Abstract
AS-48 is a 70-residue, α-helical, cationic bacteriocin produced by Enterococcus faecalis and is very singular in its circular structure and its broad antibacterial spectrum. The AS-48 preprotein consists of an N-terminal signal peptide (SP) (35 residues) followed by a proprotein moiety that undergoes posttranslational modifications to yield the mature and active circular protein. For the study of the specificity of the region of AS-48 that is responsible for maturation, three single mutants have been generated by site-directed mutagenesis in the as-48A structural gene. The substitutions were made just in the residues that are thought to constitute a recognition site for the SP cleavage enzyme (His-1, Met1) and in those involved in circularization (Met1, Trp70). Each derivative was expressed in the enterococcal JH2-2 strain containing the necessary native biosynthetic machinery for enterocin production. The importance of these derivatives in AS-48 processing has been evaluated on the basis of the production and structural characterization of the corresponding derivatives. Notably, only two of them (Trp70Ala and Met1Ala derivatives) could be purified in different forms and amounts and are characterized for their bactericidal activity and secondary structure. We could not detect any production of AS-48 in JH2-2(pAM401-81His-1Ile) by using the conventional chromatographic techniques, despite the high efficiency of the culture conditions applied to produce this enterocin. Our results underline the different important roles of the mutated residues in (i) the elimination of the SP, (ii) the production levels and antibacterial activity of the mature proteins, and (iii) protein circularization. Moreover, our findings suggest that His-1 is critically involved in cleavage site recognition, its substitution being responsible for the blockage of processing, thereby hampering the production of the specific protein in the cellular culture supernatant.
Bacteriocins constitute a family of proteins of ribosomal synthesis for microbial defense that show a variable spectrum, mode of action, molecular weight, genetic origin, and biochemical properties (44, 45). Circular bacteriocins comprise a unique group of active posttranslationally modified proteins in which their N and C ends are linked by a peptide bond to form a circular polypeptide chain. Recently, they have been grouped as a new class of bacteriocins (class IV) (28), which has been divided into two subclasses according to their sequences and predicted secondary structures (32): one group including the disparate bacteriocins AS-48 (14, 17), subtilosin A (23), carnocyclin A (31), lactocyclicin Q (50), circularin A (24), and uberolysin (60) and one including butyrivibriocin AR10 (21) and the homologous gassericin A group (gassericin A and reutericin 6, which are identical molecules, and acidocin B) (3, 22, 53, 55). The gene sequences for most of the precursors are known, but putative cleaving and/or circularizing enzymes have not yet been reported. A general trend in the biosynthesis of the circular proteins is the appearance of an N-terminal-extended prepeptide that undergoes posttranslational circularization after proteolytic cleavage to release the mature bioactive molecules (28, 50). Signal sequences containing information specifying the choice of the targeting pathway, translocation efficiency, cleavage timing, and even postcleavage functions have been proposed (20). Recently, Oman and van der Donk (38) reviewed the different roles for the signal peptides (SPs). The most common function is that of a secretion signal, but these SPs also have been postulated as a recognition motif for the posttranslational modification enzymes or as a cis-acting chaperone in which the signal actively assists during the posttranslational modification process. The comparative analysis of the SPs of the circular bacteriocins, however, reflects remarkable differences in length and sequence as well as the absence of conserved motifs among them (28, 50), hindering a consensus sequence between their cleavable sites. These signals are unusually short for subtilosin A, uberolysin, carnocyclin A, circularin A, and lactocyclicin Q formation (8, 6, 4, 3, and 2 amino acids, respectively) compared to the extended signal for AS-48 (35 residues) or acidocin B/gassericin A/reutericin 6 (33 residues).
The AS-48 character (production and immunity) depends upon the coordinated expression of the as-48A, as-48B, as-48C, as-48C1, as-48D, as-48D1, as-48E, as-48F, as-48G, and as-48H genes (reviewed in reference 27). The structural gene as-48A is cotranscribed with as-48BC genes, with essential functions in AS-48 biogenesis and immunity (29). The posttranscriptional regulation of as-48ABC, recently identified by Fernández et al. (10), provides producer cells with the maximized production of functional AS-48 without deleterious effect before the entire immunity machinery (as-48D1 and as-48EFGH determinants, which are located in an independent operon) begins to work (6, 30). In addition, the as-48 cluster contains an essential ABC transporter, As-48C1D, the absence of which renders a nonproducer phenotype, which is devoted to the secretion of newly synthesized enterocin (28, 30). Understanding the precise mechanism underlying the AS-48 maturation and its transport to the cell surface is still our main challenge, because no products with the necessary specific activities and abilities to produce the circular polypeptide backbone have been identified yet. To unravel critical positions of the AS-48 maturation reactions, three single derivatives of AS-48 have been designed: the mutated residues located in the carboxyl end of the SP (His-1Ile) and those at both ends of the proprotein (Met1Ala and Trp70Ala). Comparative results concerning the functional structural features of such residues are discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and bacterial growth conditions.
The bacterial strains and plasmids used in this work and their relevant features are shown in Tables 1 and 2, respectively. Enterococcus faecalis JH2-2 was used in cloning experiments and production of AS-48 and its derivatives, and it also was an indicator strain. Enterococcus, Listeria, and Staphylococcus were propagated in buffered brain heart infusion (BHI-B) or tryptic soy broth (TSB-B) without aeration at 37°C. The Arthrobacter sp. was grown with shaking in BHI-B at 28°C. Escherichia coli strains, used as intermediate hosts for cloning, were grown with shaking in Luria-Bertani (LB) medium at 37°C. Culture media were purchased from Scharlau (Chemie, Barcelona, Spain). Agar plates contained 1.5% (wt/vol) agar, and soft agar was made with 0.75% (wt/vol) agar. If necessary, media were buffered in 0.1 M sodium phosphate buffer, pH 7.2. When appropriate, the antibiotics ampicillin (Ap), 50 μg/ml, and chloramphenicol (Cm), 20 μg/ml, from Sigma-Aldrich (Madrid, Spain) were added as selective agents.
TABLE 1.
Microorganisms used in this work
| Microorganism | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Escherichia coli JM109 | e14-(MrcA-) recA1 endA1 gyrA96 thi-1 hsdR17 (rk− mk+) supE44 relA1 Δ(lac-proAB) (F′ traD36 proAB lacIqZΔM15) | Stratagene |
| Arthrobacter sp. | AS-48 sensitive | Universidad de Granada collection |
| Bacillus cereus LWL1 | AS-48 sensitive | 9 |
| B. megaterium | AS-48 sensitive | CECT 44 |
| Enterococcus faecalis S-47 | AS-48 sensitive | 15 |
| E. faecalis JH2-2 | Plasmid free, Rifr Fusr AS-48s | 62 |
| Listeria innocua | AS-48 sensitive | CECT 4030 |
| L. monocytogenes | AS-48 sensitive | CECT 4032 |
| Staphylococcus aureus | AS-48 sensitive | CECT 976 |
TABLE 2.
