Abstract
Listeria monocytogenes is a Gram-positive, psychrotrophic, facultative intracellular food-borne pathogen responsible for severe illness (listeriosis). The bacteria can grow in a wide range of temperatures (1 to 45°C), and low-temperature growth contributes to the food safety hazards associated with contamination of ready-to-eat foods with this pathogen. To assess the impact of oxidative stress responses on the ability of L. monocytogenes to grow at low temperatures and to tolerate repeated freeze-thaw stress (cryotolerance), we generated and characterized a catalase-deficient mutant of L. monocytogenes F2365 harboring a mariner-based transposon insertion in the catalase gene (kat). When grown aerobically on blood-free solid medium, the kat mutant exhibited impaired growth, with the extent of impairment increasing with decreasing temperature, and no growth was detected at 4°C. Aerobic growth in liquid was impaired at 4°C, especially under aeration, but not at higher temperatures (10, 25, or 37°C). Genetic complementation of the mutant with the intact kat restored normal growth, confirming that inactivation of this gene was responsible for the growth impairment. In spite of the expected impact of oxidative stress responses on cryotolerance, cryotolerance of the kat mutant was not affected.
Listeria monocytogenes is a Gram-positive, facultative intracellular food-borne pathogen that has the ability to cause a severe disease (listeriosis) in humans and animals (13, 28, 30). L. monocytogenes is ubiquitously distributed in the environment and has the ability to grow over a wide range of temperatures (between 1 and 45°C) (13). Growth at low temperature has important implications for environmental persistence of the organism and for contamination of cold-stored, ready-to-eat foods, thus contributing to the food safety hazards associated with L. monocytogenes (19).
L. monocytogenes is subjected to oxidative stress during both extracellular and intracellular growth and has evolved several responses to minimize the impact of reactive oxygen species (ROS). Catalase and superoxide dismutase (SOD) work synergistically in detoxification of ROS: superoxide anions are converted to H2O2 by SOD, with subsequent conversion of H2O2 into water and oxygen by catalase (22). Exposure to ROS may be especially acute during intracellular infection as well as under certain environmental conditions, such as those involved in repeated freezing and thawing (15, 16, 23, 29, 33).
Previous studies revealed that the ability of L. monocytogenes to survive repeated freezing and thawing (cryotolerance) was markedly dependent on growth temperature, with bacteria grown at 37°C having significantly higher cryotolerance than those grown at either 4 or 25°C (1). However, mechanisms underlying Listeria's cryotolerance have not been identified. Since oxidative damage is considered to take place during freezing and thawing, determinants such as catalase may be involved in cryotolerance.
The catalase of L. monocytogenes has been investigated primarily in terms of its potential role in pathogenesis, with somewhat conflicting results. The isolation of catalase-negative strains from human listeriosis patients has led to the speculation that catalase is not required for human virulence (4, 8, 12, 31). On the other hand, under certain conditions (e.g., reduced serum levels), catalase-negative strains were impaired in their ability to survive in activated macrophages in comparison to catalase-positive strains (32). Furthermore, the catalase gene kat was among those for which expression was induced in infected cell cultures and in the spleens of mice infected with L. monocytogenes EGD-e, suggesting possible contributions to pathogenesis (5, 9).
The potential role of catalase in environmental adaptations of L. monocytogenes such as growth at low temperature and cryotolerance was not addressed in these earlier investigations. In this study, we have characterized an isogenic mutant of L. monocytogenes F2365 to determine the involvement of catalase in growth at different temperatures, survival in selected foods, and cryotolerance of L. monocytogenes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. L. monocytogenes F2365 was implicated in the California outbreak of 1985 and has had its genome completely sequenced (24). Before each experiment bacteria were grown on Trypticase soy agar with 5% sheep blood (Remel, Lenexa, KS) at 37°C for 36 h. A single colony was transferred into 5 ml of tryptic soy broth (TSB; BBL, Sparks, MD) supplemented with 0.7% yeast extract (TSBYE; BBL), and the cultures were incubated at 37°C overnight. Motility assays utilized soft agar medium (TSBYE with 0.4% agar [TSAYE]). For growth under microaerobic conditions, we employed a CampyPak Microaerophilic System (BBL, Sparks, MD) at 37°C for 36 h and at 25°C for 48 h. The gas mix generated by these packs consist of 80% nitrogen, 7.5% hydrogen, 7.5% carbon dioxide, and 5% oxygen (2). Growth in liquid was monitored (optical density at 600 nm [OD600]) using a spectrophotometer (Bio-Rad SmartSpec 3000; Hercules, CA) with ca. 106 CFU/ml as the inoculum. The OD600 readings were taken every 1 h (37°C), every 24 h (10°C), and every 3 days (4°C). In addition, we monitored growth of the mutant and the wild type in liquid at 4°C with aeration (150 rpm).
