Abstract
Polyoxins consist of 14 structurally variable components which differentiate at three branch sites of the carbon skeleton. Open reading frame (ORF) SAV_4805 of Streptomyces avermitilis, showing similarity to thymine-7-hydroxylase, was proved to enhance the diversity of polyoxins at the C-5 site of the 1-(5′-amino-5′-deoxy-β-d-allofuranuronosyl) pyrimidine moiety.
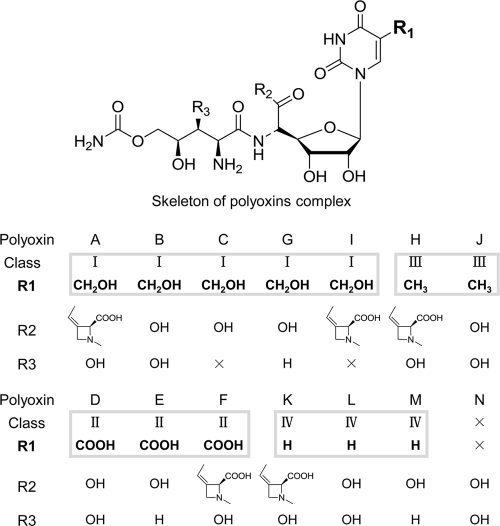
The antifungal nucleoside antibiotic polyoxins synthesized naturally by Streptomyces cacaoi subsp. asoensis (S. cacaoi here) consist of a mixture of at least 14 different compounds called polyoxins A to N (Fig. 1) (6, 13, 25). Most of the polyoxins have a common nucleoside skeleton that is attached with variable side groups at three different places (polyoxins C, I, and N have a modified skeleton). Polyoxins are grouped into four different classes according to the identity of the side group R1 attached at C-5 of the 1-(5′-amino-5′-deoxy-β-d-allofuranuronosyl) pyrimidine moiety. Different classes of polyoxins differ markedly in their activity spectra against plant pathogenic fungi (1, 10, 20).
FIG. 1.
Chemical structure of polyoxin complex. Class is determined by R1. The × indicates a different skeleton that does not have this particular residue.
It was deduced that the nucleoside moiety originated from the condensation of uridine with phosphoenolpyruvate (PEP) to generate octosyl acid as the intermediate (7, 12, 14). Then, a subsequent oxidative elimination of the two terminal carbons would create the nucleoside moiety (12). The detailed biosynthetic pathway of nucleoside moiety of polyoxins remains obscure. Our previous work showed that heterologous expression of the polyoxin biosynthetic gene cluster pol in Streptomyces lividans TK24 produces only thymine-derived polyoxin H (class III) (5). This prompted us to investigate the origin of a gene(s) related to the structural variation of polyoxins at the R1 site, which could be located outside the polyoxin biosynthetic gene cluster in the native producer.
The pyrimidine rings of polyoxins correspond one to one to the intermediates of the thymidine salvage pathway, which was characterized only for some fungi (see Fig. S1 in the supplemental material) (22). In the prime pathway of nucleotide metabolism, dUMP could be converted to dTMP under the catalysis of thymidylate synthase (TS) with tetrahydrofolic acid as a methyl donor while dTMP could not be converted back to dUMP reversibly (4, 17). In the 1970s, Shaffer et al. separated two enzymes, thymine-7-hydroxylase (THase; official name thymine dioxygenase) and isoorotate decarboxylase (IDCase), from fungal sources which showed potential ability to convert thymine to uracil (22). THase is a trifunctional oxygenase and catalyzes three enzymatic reactions: thymine to 5-hydroxymethyluracil, 5-hydroxymethyluracil to 5-formyluracil, and 5-formyluracil to uracil-5-carboxylic acid (also designated isoorotate) (18, 26). IDCase catalyzes the subsequent decarboxylation reaction, and uracil-5-carboxylic acid is converted to uracil in the end (21). This process is catalyzed by THase, and IDCase is the core step of the thymidine salvage pathway in fungi.