Relevant features of the plasmids used in this work
| Plasmid | Characteristics | Reference or source |
|---|---|---|
| pBgD12S | Apr, as-48A cloned into pSL1180 (5.4 kb) | 48 |
| pBgC | Apr, BglII-BglII (fragment C) from as-48 cluster cloned into pSL1180 (9.2 kb) | 8 |
| pBgD12SMut | Apr, pBgD12S with the His-1Ile, Met1Ala, or Trp70Ala substitution in as-48A (5.4 kb) | This work |
| pAM401 | Cmr, Tcr, cloning expression bifunctional E. coli-E. faecalis vector (10.4 kb) | 61 |
| pAM401-76 | Cmr, D and B fragments (as-48A, as-48G, as-48H genes) from pMB2 cloned into pAM401 (18.9 kb) | 8 |
| pAM401-76Mut | Cmr, pAM401-76 with the His-1Ile, Met1Ala, or Trp70Ala substitution in as-48A (18.9 kb) | This work |
| pAM401-81 | Cmr, as-48 cluster cloned in pAM401 (25 kb) | 8 |
| pAM401-81Mut | Cmr, pAM401-81 with the His-1Ile, Met1Ala, or Trp70Ala substitution in as-48A (25 kb) | This work |
Molecular techniques.
DNA cloning techniques and transformations were performed according to the standard protocols (46). E. coli and E. faecalis plasmid DNA were extracted according to Birnboim and Doly (4) and O'Sullivan and Klaenhammer (40), respectively. E. faecalis was transformed by electroporation according to Fiedler and Wirth (11). Restriction enzymes, T4, DNA ligase shrimp phosphatase, and other DNA-modifying enzymes are from Fermentas (Quimigen, Madrid, Spain) and Roche Diagnostics (San Cugat del Vallés, Spain); they were used as recommended by the suppliers. DNA was sequenced using an ABI PRISM dye terminator cycle sequencing ready reaction kit (Perkin Elmer, Applied Biosystems).
Mutagenesis procedure and pAM401-81Mut plasmid construction.
Three single mutations (producing His-1Ile, Met1Ala, and Trp70Ala) were introduced in the structural as-48A gene as described elsewhere (10). The oligonucleotides used as primers for site-directed mutagenesis (Table 3) were from Amersham Biosciences Europe (Barcelona, Spain). Once we confirmed the accuracy of the sequences, the 5.4-kb amplified products (pBgD12SMut) were used to transform competent E. coli JM109 cells. To restore the as-48 cluster, each pBgD12SMut construction containing the required substitution was SphI-BglII digested and cloned into pAM401-76 (8) by replacing the equivalent fragment (pAM401-76Mut) according to Fernández et al. (10). The proper orientation of the fragments was established by analysis with EcoRI and PCR analysis using the As48-1- and As48-B4-specific primers (Table 3).
TABLE 3.
Oligonucleotides used in this worka
| Name | Sequence 5′-3′ |
|---|---|
| His-1Ile-rv | GAACTCTTTAGCCATGATTGCAATCGGCAAAAACTGAAGACTTGC |
| His-1Ile-fw | TTGCCGATTGCAATCATGGCTAAAGAGTTCGGTATACCAGCA |
| Met1Ala-rv | CCGAACTCTTTAGCGGCATGTGCAATCGGCAAAAACTGAAGACTTGC |
| Met1Ala-fw | TTGCCGATTGCACATGCCGCTAAAGAGTTCGGTATACCAGCACCAG |
| Trp70Ala-rv | TATTGTTAAATTAGGCAGCAATAACTGCCTCTTTTTC |
| Trp70Ala-fw | AGAGCCAGTTATTGCTGCCTAATTTAACAATATGATAA |
| As48-5 | CCAAGCAATAACTGCTCTTT |
| As48-6 | GAGTATCATGGTTAAAGAAA |
| As48-1 | AATAAACTACATGGGT |
| As48-B4 | AATCCTATTACTACAATAA |
| W01 | AGAGTTTGATCM*TGGCTC |
| W012 | TACGCATTTCACCK″CTACA |
The mutations introduced are underlined. M* = A + C; K″ = G + T.
RNA isolation and semiquantitative RT-PCR.
Total RNA from cells grown as described above was isolated using the Fast RNA Pro Blue kit (Q-Biogen, MP Biomedicals, European Headquarters, France). DNA was digested with DNase for 1 h at 37°C and heat inactivated at 70°C for 10 min. DNA absence was verified by PCR with specific primers under the conditions described above. Semiquantitative analysis of transcript levels was performed by two-step reverse transcription-PCR (RT-PCR) assays. Primers used for this task were As48-6 and As48-5 (Table 3) for the specific amplification of the as-48AMut genes using the wild-type as-48A as a positive control and the nonproducer strain JH2-2 as a negative one. Primers W01 and W012 (37) (Table 3) were used for 16S RNA amplification. Moloney murine leukemia virus (M-MuLV) reverse transcriptase was used according to the manufacturer in a reaction mixture containing total RNA (1 μg), 4 μl of cDNA synthesis buffer, 1 mM deoxynucleoside triphosphate mix, 10 μM the appropriate gene-specific primer, and 200 U of enzyme in a final volume of 20 μl. RNA, oligonucleotides, and water were heated at 70°C in a denaturing step prior to the addition of the other components of the mixture, and cDNA synthesis was performed for 60 min at 42°C. For the amplification of each cDNA, 10% of the reaction mixture was used as the template. The cycling conditions were 1 cycle of 94°C for 4 min and 15, 20, 25, and 30 cycles of 94°C for 0.5 min, 50°C for 0.5 min, and 72°C for 1 min. Samples of 5 μl were taken at 15, 20, 25, and 30 cycles.
AS-48 engineered derivative purification.
Wild-type and modified AS-48 was obtained from Esprion 300 (E-300; DMV Int., Veghel, Netherlands) plus 1% glucose (E-300-G) cultures of E. faecalis JH2-2(pAM401-81) and JH2-2(pAM401-81Mut) transformants under the conditions established by Ananou et al. (2). When necessary, complex medium (CM) also was used (48). Purified proteins (up to 95% purity) were obtained by reversed-phase chromatography using high-performance liquid chromatography (RP-HPLC) C18 columns, as described elsewhere (48). The concentration of the purified samples was determined by measuring UV absorption at 280 nm, which was converted to protein concentration using molecular extinction coefficients (16), which were calculated from the contributions of individual amino acid residues introduced. Mass determinations were obtained by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) using a model Voyager-DE PRO spectrometer from Applied Biosystems and compared to those for the AS-48 sequence using MassXpert 1.0 software.
Endoproteinase Glu-C digestion and MS/MS analysis.