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype and/or description | Source or reference |
|---|---|---|
| L. monocytogenes | ||
| F2365 | Serotype 4b strain from 1985 California outbreak, epidemic clone I | 24 |
| ROA3 | Catalase-negative transposon mutant of F2365 with insertion in kat | This study |
| ROA3C | Genetically complemented ROA3 with integrated pPL2::kat | This study |
| ROA3E | Genetically complemented ROA3 with integrated pPL2 | This study |
| E. coli SM10 | Conjugation donor; F−thi-1 thr-1 leuB6 recA tonA21 lacY1 supE44 (MuC+) λ− [RP4-2(Tc::Mu)] Kmr Tra+ | D. Elhanafi |
When needed, antibiotics used with L. monocytogenes were chloramphenicol (6 μg/ml), erythromycin (5 μg/ml), and nalidixic acid (20 μg/ml). Escherichia coli strains were grown in Luria-Bertani (LB; BBL, Sparks, MD) broth or agar medium supplemented with chloramphenicol (25 μg/ml). Antibiotics were purchased from Sigma-Aldrich (St. Louis, MO).
Identification of catalase-negative mutant ROA3.
A transposon mutant library of L. monocytogenes F2365 was constructed with the mariner-based transposition system using pMC38 as described previously (1a, 6). About 3,400 mariner-based transposon mutants of L. monocytogenes F2365 were screened for catalase activity. The mutants were grown overnight at 37°C on brain heart infusion (BHI) agar (BBL, Sparks, MD). Hydrogen peroxide (H2O2; 2 μl of a 30% [vol/vol] solution) (Superoxol; J. T. Baker Chemical Co., Phillipsburg, NJ) was dropped on the agar cultures, and absence of bubbling indicated the loss of catalase activity. The transposon insertion site was determined by sequencing DNA fragments amplified by two-step arbitrary PCR using primers Marq207, Marq208, Marq255, and Marq269 as described by Cao et al. (6). The PCR product was sequenced (Genewiz Inc., South Plainfield, NJ), and the sequence was analyzed by nucleotide BLAST. Transposon copy number was determined by Southern blotting using digoxigenin-labeled ermC of pMC38 (6). Probe construction and hybridizations were performed as described previously (10).
Genetic complementation.
The promoter and the transcription termination regions of kat were identified earlier in Listeria seeligeri (17), and sequence comparisons revealed that the promoter and transcription termination sites were conserved between L. monocytogenes F2365 and L. seeligeri (data not shown). Primers ROA3F (5′-CTAACCCGGGCGATGAATTAGGTCGTCTGT-3′; XmaI site underlined) and ROA3R2 (5′-GTAAGAGCTCTTACTCCAATCTTCTAGCC-3′; SacI site underlined) were used to amplify a fragment that included the kat coding sequence and the putative promoter and terminator. The PCR product was digested with XmaI and SacI (New England Biolabs, Ipswich, MA) and purified using a PCR purification kit (Qiagen, Valencia, CA). The purified PCR product was ligated into the L. monocytogenes site-specific integration vector pPL2 (20), which was digested with the same restriction enzymes, yielding pPL2kat. The recombinant plasmid was electroporated into E. coli SM10 as described previously (10). Conjugation was performed to mobilize pPL2kat into ROA3 as described previously (1a, 10), yielding the genetically complemented mutant, designated ROA3C. Transconjugants were selected using chloramphenicol and nalidixic acid, as described previously (10). The empty vector pPL2 was similarly mobilized by conjugation into ROA3, yielding ROA3E.