BLAST analysis results with the amino acid sequence of THase from Rhodotorula glutinis and IDCase from Neurospora crassa as queries in the 25 sequenced Streptomyces strains (5 finished and 20 in process) of the genome database (from NCBI GenBank, accessed 24 July 2010) showed that the THase and IDCase gene homologs are distributed extensively in Streptomyces, such as open reading frame (ORF) SAV_4805 (sharing 27% identity and 45% similarity with the THase gene) from S. avermitilis strain MA4680 and SCO_6305 (sharing 25% identity and 44% similarity with the IDCase gene) from S. coelicolor strain A3(2) (2, 11, 23). The results indicated a potential thymidine salvage pathway in Streptomyces, and the salvage pathway provides alternative nucleoside monophosphates as precursors for polyoxin biosynthesis.
ORF SAV_4805 enhanced the structural diversity of polyoxins.
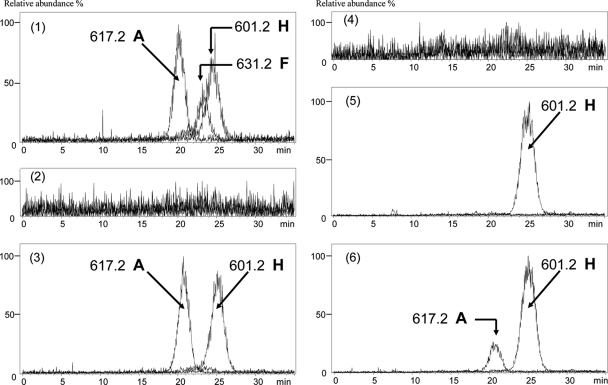
To investigate the deduced influence of this potential thymidine salvage pathway to the component variation of polyoxins, heterologous expression of the pol cluster in S. avermitilis strain NRRL8165 was conducted (Table 1). Cosmid m5A7 bearing the pol gene cluster was transferred into S. avermitilis strain NRRL8165 to give strain SJTU5003 by the method described in our previous work (5) (Table 1). Streptomyces was cultured in liquid fermentation medium [containing 20 g soy flour, 15 g corn flour, 10 g glucose, 10 g yeast extract, 4 g CaCO3, 2 g KH2PO4, 2 g NaCl, and 0.08 g FeSO4·(NH4)2SO4·6H2O per liter] at 30°C for polyoxin production. Fermentation supernatant was harvested at 72 h. The polyoxin complex was purified by IRC-50 cation exchange resin (Sigma) from the fermentation supernatant of recombinant strains and detected by liquid chromatography-mass spectrometry (LC-MS) as described previously (5). In contrast to the result with S. lividans TK24/m5A7, polyoxin A (hydroxymethyl at the R1 site) was detected from the fermentation supernatant of recombinant strain SJTU5003 apart from polyoxin H (methyl at R1 site) (Fig. 2, panel 3). This result indicated that some enzyme(s) in S. avermitilis NRRL8165 must have played a role in polyoxin structural variation at the R1 site.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain, plasmid, or cosmid | Description | Reference(s) |
|---|---|---|
| Strains | ||
| S. cacaoi subsp. asoensis | Polyoxin producer | 5 |
| S. coelicolor A3(2) | Wild-type strain | 2 |
| S. avermitilis NRRL8165 | Wild-type strain | 9 |
| S. lividans TK24/m5A7 | Recombinant strain for polyoxins heterologous producing | 5 |
| Streptomyces sp. strain SJTU5001 | S. lividans TK24/m5A7 harboring pJTU5001 | This study |
| Streptomyces sp. strain SJTU5003 | S. avermitilis NRRL8165 harboring cosmid m5A7 | This study |
| Streptomyces sp. strain SJTU5008 | S. avermitilis NRRL8165 harboring pJTU5007 | This study |
| Streptomyces sp. strain SJTU5009 | pyrC mutation derived from S. avermitilis NRRL8165 | This study |
| Streptomyces sp. strain SJTU5013 | Strain SJTU5009 harboring pJTU5010 | This study |
| Streptomyces sp. strain SJTU5014 | Strain SJTU5009 harboring pJTU5011 | This study |
| E. coli BW25113/pIJ790 | Host for PCR targeting | 8, 16 |
| E. coli ET12567/pUZ8002 | Host for E. coli/Streptomyces conjugation | 16, 19 |
| Plasmids and cosmids | ||
| pJTU968 | E. coli plasmid harboring a constitutive promoter of gene ermE* | 27 |
