113 by 17 nm) (1, 21, 34).
FIG. 1.
TEM of phage phi PVP-SE1.
Phage production in a nonpathogenic host.
To assess whether the phage maintains its lytic profile when grown in an alternative nonpathogenic host, the phage was produced for six generations in E. coli BL21. Phages were amplified by plate lysis as described by Sambrook and Russell (26). Generation one (G1) of phage phi PVP-SE1 in E. coli BL21 was produced using the phage stock suspension obtained for phi PVP-SE1 in Salmonella S1400/94 (G0). This production was then used as a stock suspension to produce G2 of phage phi PVP-SE1 in E. coli BL21. G3 to G6 were obtained using the same procedure; i.e., each generation was obtained using phages from the previous generation. It was observed that the lytic spectrum of the phage was maintained from G0 until G6. This indicates that replication of the phage in this alternative host will not narrow its lytic spectrum. Taking into account these characteristics, a broad lytic spectrum and the ability to be amplified in a nonpathogenic strain without modification of its lytic spectrum, phage phi PVP-SE1 was further characterized and G6 was compared with G0.
Phage infection parameters.
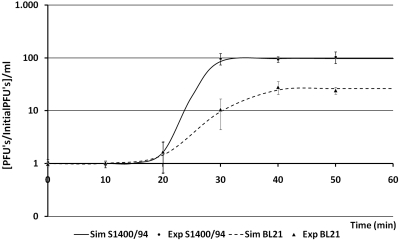
The phage infection parameters of G0 and G6 were determined through single-step growth curve experiments carried out at 37°C in LB medium as described by Sambrook and Russell (26), using an overnight preinoculum of the bacteria in the same medium. The experiments were done in duplicate and repeated on two different occasions. The values of the parameters were determined by fitting the experimental data to a four-parameter sigmoid curve using least-squares fitting. Figure 2 shows the increase in phage concentration through time of G0 in S1400/94 and G6 in BL21. Three phage infection parameters were determined, i.e., burst sizes of 102 and 28 phages per infected host cell; rise periods of 11 and 21 min, respectively, for G0 and G6; and the same latency period of 19 min. The differences between the burst sizes and rise periods are attributed to the use of a different host to replicate the phage once it was determined that the phage had not suffered any modification, and the only variable in the experiments (besides the phage generation) was the host used. To corroborate this fact, we repeated the experiment using phage from G0 and E. coli BL21 as the host and obtained the same result. The increased safety, and consequently the lower cost of producing a safer phage product, obtained when producing phi PVP-SE1 in a nonpathogenic host can overcome the disadvantages of a smaller burst size and a longer rise period.
FIG. 2.
Sim-fitted one-step growth curve of phage phi PVP-SE1 G0 in Salmonella S1400/94 and phi PVP-SE1 G6 in E. coli BL21. Exp, experimental values (average values ± standard deviations).
Phage restriction fragment length polymorphism (RFLP).
Further characterization of phage phi PVP-SE1 was carried out by determining the restriction profile of phage DNA (Fig. 3). Phage DNA was extracted using the Wizard DNA Clean-Up System from Promega Corporation (Madison, WI) and digested with HindIII and EcoRV from Sigma-Aldrich Inc. (St. Louis, MO) according to the manufacturer's instructions. Fragments were separated on a 1% agarose (Bio-Rad Laboratories Inc., Hercules, CA) gel at 70 V for 3 h (20 V for 16 h when DNA was digested with HindIII) and stained with ethidium bromide (1 μg/ml; Bio-Rad Laboratories Inc., Hercules, CA). Based on the restriction profile, it was possible to estimate a genome size of approximately 146 kb. In order to assess if replication of the phage in E. coli BL21 could cause significant mutations or modifications in the phage DNA, the DNA from G6 was extracted and digested with EcoRV, which produces a great number of bands with different sizes. Comparison of its restriction profile with that of G0 (phage grown in its natural host, Salmonella S1400/94) showed no difference.
FIG. 3.