Assessments of cryotolerance.
ROA3 and the wild-type parental strain F2365 were grown in TSBYE medium at 25°C (48 h) or 37°C (36 h), and samples (1.5 ml) were subjected to 18 cycles of repeated freezing (−20°C) and thawing, as described previously (1). Cryotolerance was assessed by enumeration of cells surviving following freezing and thawing, as described previously (1).
Survival and growth of catalase mutant in selected foods.
Pasteurized and raw milk were provided from the Food Science Dairy and Process Applications Laboratory at North Carolina State University. Overnight 37°C cultures of F2365 and the catalase mutant ROA3 were inoculated in raw or pasteurized milk (1 ml of culture into 99 ml of milk) and incubated at 4°C and 25°C for 21 days. To assess survival/growth of the bacteria in the milk, samples of appropriate dilutions were plated on modified Oxford selective medium (Oxoid, Basingstoke, England), and the numbers of CFU were determined following incubation at 37°C for 36 h. Soft cheese (queso fresco, made with pasteurized milk) was purchased at retail, cut aseptically into rectangular fragments, and surface inoculated with 100 μl of a cell suspension (ca. 105 CFU) of the bacteria. The inoculated fragments were incubated at 10°C over 10 days. At selected time points (immediately after inoculation and at 4 and 10 days) four samples inoculated with each strain were added to 100 ml of phosphate-buffered saline (PBS) and shaken vigorously, and viable bacteria in the liquid were enumerated as described above. All food inoculations were done in at least two independent trials.
Chicken embryo infections.
Overnight cultures were serially diluted in PBS, and 0.1 ml of the 10−5 dilution was inoculated into 10-day-old embryonated hen eggs via the chorioallantoic membrane, as described previously (25). Five embryos were inoculated for each strain. Listeria innocua LO718 and PBS were used as negative controls. Inoculated embryos were incubated at 37°C horizontally. Survival of embryos was monitored daily over 3 days, using transillumination.
Sequence comparison of inactivated genes between L. monocytogenes F2365 and EGD-e.
The kat genomic regions were compared by using the online sequence comparison tool WebACT. WebACT uses the Artemis Comparison Tool (ACT) developed by the Sanger Institute (7).
Statistical analysis.
All treatment combinations were replicated at least twice. For the comparison of the cryotolerance of the mutants, a log reduction of each sample was determined following 18 freeze-thaw cycles. The growth behaviors of ROA3 and F2365 were compared by growth rates and final cell densities. The differences between each strain and treatment combination were analyzed by performing analyses of variance. The significance was determined at an unadjusted level of an α of 0.05. All statistical analyses were performed using SAS, version 9.1 (Cary, NC).
RESULTS
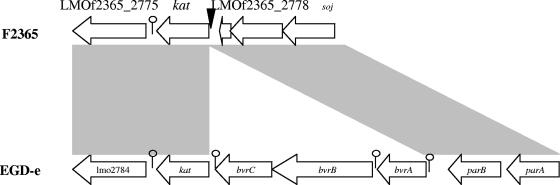
Of the ca. 3,400 transposon mutants, one (ROA3) was presumed to be catalase negative based on the absence of gas bubbles upon exposure to H2O2. Southern blotting with the erythromycin resistance gene (ermC) of pMC38 as the probe revealed the presence of a single transposon insertion in ROA3 (data not shown). Arbitrary PCR indicated that the transposon insertion was in the coding sequence (nucleotide [nt] 2833280 in the genome of F2365) of kat, the gene encoding catalase (Fig. 1). Sequence alignments of the kat genomic region between F2365 and EGD-e (serotype 1/2a) revealed that in EGD-e kat is preceded by a cassette of three genes (bvrABC) involved in β-glucoside metabolism, whereas F2365 lacks this cassette (Fig. 1). The serotype 4b strain H7858 had a kat region similar to that of F2365 (data not shown). These findings are in agreement with a previous report according to which the bvrABC locus was harbored only by strains of lineage II (serotypes 1/2a, 1/2c, 3a, and 3c) and serotype 4c (11).