| pPM927 | Streptomyces integrating vector, ori(pBR322) int(pSAM2) attP(pSAM2) addA tsr oriT RK2 | 24 |
| pOJ260 | E. coli colon vector, ori(pUC18) addC(3)IV oriT RK2 | 3 |
| pSET152 | Streptomyces integrating vector, ori(pUC18) int ΦC31, attP ΦC31 aac(3)IV oriT RK2 | 3 |
| pJTU1278 | Streptomyces replicating vector, ori ColEI rep(pIJ101) bla tsr oriT RK2 | 9 |
| pJTU5001 | pPM927 harboring PermE*-SAV_4805 MfeI/EcoRI fragment | This study |
| pJTU5005 | pOJ260 harboring a 6.7-kb PstI DNA fragment which covers pyrC gene and the flank sequence | This study |
| pJTU5006 | pyrC gene of pJTU5005 replaced by nptII gene | This study |
| pJTU5007 | pJTU1278 harboring 8.1-kb EcoRI/SpeI fragment (covering nptII gene and the flank sequence) from pJTU5006 | This study |
| pJTU5010 | pSET152 harboring PermE*-SCO_6305 MfeI/EcoRI fragment | This study |
| pJTU5011 | pSET152 harboring PermE*-pyrC MfeI/EcoRI fragment | This study |
| Cosmid m5A7 | S. cacaoi genome library cosmid covers polyoxin biosynthetic gene cluster pol | 5 |
| Cosmid 20E4 | S. avermitilis NRRL8165 genome library cosmid covers pyrC gene | 9 |
FIG. 2.
Extracted ion graph from LC-MS analysis of polyoxin samples. Panels: 1, S. cacaoi; 2, S. avermitilis NRRL8165/pSET152; 3, S. avermitilis strain SJTU5003; 4, S. lividans TK24/pSET152; 5, S. lividans TK24/m5A7/pPM927; 6, strain SJTU5001. A, polyoxin A; H, polyoxin H; F, polyoxin F.
ORF SAV_4805 was amplified with primer pair 480501/480502 (GCCATATGAGTGACGCCCGTACCC/GGAATTCACGAAAGTTGCCCGCTGA; underlining indicates engineered sites introduced into the primers) with S. avermitilis strain NRRL8165 genome DNA used as a template and then digested with NdeI/EcoRI and inserted into the NdeI/EcoRI site of plasmid pJTU968 (a pRSET B derivative with PermE* promoter and a polylinker) (27). The PermE*-SAV_4805 MfeI/EcoRI fragment was separated out and inserted into the EcoRI site of plasmid pPM927 to create plasmid pJTU5001 (24). Then, plasmid pJTU5001 was transferred into S. lividans TK24/m5A7 to give strain SJTU5001 (Table 1). In the supernatant of PermE*-SAV_4805 and the pol gene cluster-coexpressing strain SJTU5001, polyoxin A was additionally detected, although its relative content was lower than that of S. cacaoi (Fig. 2, panel 6).
Unexpectedly, polyoxins harboring carboxyl at the R1 site (such as polyoxin F, shown in Fig. 2, panel 1) have not been detected from recombinant strain fermentation supernatants yet, which might be attributed to the weaker hydroxymethyl-to-carboxyl catalytic ability of enzyme encoded by SAV_4805 relative to that of THase from a fungal source.
The thymidine salvage pathway is not functional in S. avermitilis.
UMP is de novo biosynthesized from carbamoylphosphate and aspartate in organisms (15). The thymidine salvage pathway is a UMP compensatory pathway which was characterized in fungi only, not in mammals or prokaryotic bacteria. If the SAV_4805 protein works like THase and the SCO_6305 protein works like IDCase, it is possible to construct a complete thymidine salvage pathway in Streptomyces by recombining these two ORFs in one host, such as S. avermitilis NRRL8165. The recombinant S. avermitilis should exhibit a normal phenotype when thymine is fed, even though its UMP de novo biosynthetic pathway is interrupted, since the thymidine salvage pathway could convert thymine to uracil in vivo.