The samples were incubated at 37°C for 18 h with endoproteinase Glu-C from Staphylococcus aureus V8 (sequencing grade; Sigma Chemical, St. Louis, MO) at a final concentration of 20 ng/μl in 50 mM ammonium bicarbonate (99.5% purity). Digested samples were analyzed in an Ultraflex MALDI-TOF/TOF mass spectrometer (Bruker Daltonik) (52), using internal mass calibration under the control of flexControl 2.2 software (Bruker Daltonik). Fragment ions generated by the laser-induced decomposition of the precursor were further accelerated by 19 kV in the LIFT cell, and their masses were analyzed after passing the ion reflector to an average of 1,000 spectra. For MALDI-MS/MS, calibrations were performed with fragment ion spectra obtained for the proton adducts of a peptide mixture covering the 800 to 3,200 m/z region. The analysis of mass data was performed using flexAnalysis 2.2 and BioTools 3.0 software (Bruker Daltonik) (51).
CD.
Circular dichroism (CD) spectra of the domain were collected on a Jasco J810 spectropolarimeter (Japan) fitted with a thermostated cell holder and interfaced with a Peltier cell unit. The instrument was periodically calibrated with (+)10-camphorsulfonic acid. The steady-state measurements at pH 7.0 (10 mM phosphate) and 25°C were carried out with the same experimental set as that described by Muro-Pastor et al. (35) and Neira et al. (36). To estimate the helical content, the deconvolution of the far-UV CD spectrum of the different AS-48 variants was performed using the DICHROWEB website (57, 58). The thermal denaturation of the purified samples also was carried out as described by Neira et al. (36).
Antimicrobial assays: determination of antimicrobial activities and MICs.
The production of AS-48 and derivatives by E. faecalis JH2-2 transformants was determined according to Sánchez-Hidalgo et al. (48). The determinations of the range of antimicrobial activity and the MIC for the purified preparations (defined as the minimal concentration of bacteriocin that inhibited the growth of the indicator strain) were carried out essentially as described previously (48).
RESULTS AND DISCUSSION
As a means of gaining insights into the AS-48 maturation reactions, three single-mutation AS-48 derivatives were constructed in residues directly involved in the signal cleavage of the preprotein (His-1Ile and Met1Ala) and the circularization of the proprotein (Met1Ala and Trp70Ala). Supernatants of the JH2-2(pAM401-81Mut) transformant exhibiting the modified sequence were obtained in E-300-G by applying the optimized conditions established by Ananou et al. (2) and analyzed regarding the synthesis of the AS-48 derivatives after purification by cation exchange, C18, and RP-HPLC chromatography.
Analysis of the antibacterial activity and resistance conferred by the new AS-48 derivatives.
The activity spectra of E. faecalis JH2-2(pAM401-81Mut) transformants, with that of JH2-2(pAM401-81) as a control, are compared in Table 4 by spotting assays against different Gram-positive bacteria of known AS-48 sensitivity. We noted that both Trp70 and Met1 replacements by Ala showed lower antagonistic activity than that of the reference. This was particularly pronounced for the Met1Ala derivative, which had reduced activity against the majority of the bacteria tested. In contrast, the JH2-2(pAM401-81His-1Ile) transformant exhibited no activity against any of the Gram-positive species tested.
TABLE 4.
Diameters of inhibition halos produced by the JH2-2(pAM401-81Mut) transformants against different Gram-positive strains, determined using the spot assay techniquea
| Gram-positive strain | Halo diam (mm) |
|||
|---|---|---|---|---|
| AS-48 | His-1Ile | Met1Ala | Trp70Ala | |
| Arthrobacter sp. | 31 | — | 17 | 21 |
| Bacillus cereus LWL1 | 16.5 | — | 5 | 13.5 |
| Bacillus megaterium CECT 44 | 25 | — | 11 | 19 |
| Enterococcus faecalis JH2-2 | 21 | — | 13 | 18 |
| Enterococcus faecalis S-47 | 21 | — | 12 | 17 |
| Listeria monocytogenes CECT 4032 | 27 | — | 14 | 21.5 |
| Listeria innocua CECT 4030 | 23 | — | 12 | 17 |
| Staphylococcus aureus CECT 976 | 16 | — | — | 12 |
The activity of JH2-2(pAM401-81) was determined as a control. —, no activity.
It also was found that the introduction of a neutral residue at both ends of the proprotein and at the end of the SP resulted in the same resistance level as that of the parental producer wild-type strain when they were assayed against 5-μl spots containing 18 μg of purified wild-type AS-48 (data not shown). The absence of inhibition halos after incubation indicated that the intact immunity machinery protected these transformants against AS-48, even in the case of JH2-2(pAM401-81His-1Ile), where no production could be detected later.
Single-site-directed mutagenesis of the SP.
It has long been acknowledged that some specificity of the cleavage reaction resides in the last residue of the signal sequence (43, 54). Moreover, the ability of positively charged amino-terminal residues in the SP to promote protein export seems to be based on their electrostatic interaction with negatively charged phospholipid headgroups (41). Thus, we targeted the expected processing site and mutated the positively charged His-1 to Ile, removing the aromatic side chain to test the function of this unique residue of the AS-48 preprotein, avoiding a charged residue in this position common to some other circular bacteriocins (28). The results, completely unexpected, are depicted in Fig. 1, where no active peaks with characteristic retention times could be isolated during the purification process, either in E-300-G or in complex medium (data not shown). The full loss of the activity of the JH2-2(pAM401-81His-1Ile) transformant (Table 4) is consistent with the fact that the His-1Ile substitution hinders a functional production of the AS-48 enterocin as well as of a preprotein truncated product or a lineal proprotein, although the RT-PCR assays confirmed the presence of a specific mRNA transcript (see below). These results also support the absence of potential alternative signal protease cleavage sites adjacent to the normal sites in the preprotein, as has been described formerly for E. coli during the maturation of the maltose-binding protein (12). Furthermore, a proper processing requires cleavage sites located in close proximity to the central hydrophobic and carboxyl-terminal hydrophilic border (reviewed in reference 41). Moreover, those proteins processed by proteases tend to be cleaved between elements of secondary structure rather than within helical, turn, or sheet regions (26). In this sense, the secondary-structure prediction of the native AS-48 preprotein using the PHD algorithm (http://www.predictprotein.org/) suggests that the SP (35 residues) has a propensity to form an α-helical conformation (positions 13 to 31) in wild-type AS-48. However, the His-1Ile mutant was predicted to have a larger transmembrane helical domain (positions 14 to 34), the enlargement of which could affect the cleavage reaction (data not shown). Notably, the occurrence of the His-1Met1 residues is rarely observed as an SP cleavage site; in fact, only capistruin, a 19-residue ribosomally synthesized lasso peptide produced by Burkholderia thailandensis, has His-1Gly1 residues, although the replacement of His-1 by Ala did not alter capistruin production (25, 42). It also is remarkable that the M16 family of zinc peptidases has a preference for aromatic residues at P-1 (upstream of the cleavage site) and a broader preference at P1, including hydrophobic/aromatic residues and arginine (7).