FIG. 1.
Comparison of the kat genomic regions between L. monocytogenes F2365 and L. monocytogenes EGD-e. Transposon insertion in ROA3 is shown (▾). The putative rho-independent terminators are shown by a lollipop symbol.
Absence of catalase impacts growth on agar (TSAYE) under aerobic conditions, especially at low temperatures.
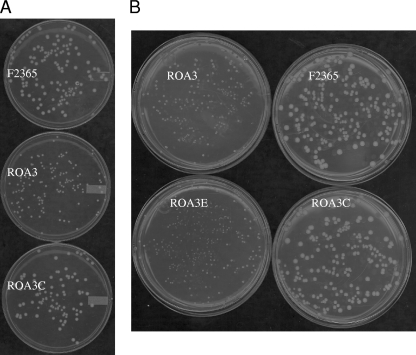
When bacteria were grown on TSAYE medium and incubated aerobically at 37°C, it was noted that colony size of the catalase mutant ROA3 was about half that of F2365 (Fig. 2 A). The impact of the catalase mutation on colony size was more pronounced with decreasing temperature: colony size of ROA3 was further reduced (in comparison to F2365) at 25°C (data not shown) and even more so at 10°C (Fig. 2B). At 4°C ROA3 failed to form colonies following 28 days of incubation (data not shown).
FIG. 2.
Impact of temperature on colony size of kat mutant ROA3. (A) Growth of F2365, ROA3, and ROA3C on TSAYE medium following 48 h of incubation at 37°C. (B) Growth of F2365, ROA3, ROA3C, and ROA3E on TSAYE medium following 7 days of incubation at 10°C.
Genetic complementation confirmed that the decreased growth of ROA3 on agar was due to the inactivation of kat. The genetically complemented mutant ROA3C, harboring an integrated intact kat, exhibited normal colony size at all tested temperatures (Fig. 2), whereas ROA3E harboring the empty integration vector was indistinguishable from ROA3 (Fig. 2B).
In contrast to the colony size impact observed with growth on TSAYE medium under aerobic conditions, colony size of ROA3 was not impacted by growth on TSAYE medium incubated microaerobically (data not shown). Furthermore, colony size of ROA3 was indistinguishable from that of F2365 when the bacteria were grown aerobically on blood agar plates at 4, 10, 25, or 37°C (data not shown).
Absence of catalase results in impaired growth in liquid at 4°C, but not at 10, 25, or 37°C.
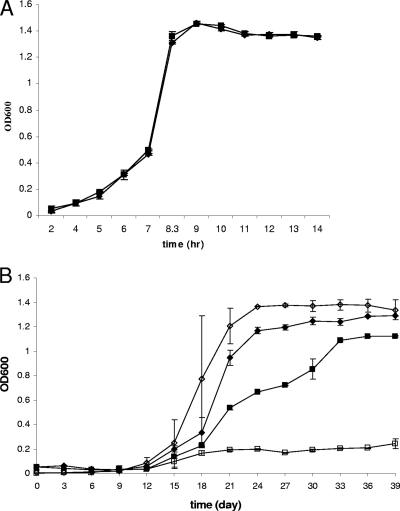
In liquid medium (TSBYE) under static conditions, growth curves of ROA3 were not significantly different from those of F2365 at 10, 25, or 37°C (Fig. 3 A; also data not shown). At 4°C, however, ROA3 grew significantly more slowly than F2365 (P < 0.05). Furthermore, final cell density of ROA3 grown at 4°C was significantly lower than that of F2365 (P < 0.05). Growth of ROA3 was inhibited even further when the cells were grown at 4°C with agitation (Fig. 3B). Growth at 4°C in liquid was completely restored in the genetically complemented mutant ROA3C (data not shown).