To knock out gene pyrC, which encodes dihydroorotase, to interrupt the UMP de novo biosynthetic pathway in S. avermitilis NRRL8165, cosmid 20E4 was selected out by PCR screening with primer pair validate1/validate2 (TTGGCGGCAACGAACCC/TGAGGGTGGTGGTGGGAGA) from the genome library of S. avermitilis strain NRRL8165. According to the genome sequence of S. avermitilis strain MA4680, a 6.7-kb PstI DNA fragment which covers the pyrC gene and its flanking sequence (2.4 kb upstream and 2.9 kb downstream) was separated out from cosmid 20E4 and inserted into the PstI site of plasmid pOJ260 to give recombinant plasmid pJTU5005 (3). The nptII gene fragment with 39-nucleotide (nt) homology extensions (underlined sequence) amplified by primer pair targeting1/targeting2 (CGTCAGCCACACCCGTATCGAGGAGAAGTAAGACAGATGAGCTATTCCAGAAGTAGTGAGG/TCTGTCCGGCGGCAAGGTACGTCTGATGCAGTGATGTCAGCTCTGGATGCCGACGGATTTG) was used to target plasmid pJTU5005 in Escherichia coli strain BW25113/pIJ790 (8, 16). Recombinant plasmid pJTU5006 was created when the pyrC gene (from ATG to TGA) was replaced by the nptII gene. The 8.1-kb EcoRI/SpeI fragment was separated out from plasmid pJTU5006 and inserted into the EcoRI/SpeI site of plasmid pJTU1278 to construct recombinant plasmid pJTU5007 (9). Plasmid pJTU5007 was transferred by conjugation into S. avermitilis strain NRRL8165 to give recombinant strain SJTU5008 (16, 19). After antibiotic-free culture and kanamycin selection, the double-crossover pyrC mutant strain SJTU5009 was picked up and confirmed by PCR with primer pair validate1/validate2 (Table 1).
The coding sequence of pyrC gene was amplified with primer pair dih1/dih2 (CACGACATATGAGCAAGATCCTGATCCGTGGTG/GGAATTCACTTCTGTCCGGCGGCAAGG), and fusion gene PermE*-pyrC was created similarly to the construction of PermE*-SAV_4805. The PermE*-pyrC MfeI/EcoRI fragment was inserted into the EcoRI site of plasmid pSET152 to create plasmid pJTU5011, and pJTU5011 was then introduced into strain SJTU5009 to give recombinant strain SJTU5014 (Table 1) (3).
ORF SCO_6305 was amplified by primer pair IDCase05/IDCase04 (GACATATGCCCACTCCAGCCGTGCCC/GCGAATTCAGTCGAGCTGCTTGTCGTAGTCC) with S. coelicolor strain A3(2) total DNA used as a template. Fusion gene PermE*-SCO_6305 was created and inserted into the EcoRI site of plasmid pSET152 to create plasmid pJTU5010; then pJTU5010 was introduced into strain SJTU5009 to give recombinant strain SJTU5013.