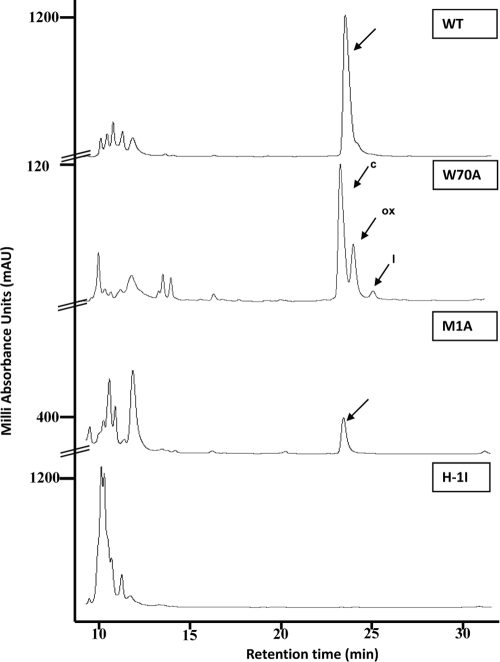
FIG. 1.
RP-HPLC spectra found in a C18 semipreparative column from E-300-G E. faecalis JH2-2 transformants expressing wild-type AS-48 and the three site-directed derivatives. The amounts of protein loaded in the runs were different. The symbols c, ox, and l indicate the circular, oxidized, and linear forms of Trp70Ala, respectively.
Consequences of Trp70 and Met1 residue substitution for AS-48 circularization.
Roughly 50% of circular bacteriocins have an aromatic residue (Trp or Tyr) at the carboxyl end of the lineal proprotein, either as the last amino acid (AS-48, lactocyclicin Q, uberolysin, and circularin A) or in a subterminal position (subtilosin A) (28, 50), although none has Met at the P1 position. These data suggest that this type of residue is important during secretion/transport or for acting as a recognition site for the enzymes involved in their maturation. Thus, the effect of both substitutions on AS-48 biosynthesis was evaluated using bioassays for the production of the derivatives against the most-sensitive bacterium, Bacillus megaterium CECT44. The two derivatives were quantitatively analyzed according to the total amount of protein obtained from cultures using the usual combination of chromatographic techniques. Indeed, JH2-2 cells harboring pAM401-81Met1Ala or pAM401-81Trp70Ala plasmid harboring the wild-type as-48BCC1DD1EFGH genes produced mature and mainly processed AS-48 derivatives, as judged by the appearance of the characteristic peaks after reverse-phase HPLC from supernatant cultures in E-300-G medium (Fig. 1).
Met1 is the N-terminal amino acid of the AS-48 proprotein, and as such it is directly involved in cleavage and propeptide circularization. In this case, a single active fraction with a retention time of 22.4 min (Fig. 1) was purified from supernatant of cells harboring the pAM401-81Met1Ala plasmid. The exact mass of 7,092.23 Da was assigned by MALDI-TOF MS, in agreement with the mass of the circular Met1Ala derivative form. This result also was confirmed by fingerprinting with endoproteinase Glu-C digestion (data not shown). Remarkably, protein yield in culture decreased to values of approximately 2% (0.032 mg·liter−1 compared to 1,555 mg·liter−1 for wild-type AS-48). The strikingly low expression of this mutant could be caused by an inefficient processing of the preprotein induced by the absence of a Met in such a critical position. It bears noting that no linear forms could be detected under any of the conditions assayed, and in the absence of a Met residue, no oxidized form was predictable. The structure prediction using the Web server http://www.compbio.dundee.ac.uk/www-jpred/ (6) shows a minor tendency to fold in the α-helix for this derivative (data no shown). However, it is important to point out that due to the absence of computer programs for circular proteins, we have considered the AS-48 molecule as beginning in the turn connecting the fifth and first helices.
During Trp70Ala mutant purification, three active peaks against B. megaterium CECT44, with retention times of 22.6, 23.3, and 24.3 min, were found (Fig. 1). Each fraction was subjected to a second purification step in an analytical RP-HPLC on a C18 column. The masses for these active samples were determined by MALDI-TOF (yielding values of 7,035.42, 7,050.93, and 7,053.35 Da) and agreed with the theoretical masses of the circular, oxidized, and linear forms of the Trp70Ala derivative, respectively. The protein yield was in the range of 185 mg liter−1 (approximately 12%), which is significantly lower than that of wild-type AS-48. The presence of a naturally oxidized Trp70Ala form (due to the addition of oxygen to Met) is not unusual, because the oxidation of this residue is a common phenomenon in proteins (56). In fact, such oxidized forms currently are arising during the purification of wild-type AS-48 in different amounts depending of the conditions of purification used (unpublished results). The subsequent bioassays revealed that the naturally oxidized form had a partial loss of activity (data not shown), as has been described for oxidized nisin (59). However, the similar retention times for native and oxidized forms imply that the hydrophobicity features and the protein structure were practically identical upon oxidation.
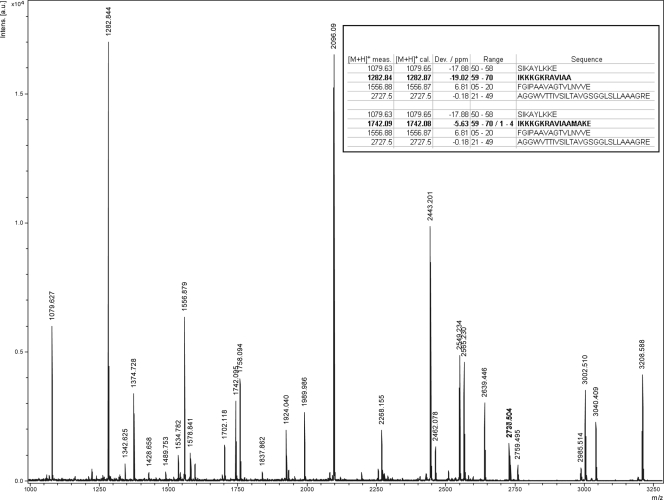
The identification of a linear form in the third little peak with a delayed retention time of 24.3 min is noteworthy (Fig. 1). The fragmentation of this peak with endoproteinase Glu-C led to an MS spectrum that shows an ion with an m/z value of 1,282.84 (Fig. 2), corresponding to the 12-residue polypeptide patch (IKKKGKRAVIAA) that could be derived only from a lineal Trp70Ala proprotein. Moreover, MS/MS analysis of this fragment unambiguously identifies this peak as linear Trp70Ala. This is the most significant finding of this work, since linear species have never before been described in AS-48 or in other circular proteins. Unfortunately, the specific activity of the linear forms could not be properly established due to the contamination with circular and oxidized species detected in the longest polypeptide region (IKKKGKRAVIAAMAKE), with m/z values of 1,742.09 and 1,758.09, respectively (Fig. 2).
FIG. 2.
Endoproteinase Glu-C digestion and MS/MS analysis of the linear Trp70Ala derivative. For clarity, each major peak was labeled with the mass and the corresponding single-amino-acid code of the ending residue.