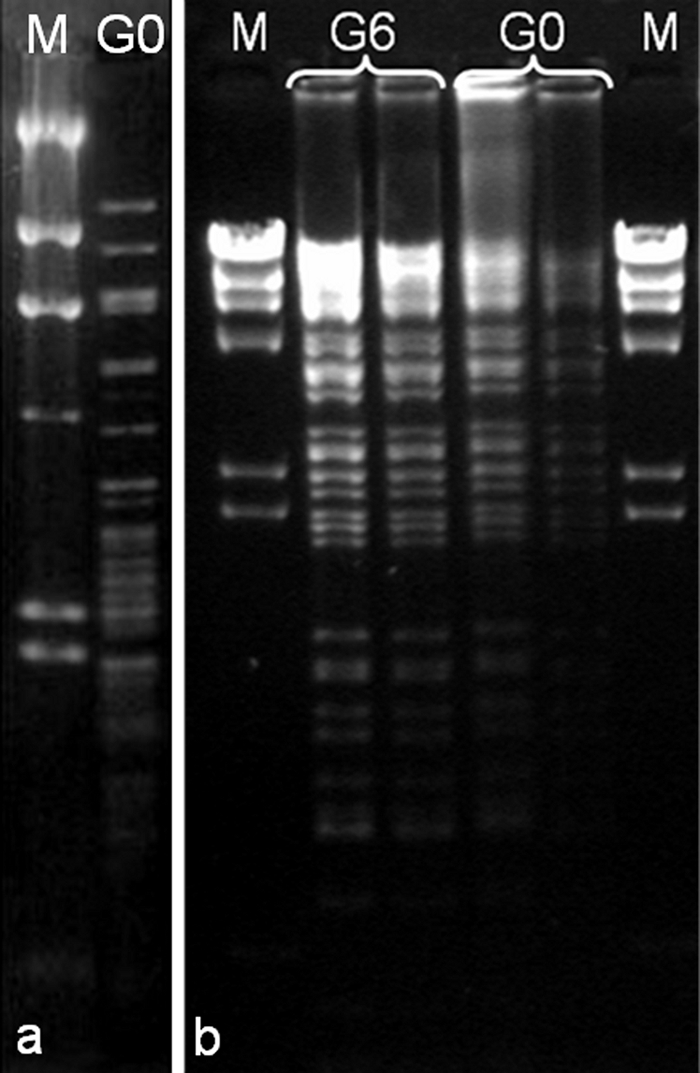
Restriction profile of phage phi PVP-SE1 DNA. (a) phi PVP-SE1 G0 DNA digested with HindIII. (b) phi PVP-SE1 DNA (G0 and G6) digested with EcoRV. Lane M (marker) is a HindIII digest of lambda DNA.
Influence of the last host on the phage phenotype.
The genotype of the host in which a virus reproduces affects the phenotype of the newly formed virus progeny (22). In contrast to mutation, antagonist pleiotropy may be applied simultaneously to almost all of the members of a developing phage population and generally is determined only by the nature of the last host in which the phage was replicated, being independent of the previous phage history (13). As a consequence of this ubiquitous phenomenon, the growth and adaptation of the virus to the new host will lead to the achievement of a new stock that may be divergent from the original (11, 12). Taking into account this science-based knowledge, it would be expected that the replication of a phage in a different host would lead to progeny with different characteristics. In fact, the phenomenon of host-controlled variation has already been reported for phages and is usually produced in a few growth cycles; in some cases, the phenotypic change is evident right after a single growth step where phages lose their ability to growth and lyse their original host (5, 8, 11, 13). Therefore, the use of a nonpathogenic host to produce phages for therapeutic purposes could be compromised by induced changes in the phage lytic spectrum.
In this study, the lytic activity of the phage after its replication in the nonpathogenic strain E. coli BL21 (G1 to G6) showed an efficiency equal to that of phi PVP-SE1 produced in Salmonella (G0), and thus, no host-induced modification of the phage or restriction by the pathogenic target bacteria occurred; i.e., there was no increase in the specificity of the phage to that strain with consequent narrowing of the host range. Therefore, in this case, it is expected that there will be no risk of treatment failure, in contrast to what happened in the past when phages were produced in laboratory strains and then used in therapy (36). Moreover, this work shows that the production of Salmonella phage phi PVP-SE1 in nonpathogenic E. coli BL21 maintained its ability to lyse Salmonella strains and did not induce DNA modifications visible by RFLP.
Importance of broad-host-range phages.
The aims of this work were to select and characterize a phage with a broad host range of Salmonella strains and to investigate the possibility of producing this phage in a nonpathogenic strain. The highly specialized nature of phages and the uncommon existence in nature of broad-host-range phages (with a host range over species borders) make the isolation of such phages a rare situation. In particular, the narrow host specificity of Salmonella phages and the relatively large number of pathogenic Salmonella strains exacerbate this difficulty (16, 29). The phages used in this study might thus have added value in phage therapy since all of them present a broad host range among Salmonella strains. Of particular interest is phage phi PVP-SE1, which was able to lyse almost all of the Salmonella strains used in this work and also bacteria other than salmonellae, e.g., the nonpathogenic strain E. coli BL21. It is believed that the broader the host range of a phage, the higher the probability of success in phage therapy and also of finding alternative hosts in which to reproduce the phage (18).
Comparing the lytic spectra of this phage and the well-known phage Felix-O1 (a virulent phage originally isolated by Felix and Callow in 1943 [20]) among the isolates tested, it was observed that phi PVP-SE1 presents a broader host range than Felix-O1. Felix-O1 is used routinely as a diagnostic tool in the identification of salmonellae due to its ability to lyse up to 99.5% of Salmonella strains (17). Thus, like Felix-O1, newly isolated phage phi PVP-SE1 can also be an excellent diagnostic tool.