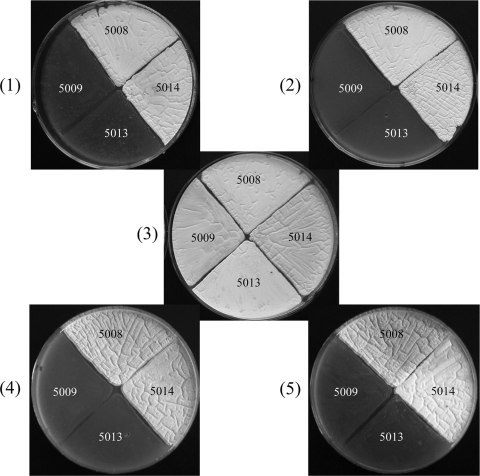
As shown in Fig. 3, the pyrC mutant strain SJTU5009 showed abnormal morphological differentiation under the growth conditions investigated while resuming a normal morphology when exogenous uracil was supplied (Fig. 3, panel 3). Plasmid pJTU5011 bearing fusion gene PermE*-pyrC also absolutely reversed the mutant strain SJTU5009 back to the wild type (Fig. 3, panel 1). The mutation in SJTU5009 was used to test if the potential thymidine salvage pathway works in Streptomyces. Unfortunately, strain SJTU5013 still showed an abnormal phenotype when thymine was fed (Fig. 3, panel 2). In addition, neither 5-hydroxymethyluracil nor uracil-5-carboxylic acid could restore strain SJTU5013 to the normal phenotype (Fig. 3, panels 4 and 5). These results indicated that thymine and its derivatives could not be converted to uracil in strain SJTU5013. In other words, the constructed thymidine salvage pathway does not work in S. avermitilis, although SAV_4805 and SCO_6305 proteins shared certain sequence similarities with fungal THase and IDCase, respectively.
FIG. 3.
The pyrC mutation restored by ORF SCO_6305, pyrC, and different pyrimidine derivatives. Panels: 1, SFM medium; 2, SFM medium with 5 mM thymine; 3, SFM medium with 5 mM uracil; 4, SFM medium with 5 mM 5-hydroxymethyluracil; 5, SFM medium with 5 mM uracil-5-carboxylic acid. 5008, recombinant S. avermitilis strain SJTU5008; 5009, recombinant S. avermitilis strain SJTU5009; 5013, recombinant S. avermitilis strain SJTU5013; 5014, recombinant S. avermitilis strain SJTU5014.
Despite the disability which might be due to the limited capability of ORF SCO_6305, it seems more likely that SAV_4805 protein is not a full THase and cannot catalyze the oxidation reaction from hydroxymethyl to carboxyl (attached on the C-5 of the pyrimidine ring), with the result that only polyoxins H (methyl at R1 site) and A (hydroxymethyl at R1 site), but not polyoxin F (carboxyl at R1 site), were detected when the pol gene cluster and PermE*-SAV_4805 fusion gene were coexpressed in polyoxin-nonproducing strain S. lividans TK24.
Supplementary Material
Acknowledgments
We are very grateful to Tobias Kieser for critical reading of the manuscript and many valuable comments.
We also thank the Ministry of Science and Technology [973 (2003CB114205) and 863 programs], the National Science Foundation of China, China Postdoctoral Science Foundation of the Chinese Ministry of Education, and the Shanghai Municipal Council of Science and Technology for research support.
Footnotes
Published ahead of print on 3 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Archer, D. B. 1977. Chitin biosynthesis in protoplasts and subcellular fractions of Aspergillus fumigatus. Biochem. J. 164:653-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 4.Carreras, C. W., and D. V. Santi. 1995. The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem. 64:721-762. [DOI] [PubMed] [Google Scholar]
- 5.Chen, W., T. Huang, X. He, Q. Meng, D. You, L. Bai, J. Li, M. Wu, R. Li, Z. Xie, H. Zhou, X. Zhou, H. Tan, and Z. Deng. 2009. Characterization of the polyoxin biosynthetic gene cluster from Streptomyces cacaoi and engineered production of polyoxin H. J. Biol. Chem. 284:10627-10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endo, A., K. Kakiki, and T. Misato. 1970. Mechanism of action of the antifungal agent polyoxin D. J. Bacteriol. 104:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginj, C., H. Rüegger, N. Amrhein, and P. Macheroux. 2005. 3′-Enolpyruvyl-UMP, a novel and unexpected metabolite in nikkomycin biosynthesis. Chembiochem. 6:1974-1976. [DOI] [PubMed] [Google Scholar]