Another outstanding result was the high activity of mutant Trp70Ala despite the fact that the Trp residues are supposed to be strongly involved in the interaction with the membranes (13). The importance of the Trp70 and Trp24 residues in the activity of AS-48 seems to be influenced by the diverse roles that the different domains have in the approach to and insertion into the membranes of sensitive cells. Thus, it is clear that the α1-α2 helices, where Trp24 is located, interact with the hydrophobic part of the membrane, leading to its permeabilization (49), whereas the most-amphiphilic helices, α4-α5 in the AS-48 molecule, where Trp70 is situated, are involved in access to the membrane-water interface, being responsible for the union to the host cells (47, 49). Similar results have been found with the genetically modified bacteriocins mesentericin Y105 (34), sakacin P (13), lactococcin G (38), and curvacin A (18). In fact, the determination of the MICs against different indicator strains confirmed that the circular Trp70Ala activity was very similar to that of the wild-type AS-48, whereas the effect of replacing Met with Ala rendered MICs much higher against the different strains assayed (Table 5). Since the mutation is the same (Ala in both cases) and at nearby positions, the effect observed must be due to structural questions, as discussed below.
TABLE 5.
MIC values for purified preparations of the circular Trp70Ala and Met1Ala derivativesa
| Indicator strain | AS-48 MIC (mg/liter) for: |
||
|---|---|---|---|
| WT | Met1Ala | Trp70Ala | |
| Arthrobacter sp. | 1.56 | 6.25 | 2 |
| B. cereus LWL1 | 12.5 | 25 | 12.5 |
| B. megaterium CECT 44 | 1.56 | 3.12 | 1.56 |
| E. faecalis JH2-2 | 6.25 | 12.5 | 6.25 |
| E. faecalis S-47 | 6.25 | 12.5 | 6.25 |
| L. innocua CECT 4030 | 2 | 8 | 2 |
| L. monocytogenes CECT 4032 | 1.56 | 3.12 | 1.56 |
| S. aureus CECT 976 | 6.25 | 12.5 | 6.25 |
Samples were serially diluted in sterile 10 mM sodium phosphate buffer at pH 7.0 and assayed against different susceptible strains. The activity of purified wild-type AS-48 (WT) was determined as a control.
RT-PCR analysis of the derivative genes.
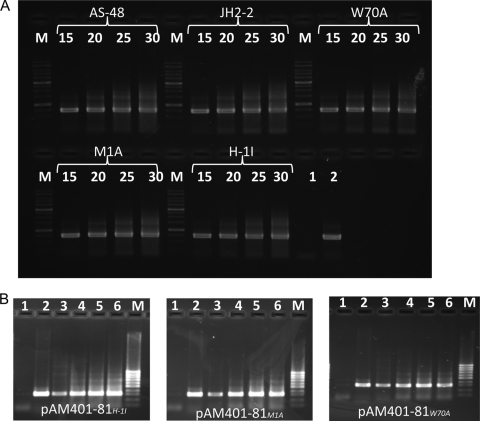
To investigate the reasons behind the negative results found with His-1Ile and to evaluate the different protein levels in the other cases, we performed an RT-PCR experiment. The results using specific primers (Table 3) confirmed the presence and approximate quantity of the transcripts. Notably, the expected 321-nucleotide (nt) amplicons were detected in the transformants after just 15 PCR cycles (Fig. 3 B). The controls carried out with rRNAs of the wild type and derivatives were intact, and the amplification occurred in all cases in similar ways (Fig. 3A). Therefore, these results confirmed that no transcriptional block could have been responsible for the lack of protein in the His-1Ile derivative, but they do not explain the differences in the expression levels of proteins found between Met1Ala and Trp70Ala derivatives.
FIG. 3.
Semiquantitative RT-PCR analysis of the transcription of the as-48A permuted genes. RNA was isolated from E. faecalis JH2-2 carrying the recombinant pAM401-81Mut constructions. Samples of 5 μl were taken at 15, 20, 25, and 30 cycles of control rRNA (A) and specific genes (B). Lane 1, negative control; lane 2, positive control; lanes 3, 4, 5, and 6, 15, 20, 25, and 30 cycles. Lanes M indicate the molecular size markers, 1 kb (A) and 100 bp (B).
Secondary-structure analysis and stability as measured by CD.
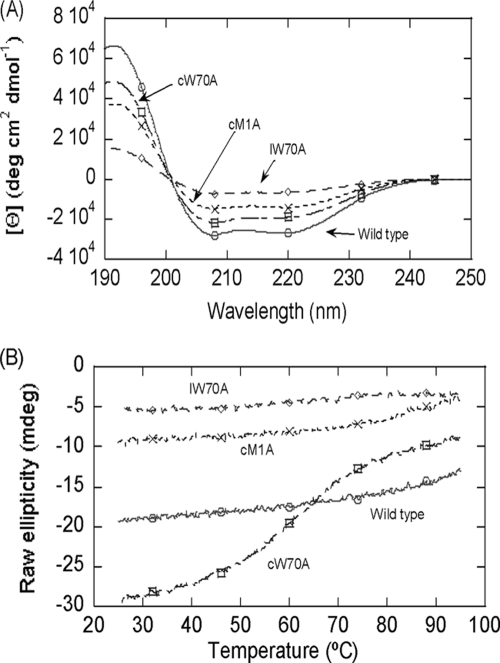
Met1Ala and Trp70Ala derivatives were examined by CD spectroscopy to evaluate the impact of the mutations introduced on the global conformation of these molecules. The far-UV spectra of the wild type and the different forms of the AS-48 derivatives are shown in Fig. 4 A. The protein with the largest molar ellipticity values is the wild-type species, and that with the lowest corresponds to the Trp70Ala linear derivative. The ellipticity and the shape of the far-UV CD spectrum of the wild type are identical to those measured previously for the wild type under similar conditions (48). In the same way, the deconvolution of the spectra (Table 6) according to the DICHROWEB website (57, 58) indicates that the highest percentage of helical structure corresponds to wild-type AS-48 (80%). As expected, the linear Trp70Ala species has the lowest percentage of helical structure of all the AS-48 derivatives found so far (48, 49), although this helical percentage is similar to those of the linear derivatives obtained in vitro (33). It is worth mentioning that although the two circular Trp70Ala and Met1Ala derivatives have a smaller percentage of helical structure than the wild-type AS-48 molecule, this value was even lower in the Met1Ala derivative (Table 6) despite the absence of the indole moiety in the circular Trp70Ala derivative, which also absorbs at 222 nm (57, 58). Thus, it appears that upon mutation, there is a marked helical fraying toward a less-rigid turn-like structure. This agrees with the fact that the Met1Ala derivative shows weaker membrane-protein interactions, as suggested by the higher MICs shown in Table 5. However, is this lower percentage of helical structure mirrored by a corresponding decrease in stability? To address this question, we conducted thermal denaturizing experiments. As has been described previously, the wild-type protein is extremely stable (5), with no sigmoidal transition between 25 and 95°C (Fig. 4B; Table 6). The circularized Met1Ala species also had high stability without any observable sigmoidal transition, which was similar to the wild type (Fig. 4B), even though its helical content was reduced (Table 6). We suggest that the presence of a higher population of turns (∼15% in circular Met1Ala versus 2% in the wild-type protein), which are structurally similar to α-helices, helps to stabilize the protein and thus to increase the melting point (Tm) (Table 6). Notably, the replacement of Trp70 with Ala (situated in the most-amphiphilic helices of the AS-48 molecule) clearly affects the stability of the circular and linear species, with Tms of 59 and 61°C, respectively, which is in accordance with the decrease in helical content observed in each variant (Fig. 4B; Table 6). This behavior is, however, very different from that shown by Trp24, whose replacement by Ala clearly favored stability, although it was extremely detrimental to the activity of the mutant (49). Thus, the effect of Trp replacements in AS-48 is strongly dependent on their position in the molecule, confirming that these aromatic residues can interact with different environments in the target cell membrane.