FIG. 3.
Impact of temperature on growth of kat mutant ROA3 in liquid medium. L. monocytogenes F2365 (⧫) and ROA3 (▪) were grown at 37°C (A) and 4°C (B) statically; F2365 (⋄) and ROA3 (□) were grown at 4°C with agitation (150 rpm) in BHI broth, and growth was monitored as described in Materials and Methods.
Catalase is not required for cryotolerance of L. monocytogenes.
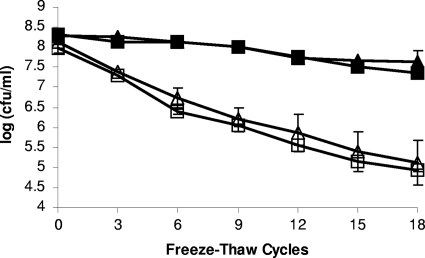
To determine the impact of catalase on the cryotolerance of L. monocytogenes, we screened the survival of F2365 and ROA3 grown in liquid medium at 25°C and 37°C following 18 freeze-thaw cycles. As previously observed (1), cells grown at 37°C were more cryotolerant than those grown at 25°C. However, cryotolerance of ROA3 was indistinguishable from that of F2365, regardless of whether the cells were grown at 25°C or 37°C (Fig. 4). Freeze-thaw tolerance was also assessed over 18 freeze-thaw cycles with F2365 and ROA3 grown on agar at 25°C. Survival following repeated freezing and thawing was higher than with liquid-grown cells, as previously observed (1), but no significant differences in survival were noted between F2365 and ROA3; both strains exhibited ca. 2-log reductions following 18 cycles (data not shown).
FIG. 4.
Freeze-thaw tolerance of kat mutant ROA3. L. monocytogenes F2365 (triangles) and ROA3 (squares) were grown at 37°C (filled symbols) or at 25°C (open symbols) and exposed to 18 repeated freeze-thaw cycles. Survival was assessed as described in Materials and Methods.
Survival or growth in selected foods was not affected in the catalase mutant.
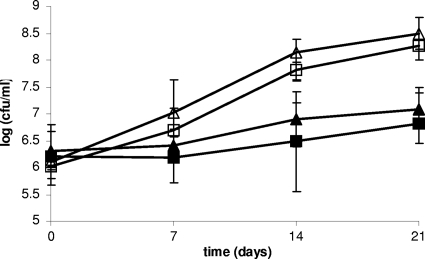
Both F2365 and ROA3 grew at 4°C in pasteurized milk over a 21-day period, and no significant differences were detected between the two strains (Fig. 5). Although growth in raw milk over the same time period was suppressed, bacteria survived, and there were no detectible differences between F2365 and ROA3 (Fig. 5). F2365 and ROA3 grew similarly on the inoculated pasteurized soft cheese samples at 10°C, with ca. a 2-log increase over the 10-day period (data not shown).
FIG. 5.
Growth of kat mutant ROA3 in milk. Raw milk (filled symbols) and pasteurized milk (open symbols) were inoculated with L. monocytogenes F2365 (triangles) and ROA3 (squares) and incubated at 4°C over 21 days. Growth was monitored as described in Materials and Methods.
Catalase-negative bacteria remain virulent in the chicken embryo model.
No significant differences were noted in survival rates of embryonated eggs inoculated with F2365 or mutant ROA3. By day 2, all embryos were killed, regardless of whether the inoculum was F2365 or ROA3. No embryo mortalities were seen for eggs inoculated with L. innocua or PBS (data not shown).
DISCUSSION
In this study, the role of catalase in the growth of L. monocytogenes was assessed by using a catalase-negative transposon mutant of L. monocytogenes F2365 and its genetically complemented derivative. The catalase-negative mutant showed impaired growth on solid medium at all temperatures tested, except when the bacteria were grown microaerobically or on blood agar. As the temperature decreased, there was a pronounced increase in the impact of the mutation on growth on solid medium, with no growth on TSAYE agar plates incubated aerobically at 4°C.