- 8.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He, Y., Y. Sun, T. Liu, X. Zhou, L. Bai, and Z. Deng. 2010. Cloning of separate meilingmycin biosynthesis gene clusters by use of acyltransferase-ketoreductase didomain PCR amplification. Appl. Environ. Microbiol. 76:3283-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hori, M., J. Eguchi, K. Kakiki, and T. Misato. 1974. Studies on the mode of action of polyoxins. VI. Effect of polyoxin B on chitin synthesis in polyoxin-sensitive and resistant strains of Alternaria kikuchiana. J. Antibiot. (Tokyo) 27:260-266. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 12.Isono, K. 1978. Biosynthesis of the nucleoside skeleton of polyoxins. J. Am. Chem. Soc. 100:3937-3939. [Google Scholar]
- 13.Isono, K. 1988. Nucleoside antibiotics: structure, biological activity, and biosynthesis. J. Antibiot. (Tokyo) 41:1711-1739. [DOI] [PubMed] [Google Scholar]
- 14.Isono, K., P. F. Crain, and J. A. McCloskey. 1975. Isolation and structure of octosyl acids. Anhydrooctose uronic acid nucleosides. J. Am. Chem. Soc. 97:943-945. [Google Scholar]
- 15.Jones, M. E. 1980. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Annu. Rev. Biochem. 49:253-279. [DOI] [PubMed] [Google Scholar]
- 16.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 17.Koehn, E. M., T. Fleischmann, J. A. Conrad, B. A. Palfey, S. A. Lesley, I. I. Mathews, and A. Kohen. 2009. An unusual mechanism of thymidylate biosynthesis in organisms containing the thyX gene. Nature 458:919-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, C. K., P. M. Shaffer, R. S. Slaughter, R. P. McCroskey, and M. T. Abbott. 1972. Stoichiometry of the pyrimidine deoxyribonucleoside 2′-hydroxylase reaction and of the conversions of 5-hydroxymethyluracil to 5-formyluracil and of the latter to uracil-5-carboxylic acid. Biochemistry 11:2172-2176. [DOI] [PubMed] [Google Scholar]
- 19.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 20.Naider, F., P. Shenbagamurthi, A. S. Steinfeld, H. A. Smith, C. Boney, and J. M. Becker. 1983. Synthesis and biological activity of tripeptidyl polyoxins as antifungal agents. Antimicrob. Agents Chemother. 24:787-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmatier, R. D., R. P. McCroskey, and M. T. Abbott. 1970. The enzymatic conversion of uracil 5-carboxylic acid to uracil and carbon dioxide. J. Biol. Chem. 245:6706-6710. [PubMed] [Google Scholar]
- 22.Shaffer, P. M., C. A. Hsu, and M. T. Abbott. 1975. Metabolism of pyrimidine deoxyribonucleosides in Neurospora crassa. J. Bacteriol. 121:648-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smiley, J. A., M. Kundracik, D. A. Landfried, V. R. Barnes, Sr., and A. A. Axhemi. 2005. Genes of the thymidine salvage pathway: thymine-7-hydroxylase from a Rhodotorula glutinis cDNA library and iso-orotate decarboxylase from Neurospora crassa. Biochim. Biophys. Acta 1723:256-264. [DOI] [PubMed] [Google Scholar]
- 24.Smokvina, T., P. Mazodier, F. Boccard, C. J. Thompson, and M. Guérineau. 1990. Construction of a series of pSAM2-based integrative vectors for use in actinomycetes. Gene 94:53-59. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki, S., K. Isono, J. Nagatsu, T. Mizutani, Y. Kawashima, and T. Mizuno. 1965. A new antibiotic, polyoxin A. J. Antibiot. (Tokyo) 18:131. [PubMed] [Google Scholar]
- 26.Watanabe, M. S., R. P. McCroskey, and M. T. Abbott. 1970. The enzymatic conversion of 5-formyluracil to uracil 5-carboxylic acid. J. Biol. Chem. 245:2023-2026. [PubMed] [Google Scholar]
- 27.Xu, H., Y. Zhang, J. Yang, T. Mahmud, L. Bai, and Z. Deng. 2009. Alternative epimerization in C(7)N-aminocyclitol biosynthesis is catalyzed by ValD, a large protein of the vicinal oxygen chelate superfamily. Chem. Biol. 16:567-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.