FIG. 4.
Far-UV CD spectra of wild-type AS-48 and derivatives. (A) Spectra were acquired in 10 mM phosphate buffer at pH 7.0 and 25°C. (B) Thermal denaturation of wild-type AS-48 and derivatives. The data have been scaled on the y axis for ease of reading. The symbols cTrp70Ala and lTrp70Ala are the circularized and linear forms of the Trp70Ala derivative, respectively, and cMet1Ala is the circularized form of the Met1Ala derivative.
TABLE 6.
Stability and percentages of secondary structure of wild-type AS-48 and derivatives as deduced from the DICHROWEBa site
| Structurec | % secondary structureb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT |
Circular Trp70Ala |
Linear Trp70Ala |
Circular Met1Ala |
|||||||||
| SCM | Contin | k2D | SCM | Contin | k2D | SCM | Contin | k2D | SCM | Contin | k2D | |
| α-Helix | 82 | 69.8 | 84 | 62.4 | 54.7 | 63 | 25.2 | 20.8 | 27 | 49.5 | 44 | 47 |
| β-Strand | 0 | 3 | 0 | 5.1 | 4.5 | 6 | 25.8 | 29.2 | 20 | 9.9 | 14.1 | 16 |
| Turn | 2 | 9.6 | 7.4 | 16.4 | 16.0 | 19.9 | 15.5 | 16.8 | ||||
| Random coil | 26 | 17.6 | 16 | 25.1 | 24.4 | 31 | 33 | 30.1 | 53 | 25.1 | 25.1 | 37 |
The following programs used were on the DICHROWEB website: Self-Consistent Method (SCM), Contin, and k2D.
Obtained as described by Neira et al. (36). Percentages of α-helix were also calculated as described elsewhere (36): 72 for WT, 52 for circular Trp70Ala, 18 for linear Trp70Ala, and 39 for circular Met1Ala.
The melting temperatures were as follows: wild type (WT), >95°C; circular Trp70Ala, 59°C; linear Trp70Ala, 61°C; and circular Met1Ala, >95°C.
Conclusions.
In the present study, we have investigated the functionality of three residues involved in the maturation of the AS-48 preprotein by expressing the respective single derivatives. Our results convincingly show that the residue His-1 should not be mutated to Ile, because this change affects the efficiency of a still-unidentified enzyme involved in the cleavage of the SP. In fact, only the His-1Ile mutation categorically restrains the cleavage of the precursor. Moreover, the protein, if synthesized, retains its leading peptide and must be rapidly degraded. Thus, we hypothesize that the proteolytic cleavage might be specific for His-1, and the imidazole ring may facilitate the protein cleavage (63). However, we cannot rule out the requirement of Met1 as a substrate, since its replacement decreases the efficiency for Met1Ala production (approximately 2%).
Another preliminary conclusion from the data presented here is the requirement of a second biosynthetic enzyme or a separate catalytic domain that is responsible for the circularization reaction, the partial specificity of which seems to reside in the last Trp70 residue of the proprotein, since linear forms could be derived only from JH2-2(pAM401-81Trp70Ala) transformants. Unmodified pre-AS-48 contains 105 amino acid residues that were targeted to modification machinery, most likely located at the cytoplasmic membrane. Posttranslational AS-48 processing by an enzyme/domain that probably requires the proximity of an NH2 group at the N-terminal end of the proprotein occurs presumably on the extracellular side of the cytoplasmic membrane because of the existence of linear Trp70Ala proteins in the supernatants. This amino group could act as a nucleophile in the circularization reaction with the specific, although not at all essential, C-terminal Trp70 residue, since circular forms also are being produced.
The C-terminal SP sequence appears to be crucial for the machinery involved in processing and transport. The candidate might be the essential ABC transporter As-48C1D, either alone or as a multicomponent system (reviewed in reference 28), able to carry out sequentially the maturation and secretion of AS-48 (involving the removal of the leader peptide and tail-head circularization). In this sense, it has been shown that some ABC exporters are able to remove leader peptides of the double-glycine type concomitantly with export (19). Further research is needed to confirm the biosynthetic maturation pathway and to identify the enzymes involved in this process.
Acknowledgments
This work was supported by the Spanish Dirección General de Investigación Científica y Técnica (projects BIO2008-01708, CSD20008-00005, and SAF2008-05742-C02-01) and Grupo de Investigación de la Junta de Andalucía (CIV 016), as well as ACOMP/2010/114 by Generalitat Valenciana and the FIPSE foundation (number 36557/06). Rubén Cebrián is the beneficiary of a grant from the Spanish Ministry of Education.
Footnotes
Published ahead of print on 10 September 2010.
REFERENCES
- 1.Abriouel, H., E. Valdivia, M. Martínez-Bueno, M. Maqueda, and A. Gálvez. 2003. A simple method for semi-preparative-scale production and recovery of enterocin AS-48 derived from Enterococcus faecalis subsp. liquefaciens A-48-32. J. Microbiol. Methods 55:599-605. [DOI] [PubMed] [Google Scholar]
- 2.Ananou, S., A. Muñoz, A. Gálvez, M. Martínez-Bueno, M. Maqueda, and E. Valdivia. 2008. Optimization of enterocin AS-48 production on a whey-based substrate. Int. Dairy J. 18:923-927. [Google Scholar]
- 3.Arakawa, K., Y. Kawai, Y. Ito, K. Nakamura, T. Chujo, J. Nishimura, H. Kitazawa, and T. Saito. 2010. HPLC purification and re-evaluation of chemical identity of two circular bacteriocins, gassericin A and reutericin 6. Lett. Appl. Microbiol. 50:406-4011. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cobos, E. S., V. V. Filimonov, A. Gálvez, E. Valdivia, M. Maqueda, J. C. Martínez, and P. L. Mateo. 2002. The denaturation of circular enterocin AS-48 by urea and guanidinium hydrochloride. Biochim. Biophys. Acta 1598:98-107. [DOI] [PubMed] [Google Scholar]
- 6.Cole, C., J. D. Barber, and G. J. Barton. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197-W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dabonné, S., C. Moallic, J. P. Sine, S. Niamké, M. Dion, and B. Colas. 2005. Cloning, expression and characterization of a 46.5-kDa metallopeptidase from Bacillus halodurans H4 sharing properties with the pitrilysin family. Biochim. Biophys. Acta 1725:136-143. [DOI] [PubMed] [Google Scholar]
- 8.Díaz, M., E. Valdivia, M. Martínez-Bueno, M. Fernández, A. S. Soler-González, H. Ramírez-Rodrigo, and M. Maqueda. 2003. Characterization of a new operon, as-48EFGH, from the as-48 gene cluster involved in immunity to enterocin AS-48. Appl. Environ. Microbiol. 69:1229-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufrenne, J., M. Bijwaard, M. te Giffel, R. Beumer, and S. Notermans. 1995. Characteristics of some psychrotrophic Bacillus cereus isolates. Int. J. Food Microbiol. 27:175-183. [DOI] [PubMed] [Google Scholar]
- 10.Fernández, M., M. Sánchez-Hidalgo, N. García-Quintáns, M. Martínez-Bueno, E. Valdivia, P. López, and M. Maqueda. 2008. Processing of as-48ABC RNA in AS-48 enterocin production by Enterococcus faecalis. J. Bacteriol. 190:240-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiedler, S., and R. Wirth. 1991. Transformation of Enterococcus faecalis and Enterococcus faecium by electroporation, p. 301. In G. M. Dunny, P. P. Cleary, and L. L. McKay (ed.), Genetics and molecular biology of streptococci, lactococci, and enterococci. American Society for Microbiology, Washington, DC.