The fact that growth was normal on blood agar plates may reflect the ability of heme to quench some of the oxygen or the presence of catalase in the blood. This, along with normal growth on TSAYE medium under microaerobic conditions, suggests that catalase contributes to protecting the cells against damaging effects of reactive oxygen species produced during aerobic growth. The observed increasing growth impairment of the catalase-negative mutant with decreasing temperature suggests that when L. monocytogenes grows aerobically on agar, production of reactive oxygen species increases with decreasing temperature, resulting in increased requirements for a functional catalase. Impact of the mutation on low-temperature growth was also observed when the bacteria were grown at 4°C in liquid medium as the lag phase was significantly increased in the mutant, and its final cell density was lower than observed with the parental strain F2365. The inhibitory impact of the mutation on growth at 4°C in liquid was markedly enhanced upon aeration of the cultures, suggesting increased damage due to oxidative stress. Interestingly, the kat mutant's survival or growth in milk at 4°C or on cheese at 10°C was not significantly different from that of the wild-type parental strain. One may speculate that components such as SOD in the foods compensated for the absence of catalase in the mutant by reducing reactive oxygen intermediates in the environment of the bacteria. SOD is present in milk and can remain active after pasteurization (18).
These findings were of special interest as they suggest that catalase may be one of the contributors to the ability of L. monocytogenes to grow aerobically at low temperatures, especially on solid medium that lacks oxygen-quenching components. The restoration of the growth impairment in the genetically complemented mutant confirmed that the effects were specifically associated with the inactivation of kat. SOD activity has been reported to be increased in kat mutants (21, 34). SOD levels of F2365 and ROA3 were not assessed in this study. Further studies will be required to assess the possibly synergistic impact of these enzymes on aerobic growth of L. monocytogenes, especially at low temperatures.
Previous studies investigated the role of catalase in virulence of L. monocytogenes (21, 34). In addition, several reports have described catalase-negative strains from human listeriosis cases, suggesting that catalase was not required for virulence (4, 8, 12, 31). This was also supported by our findings using the chicken embryo model. However, formation of smaller colonies by catalase-negative mutants or field isolates has not been reported before in Listeria. It is noteworthy that small-colony variants of other pathogens were isolated from human clinical samples and reported to be deficient in catalase (14, 27). Mutations in the heme biosynthesis pathway were implicated in the phenotype of these small-colony variants in E. coli (27).
In L. monocytogenes, impact of catalase loss on growth would have been especially noticeable if isogenic kat mutants, such as those described by Leblond-Francillard et al. (21), were grown under specific conditions, such as aerobically on blood-free agar, and at a low temperature (e.g., 4°C). Growth under such conditions was not reported in these earlier studies, possibly accounting for the lack of reports on impaired growth. Differences in the strains used may also account for impaired growth in the mutant of F2365 and possibly not in kat mutants of other strains. Previously described transposon mutants in kat were constructed in strain EGD (serotype 1/2a) (21). In this strain kat is preceded by the bvrABC locus that appears to be lacking from the genome of F2365 and other serotype 4b strains (11). However, we think that the absence of bvrABC from the genome of F2365 is not a likely reason for the impaired growth as the kat promoter region appears identical in EGD-e and F2365, and a transcriptional terminator was identified downstream of bvrC in EGD-e (3).
In L. monocytogenes inactivation of perR, implicated in defense against peroxide and ROS stress, resulted in reduced colony size and enhanced susceptibility to peroxide (26) although the possible impact of growth temperature on the perR mutant phenotype was not reported. Interestingly, catalase transcription and activity was enhanced in the perR mutant, and genetic constructs overproducing catalase also had reduced colony size. Such findings led to the speculation that the increased expression of catalase was toxic to the cells and contributed to decreased colony size (26). These data, together with the data from the current study, suggest that growth of L. monocytogenes is compromised by both increased expression and inactivation of kat, with inactivation conferring temperature-dependent inhibition of growth and loss of catalase activity.