Concluding remarks.
This work reports the selection and characterization of phi PVP-SE1, a phage with a broad host range among salmonellae, which represents an added value in phage therapy. This phage can thus be used in the creation of a small library of phages able to act as biocontrol agents in salmonellosis. Phage phi PVP-SE1 presents a broad lytic spectrum, one even broader than that of Salmonella-specific phage Felix-O1. The most important characteristic of this phage was the ability to lyse and consequently be produced in nonpathogenic E. coli, and mainly without modification of its lytic spectrum, in this way ensuring its stability when used in phage therapy. Furthermore, the use of this nonpathogenic E. coli strain in phi PVP-SE1 production will surely facilitate the production and purification processes by eliminating the risk of introducing a phage-resistant pathogenic bacterium. The consequent cost reduction and increased safety of the phage preparations will lead to easier and faster approval for commercial applications.
Acknowledgments
We acknowledge financial support through the European project Phagevet-P (project 2005-7224) and grant SFRH/BD/32278/2006 from the Fundação para a Ciência e Tecnologia (FCT-Portugal).
We are grateful to H.-W. Ackermann for the morphological characterization of the phages.
Footnotes
Published ahead of print on 3 September 2010.
REFERENCES
- 1.Ackermann, H.-W. 2007. 5500 Phages examined in the electron microscope. Arch. Virol. 152:227-243. [DOI] [PubMed] [Google Scholar]
- 2.Arlet, G., T. J. Barrett, P. Butaye, A. Cloeckaert, M. R. Mulvey, and D. G. White. 2006. Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8:1945-1954. [DOI] [PubMed] [Google Scholar]
- 3.Atterbury, R. J., M. A. P. Van Bergen, F. Ortiz, M. A. Lovell, J. A. Harris, A. De Boer, J. A. Wagenaar, V. M. Allen, and P. A. Barrow. 2007. Bacteriophage therapy to reduce Salmonella colonization of broiler chickens. Appl. Environ. Microbiol. 73:4543-4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrow, P. A., and J. S. Soothill. 1997. Bacteriophage therapy and prophylaxis: rediscovery and renewed assessment of potential. Trends Microbiol. 5:268-271. [DOI] [PubMed] [Google Scholar]
- 5.Bertani, G., and J. J. Weigle. 1953. Host controlled variation in bacterial viruses. J. Bacteriol. 65:113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bielke, L. R., S. Higgins, A. M. Donoghue, D. J. Donoghue, and B. M. Hargis. 2007. Salmonella host range of bacteriophages that infect multiple genera. Poult. Sci. 86:2536-2540. [DOI] [PubMed] [Google Scholar]
- 7.Bielke, L. R., S. E. Higgins, A. M. Donoghue, D. J. Donoghue, B. M. Hargis, and G. Tellez. 2007. Use of wide-host-range bacteriophages to reduce Salmonella on poultry products. Int. J. Poult. Sci. 6:754-757. [Google Scholar]
- 8.Brunovskis, I., and R. O. Burns. 1973. Growth of coliphage T7 in Salmonella typhimurium. J. Virol. 11:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2005. Salmonella surveillance: annual summary, 2004. Centers for Disease Control and Prevention, Atlanta, GA.
- 10.Clark, J. R., and J. B. March. 2006. Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends Biotechnol. 24:212-218. [DOI] [PubMed] [Google Scholar]
- 11.Crill, W. D., H. A. Wichman, and J. J. Bull. 2000. Evolutionary reversals during viral adaptation to alternating hosts. Genetics 154:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elena, S. F., and R. Sanjuán. 2003. Climb every mountain? Science 302:2074-2075. [DOI] [PubMed] [Google Scholar]
- 13.Eskridge, R. W., H. Weinfeld, and K. Paigen. 1967. Susceptibility of different coliphage genomes to host-controlled variation. J. Bacteriol. 93:835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.European Food Safety Authority. 2004. The community summary report on trends and sources of zoonoses, zoonotic agents and antimicrobial resistance in the European Union in 2006. European Food Safety Authority, Parma, Italy.