- 12.Fikes, J. D., G. A. Barkocy-Gallagher, D. G. Klapper, and P. Bassford, Jr. 1990. Maturation of Escherichia coli maltose-binding protein by signal peptidase I in vivo. Sequence requirements for efficient processing and demonstration of an alternate cleavage site. J. Biol. Chem. 265:3417-3423. [PubMed] [Google Scholar]
- 13.Fimland, G., V. G. H. Eijsink, and J. Nissen Meyer. 2002. Mutational analysis of the role of tryptophan residues in an antimicrobial peptide. Biochemistry 41:9508-9515. [DOI] [PubMed] [Google Scholar]
- 14.Gálvez, A., M. Maqueda, E. Valdivia, A. Quesada, and E. Montoya. 1986. Characterization and partial purification of a broad spectrum antibiotic AS-48 produced by Streptococcus faecalis. Can. J. Microbiol. 32:765-771. [DOI] [PubMed] [Google Scholar]
- 15.Gálvez, A., E. Valdivia, M. Martinez, and M. Maqueda. 1989. Effect of peptide AS-48 on Enterococcus faecalis subsp. liquefaciens S-47. Antimicrob. Agents Chemother. 33:641-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 17.González, C., G. M. Langdon, M. Bruix, A. Gálvez, E. Valdivia, M. Maqueda, and M. Rico. 2000. Bacteriocin AS-48, a cyclic polypeptide structurally and functionally related to mammalian NK-lysin. Proc. Natl. Acad. Sci. U. S. A. 97:11221-11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haugen, H. S., P. E. Kristiansen, G. Fimland, and J. Nissen-Meyer. 2008. Mutational analysis of the class IIa bacteriocin curvacin A and its orientation in target cell membranes. Appl. Environ. Microbiol. 74:6766-6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Håvarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carries out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 20.Hegde, R. S., and H. D. Bernstein. 2006. The surprising complexity of signal sequences. Trends Biochem. Sci. 31:563-571. [DOI] [PubMed] [Google Scholar]
- 21.Kalmokoff, M. L., and R. M. Teather. 1997. Isolation and characterization of a bacteriocin (butyrivibriocin AR10) from the ruminal anaerobe Butyrivibrio fibrisolvens AR10: evidence in support of the widespread occurrence of bacteriocin-like activity among ruminal isolates of B. fibrisolvens. Appl. Environ. Microbiol. 63:394-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawai, Y., R. Kemperman, J. Kok, and T. Saito. 2004. The circular bacteriocins gassericin A and circularin A. Curr. Protein Pept. Sci. 5:393-398. [DOI] [PubMed] [Google Scholar]
- 23.Kawulka, K., T. Sprules, R. T. McKay, P. Mercier, C. M. Diaper, P. Zuber, and J. C. Vederas. 2003. Structure of subtilosin A, an antimicrobial peptide from Bacillus subtilis with unusual posttranslational modifications linking cysteine sulfurs to alpha-carbons of phenylalanine and threonine. J. Am. Chem. Soc. 125:4726-4727. [DOI] [PubMed] [Google Scholar]
- 24.Kemperman, R., A. Kuipers, H. Karsens, A. Nauta, O. Kuipers, and J. Kok. 2003. Identification and characterization of two novel clostridial bacteriocins, circularin A and closticin 574. Appl. Environ. Microbiol. 69:1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knappe, T. A., U. Linne, L. Robbel, and M. A. Marahiel. 2009. Insights into the biosynthesis and stability of the lasso peptide capistruin. Chem. Biol. 16:1290-1298. [DOI] [PubMed] [Google Scholar]
- 26.Madala, P. K., J. D. A. Tyndall, T. Nall, and D. P. Fairlie. 8 April 2010, posting date. Update 1 of proteases universally recognize Beta strands in their active sites. Chem. Rev. doi: 10.1021/cr900368a. [DOI] [PubMed]
- 27.Maqueda, M., A. Gálvez, M. Martínez-Bueno, M. J. Sánchez-Barrena, C. González, A. Albert, M. Rico, and E. Valdivia. 2004. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr. Protein Pept. Sci. 5:399-416. [DOI] [PubMed] [Google Scholar]
- 28.Maqueda, M., M. Sánchez-Hidalgo, M. Fernández, M. Montalbán-López, E. Valdivia, and M. Martínez-Bueno. 2008. Genetic features of circular bacteriocins produced by Gram-positive bacteria. FEMS Microbiol. Rev. 32:2-22. [DOI] [PubMed] [Google Scholar]
- 29.Martínez-Bueno, M., M. Maqueda, A. Gálvez, B. Samyn, J. Van Beeumen, J. Coyette, and E. Valdivia. 1994. Determination of the gene sequence and the molecular structure of the enterococcal peptide antibiotic AS-48. J. Bacteriol. 176:6334-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez-Bueno, M., E. Valdivia, A. Gálvez, J. Coyette, and M. Maqueda. 1998. Analysis of the gene cluster involved in production and immunity of the peptide antibiotic AS-48 in Enterococcus faecalis. Mol. Microbiol. 27:347-358. [DOI] [PubMed] [Google Scholar]
- 31.Martin-Visscher, L. A., M. J. van Belkum, S. Garneau-Tsodikova, R. M. Whittal, J. Zheng, L. M. McMullen, and J. C. Vede. 2008. Isolation and characterization of carnocyclin A, a novel circular bacteriocin, produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 74:4756-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Visscher, L. A., X. Gong, M. Duszyk, and J. C. Vederas. 2009. The three dimensional structure of carnocyclin A reveals that many circular bacteriocins share a common structural motif. J. Biol. Chem. 284:28674-28681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montalbán-López, M., B. Spolaore, O. Pinato, M. Martínez-Bueno, E. Valdivia, M. Maqueda, and A. Fontana. 2008. Characterization of linear forms of the circular enterocin obtained by limited proteolysis. FEBS Lett. 582:3237-3242. [DOI] [PubMed] [Google Scholar]
- 34.Morisset, D., J. M. Berjeaud, D. Marion, C. Lacombe, and J. Frère. 2004. Mutational analysis of mesentericin Y105, an anti-Listeria bacteriocin, for determination of impact on bactericidal activity, in vitro secondary structure, and membrane interaction. Appl. Environ. Microbiol. 70:4672-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muro-Pastor, M. I., F. N. Barrera, J. C. Reyes, F. J. Florencio, and J. L. Neira. 2003. The inactivating factor of glutamine synthetase, IF7, is a “natively unfolded” protein. Protein Sci. 12:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neira, J. L., L. M. Contreras, O. Ruiz de los Paños, M. Sánchez-Hidalgo, M. Martínez-Bueno, M. Maqueda, and M. Rico. 2010. Structural characterization of the natively unfolded enterocin EJ97. Protein Eng. Des. Sel. 23:507-518. [DOI] [PubMed] [Google Scholar]