The absence of an impact of catalase loss on freeze-thaw tolerance of Listeria observed in this study was surprising. Cells are subjected to oxygen stress during thawing (15, 16), and SOD has been reported to be important in the freeze-thaw tolerance of other bacteria (16, 29). SOD and other oxidative stress enzymes may compensate for the absence of catalase to protect the bacteria against the damaging effects of ROS generated during freezing and thawing. This may be especially likely if SOD levels are increased in the kat mutant, as described with other catalase-deficient strains of L. monocytogenes (21, 34).
In conclusion, our findings revealed that growth impairment of the kat mutant increased with decreasing temperature and was significantly more pronounced on solid medium than in planktonic cells. The data suggest enhanced peroxide stress for aerobically grown surface-associated bacteria, especially at low temperatures. This may have important implications for environmental survival and persistence of L. monocytogenes in natural ecosystems as well as in the processing plant environment and on foods.
Acknowledgments
Funding for this research was partially provided by USDA grant 2006-35201-17377.
We thank H. Marquis for the gift of pMC38. We are grateful to J. Guy for assistance with the chicken embryo study and to G. Cartwright and K. Hollifield for the samples of raw and pasteurized milk. We are grateful to R. M. Siletzky for laboratory support and to all other members of our laboratory for discussions, encouragement, and support in the course of this project.
Footnotes
Published ahead of print on 3 September 2010.
REFERENCES
- 1.Azizoglu, R. O., J. Osborne, S. Wilson, and S. Kathariou. 2009. Role of growth temperature in freeze-thaw tolerance of Listeria spp. Appl. Environ. Microbiol. 75:5315-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Azizoglu, R. O., and S. Kathariou. 2010. Inactivation of a cold-induced putative RNA helicase gene of Listeria monocytogenes is accompanied by failure to grow at low temperatures but does not affect freeze-thaw tolerance. J. Food Prot. 73:1474-1479. [DOI] [PubMed] [Google Scholar]
- 2.Bolton, F. J., D. R. A. Wareing, and A. D. Sails. 1997. Comparison of a novel microaerobic system with three other gas-generating systems for the recovery of Campylobacter species from human faecal samples. Eur. J. Clin. Microbiol. Infect. Dis. 16:839-842. [DOI] [PubMed] [Google Scholar]
- 3.Brehm, K., M. T. Ripio, J. Kreft, and J. A. Vazquez-Boland. 1999. The bvr locus of Listeria monocytogenes mediates virulence gene repression by β-glucosides. J. Bacteriol. 181:5024-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bubert, A., J. Riebe, N. Schnitzler, A. Schönberg, W. Goebel, and P. Schubert. 1997. Isolation of catalase-negative Listeria monocytogenes strains from listeriosis patients and their rapid identification by anti-p60 antibodies and/or PCR. J. Clin. Microbiol. 35:179-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camejo, A., C. Buchrieser, E. Couvé, F. Carvalho, O. Reis, P. Ferreira, S. Sousa, P. Cossart, and D. Cabanes. 2009. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 5:e1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, M., A. P. Bitar, and H. Marquis. 2007. A mariner-based transposition system for Listeria monocytogenes. Appl. Environ. Microbiol. 73:2758-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carver, T. J., K. M. Rutherford, M. Berriman, M. A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis Comparison Tool. J. Bioinformatics 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 8.Cepeda, J. A., M. Millar, E. A. Sheridan, S. Warwick, M. Raftery, D. C. Bean, and D. W. Wareham. 2006. Listeriosis due to infection with a catalase-negative strain of Listeria monocytogenes. J. Clin. Microbiol. 44:1917-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, Y., N. Promadej, J. W. Kim, and S. Kathariou. 2008. Teichoic acid glycosylation mediated by gtcA is required for phage adsorption and susceptibility of Listeria monocytogenes serotype 4b. Appl. Environ. Microbiol. 74:1653-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doumith, M. C., Cazalet, N. Simoes, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elsner, H.-A., I. Sobottka, A. Bubert, H. Albrecht, R. Laufs, and D. Mack. 1996. Catalase-negative Listeria monocytogenes causing lethal sepsis and meningitis in an adult hematologic patient. Eur. J. Clin. Microbiol. Infect. Dis. 15:965-966. [DOI] [PubMed] [Google Scholar]