- 15.Fiorentin, L., N. D. Vieira, and W. Barioni. 2005. Oral treatment with bacteriophages reduces the concentration of Salmonella Enteritidis PT4 in caecal contents of broilers. Avian Pathol. 34:258-263. [DOI] [PubMed] [Google Scholar]
- 16.Higgins, J. P., S. E. Higgins, K. L. Guenther, W. Huff, A. M. Donoghue, D. J. Donoghue, and B. M. Hargis. 2005. Use of a specific bacteriophage treatment to reduce Salmonella in poultry products. Poult. Sci. 84:1141-1145. [DOI] [PubMed] [Google Scholar]
- 17.Hirsh, D. C., and L. D. Martin. 1983. Rapid detection of Salmonella spp. by using Felix-O1 bacteriophage and high-performance liquid chromatography. Appl. Environ. Microbiol. 45:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kropinski, A. M. 2006. Phage therapy—everything old is new again. Can. J. Infect. Dis. Med. Microbiol. 17:297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krylov, V. N., S. Miller, R. Rachel, M. Biebl, E. A. Pleteneva, M. Schuetz, S. V. Krylov, and O. V. Shaburova. 2006. Ambivalent bacteriophages of different species active on Escherichia coli K12 and Salmonella sp. strains. Russ. J. Genet. 42:106-114. [PubMed] [Google Scholar]
- 20.Kuhn, J., M. Suissa, J. Wyse, I. Cohen, I. Weiser, S. Reznick, S. Lubinsky-Mink, G. Stewart, and S. Ulitzur. 2002. Detection of bacteria using foreign DNA: the development of a bacteriophage reagent for Salmonella. Int. J. Food Microbiol. 74:229-238. [DOI] [PubMed] [Google Scholar]
- 21.Kutter, E., and A. Sulakvelidze. 2005. Bacteriophages. Biology and applications. CRC Press, Inc., Boca Raton, FL.
- 22.Luria, S. E., and M. L. Human. 1952. A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol. 64:557-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuzaki, S., L. Rashel, J. Uchiyama, S. Sakurai, T. Ujihara, M. Kuroda, M. Ikeuchi, T. Tani, M. Fujieda, H. Wakiguchi, and S. Imai. 2005. Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J. Infect. Chemother. 11:211-219. [DOI] [PubMed] [Google Scholar]
- 24.Merril, C. R., B. Biswas, R. Carlton, N. C. Jensen, G. J. Creed, S. Zullo, and S. Adhya. 1996. Long-circulating bacteriophage as antibacterial agents. Proc. Natl. Acad. Sci. U. S. A. 93:3188-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petty, N. K., T. J. Evans, P. C. Fineran, and G. P. C. Salmond. 2007. Biotechnological exploitation of bacteriophage research. Trends Biotechnol. 25:7-15. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Santander, J., and J. Robeson. 2004. Bacteriophage prophylaxis against Salmonella enteritidis and Salmonella pullorum using Caenorhabditis elegans as an assay system. Electronic J. Biotechnol. 7:208-211. [Google Scholar]
- 28.Sillankorva, S., E. Pleteneva, O. Shaburova, S. Santos, C. Carvalho, J. Azeredo, and V. Krylov. 2010. Salmonella Enteritidis bacteriophage candidates for phage therapy of poultry. J. Appl. Microbiol. 108:1175-1186. [DOI] [PubMed] [Google Scholar]
- 29.Skurnik, M., M. Pajunen, and S. Kiljunen. 2007. Biotechnological challenges of phage therapy. Biotechnol. Lett. 29:995-1003. [DOI] [PubMed] [Google Scholar]
- 30.Skurnik, M., and E. Strauch. 2006. Phage therapy: facts and fiction. Int. J. Med. Microbiol. 296:5-14. [DOI] [PubMed] [Google Scholar]
- 31.Sulakvelidze, A., Z. Alavidze, and J. G. Morris. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tinsley, C. R., E. Bille, and X. Nassif. 2006. Bacteriophages and pathogenicity: more than just providing a toxin? Microbes Infect. 8:1365-1371. [DOI] [PubMed] [Google Scholar]
- 33.Velge, P., A. Cloeckaert, and P. Barrow. 2005. Emergence of Salmonella epidemics: the problems related to Salmonella enterica serotype Enteritidis and multiple antibiotic resistance in other major serotypes. Vet. Res. 36:267-288. [DOI] [PubMed] [Google Scholar]
- 34.Villegas, A., Y. M. She, A. M. Kropinski, E. J. Lingohr, A. Mazzocco, S. Ojha, T. E. Waddell, H.-W. Ackermann, D. M. Moyles, R. Ahmed, and R. P. Johnson. 2009. The genome and proteome of a virulent Escherichia coli O157:H7 bacteriophage closely resembling Salmonella phage Felix O1. Virol. J. 6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker, K. 2006. Use of bacteriophages as novel food additives. In N. D. Fortin (ed.), Food & drug regulation: a web book of student papers. Institute for Food Laws and Regulations, East Lansing, MI.
- 36.Withey, S., E. Cartmell, L. M. Avery, and T. Stephenson. 2005. Bacteriophages—potential for application in wastewater treatment processes. Sci. Total Environ. 339:1-18. [DOI] [PubMed] [Google Scholar]