- 37.Ogier, J. C., O. Son, A. Gruss, P. Tailliez, and A. Delacroix-Buchet. 2002. Identification of the bacterial microflora in dairy products by temporal temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 68:3691-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oman, T. J., and W. A. van der Donk. 2010. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat. Chem. Biol. 6:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oppegård, C., J. Schmidt, P. E. Kristiansen, and J. Nissen-Meyer. 2008. Mutational analysis of putative helix-helix interacting GxxxG-motifs and tryptophan residues in the two-peptide bacteriocin lactococcin G. Biochemistry 47:5242-5249. [DOI] [PubMed] [Google Scholar]
- 40.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 59:2730-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paetzel, M., A. Karla, N. C. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem. Rev. 102:4549-4580. [DOI] [PubMed] [Google Scholar]
- 42.Pavlova, O., J. Mukhopadhyay, E. Sineva, R. H. Ebright, and K. Severinov. 2008. Systematic structure-activity analysis of microcin J25. J. Biol. Chem. 283:25589-25595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phan, U. T., R. L. Lackman, and P. Cresswell. 2002. Role of the C-terminal propeptide in the activity and maturation of gamma-interferon-inducible lysosomal thiol reductase (GILT). Proc. Natl. Acad. Sci. U. S. A. 99:12298-12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riley, M. A., and J. E. Wertz. 2002. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie 84:357-364. [DOI] [PubMed] [Google Scholar]
- 45.Riley, M. A., and J. E. Wertz. 2002. Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 56:117-137. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Sánchez-Barrena, M., G. Martínez-Ripoll, A. Gálvez, E. Valdivia, M. Maqueda, V. Cruz, and A. Albert. 2003. Structure of bacteriocin AS-48: from soluble state to membrane bound state. J. Mol. Biol. 334:541-549. [DOI] [PubMed] [Google Scholar]
- 48.Sánchez-Hidalgo, M., M. Martínez-Bueno, A. M. Fernández-Escamilla, E. Valdivia, L. Serrano, and M. Maqueda. 2008. Effect of replacing glutamic residues upon the biological activity and stability of the circular enterocin AS-48. J. Antimicrob. Chemother. 61:1256-1265. [DOI] [PubMed] [Google Scholar]
- 49.Sánchez-Hidalgo, M., A. M. Fernández-Escamilla, M. Martínez-Bueno, E. Valdivia, L. Serrano, and M. Maqueda. 2010. Conformational stability and activity of circular enterocin AS-48 derivatives. Protein Pept. Lett. 17:708-714. [DOI] [PubMed] [Google Scholar]
- 50.Sawa, N., T. Zendo, J. Kiyofuji, K. Fujita, K. Himeno, J. Nakayama, and K. Sonomoto. 2009. Identification and characterization of lactocyclicin Q, a novel cyclic bacteriocin produced by Lactococcus sp. QU 12. Appl. Environ. Microbiol. 75:1552-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuerenberg, M., C. Luebbert, H. Eickhoff, M. Kalkum, H. Lehrach, and E. Nordhoff. 2000. Prestructured MALDI-MS sample supports. Anal. Chem. 72:3436-3442. [DOI] [PubMed] [Google Scholar]
- 52.Suckau, D., A. Resemann, M. Schuerenberg, P. Hufnagel, J. Franzen, and A. Holle. 2003. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 376:952-965. [DOI] [PubMed] [Google Scholar]
- 53.ten Brink, B., M. Minekus, J. M. van der Vossen, R. J. Leer, and J. H. Huis in't Veld. 1994. Antimicrobial activity of lactobacilli: preliminary characterization and optimization of production of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus M46. J. Appl. Bacteriol. 77:140-148. [DOI] [PubMed] [Google Scholar]
- 54.Tyndall, J. D. A., T. Nall, and D. P. Fairlie. 2005. Proteases universally recognize beta strands in their active sites. Chem. Rev. 105:973-999. [DOI] [PubMed] [Google Scholar]
- 55.Toba, T., S. K. Samant, E. Yoshioka, and T. Itoh. 1991. Reutericin 6, a new bacteriocin produced by Lactobacillus reuteri LA6. Lett. Appl. Microbiol. 13:281-286. [Google Scholar]
- 56.Vogt, W. 1995. Oxidation of methionyl residues in proteins: tools, targets, and reversal. Free Rad. Biol. Med. 18:93-105. [DOI] [PubMed] [Google Scholar]
- 57.Whitmore, L., and B. A. Wallace. 2004. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32:W668-W673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitmore, L., and B. A. Wallace. 2008. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89:392-400. [DOI] [PubMed] [Google Scholar]
- 59.Wilson-Stanford, S., A. Kalli, K. Håkansson, J. Kastrantas, R. S. Orugunty, and L. Smith. 2009. Oxidation of lanthionines renders the lantibiotic nisin inactive. Appl. Environ. Microbiol. 75:1381-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wirawan, R. E., K. M. Swanson, T. Kleffmann, R. W. Jack, and J. R. Tagg. 2007. Uberolysin: a novel cyclic bacteriocin produced by Streptococcus uberis. Microbiology 153:1619-1630. [DOI] [PubMed] [Google Scholar]
- 61.Wirth, R., F. Y. An, and D. B. Clewell. 1986. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J. Bacteriol. 165:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yagi, Y., and D. B. Clewell. 1980. Recombination-deficient derivative of Streptococcus faecalis. J. Bacteriol. 143:966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yates, Z. M., K. Gunasekaran, H. Zhou, Z. Hu, Z. Liu, R. R. Ketchem, and B. Yan. 19 March 2010, posting date. Histidine residue mediates radical induced hinge cleavage of a human IgG1. J. Biol. Chem. doi: 10.1074/jbc.M110.108597. [DOI] [PMC free article] [PubMed]