- 13.Farber, K. R., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Funada, H., K. I. Hattori, and N. Kosakai. 1978. Catalase-negative Escherichia coli isolated from blood. J. Clin. Microbiol. 7:474-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, D., and J. K. Critser. 2000. Mechanisms of cryoinjury in living cells. ILAR J. 41:187-196. [DOI] [PubMed] [Google Scholar]
- 16.Garénaux, A., M. Ritz, F. Jugiau, F. Rama, M. Federighi, and R. de Jonge. 2009. Role of oxidative stress in C. jejuni inactivation during freeze-thaw treatment. Curr. Microbiol. 58:134-138. [DOI] [PubMed] [Google Scholar]
- 17.Haas, A., K. Brehm, J. Kreft, and W. Goebel. 1991. Cloning, characterization, and expression in Escherichia coli of a gene encoding Listeria seeligeri catalase, a bacterial enzyme highly homologous to mammalian catalases. J. Bacteriol. 173:5159-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks, C. L. 1980. Occurrence and consequence of superoxide dismutase in milk products: a review. J. Dairy Sci. 63:1199-1204. [DOI] [PubMed] [Google Scholar]
- 19.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity: a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 20.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leblond-Francillard, M., J.-L. Gaillard, and P. Berche. 1989. Loss of catalase activity in Tn1545-induced mutants does not reduce growth of Listeria monocytogenes in vivo. Infect. Immun. 57:2569-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mongkolsuk, S., and J. D. Helmann. 2002. Regulation of inducible peroxide stress response. Mol. Microbiol. 45:9-15. [DOI] [PubMed] [Google Scholar]
- 23.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 24.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Frase. 2004. Whole genome comparison of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olier, M., F. Pierre, J. P. Lemaitre, C. Divies, A. Rousset, and J. Guzzo. 2002. Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148:1855-1862. [DOI] [PubMed] [Google Scholar]
- 26.Rea, R., C. Hill, and C. G. M. Gahan. 2005. Listeria monocytogenes PerR mutants display a small-colony phenotype, increased sensitivity to hydrogen peroxide, and significantly reduced murine virulence. Appl. Environ. Microbiol. 71:8314-8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roggenkamp, A., A. Sing, M. Hornef, U. Brunner, I. B. Autenrieth, and J. Heesemann. 1998. Chronic prosthetic hip infection caused by a small-colony variant of Escherichia coli. J. Clin. Microbiol. 36:2530-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlech, W. F., III. 2000. Foodborne listeriosis. Clin. Infect. Dis. 31:770-775. [DOI] [PubMed] [Google Scholar]
- 29.Stead, D., and S. F. Park. 2000. Roles of Fe superoxide dismutase and catalase in resistance of Campylobacter coli to freeze-thaw stress. Appl. Environ. Microbiol. 66:3110-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swaminathan, B., and P. Gerner-Smidt. 2007. The epidemiology of human listeriosis. Microbes Infect. 9:1236-1243. [DOI] [PubMed] [Google Scholar]
- 31.Swartz, M. A., D. F. Welch, R. P. Narayanan, and R. A. Greenfield. 1991. Catalase-negative Listeria monocytogenes causing meningitis in an adult: clinical and laboratory features. Am. J. Clin. Pathol. 96:130-133. [DOI] [PubMed] [Google Scholar]
- 32.van Dissel, J. T., J. J. M. Stikkelbroeck, and R. van Furth. 1993. Differences in the rate of intracellular killing of catalase-negative and catalase-positive Listeria monocytogenes by normal and interferon-γ activated macrophages. Scand. J. Immunol. 37:443-446. [DOI] [PubMed] [Google Scholar]
- 33.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welch, D. F., C. P. Sword, S. Brehm, and D. Dusanic. 1979. Relationship between superoxide dismutase and pathogenic mechanisms of Listeria monocytogenes. Infect. Immun. 23:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]