Abstract
Most Ralstonia solanacearum strains are tropical plant pathogens, but race 3, biovar 2 (R3bv2), strains can cause bacterial wilt in temperate zones or tropical highlands where other strains cannot. R3bv2 is a quarantine pathogen in North America and Europe because of its potential to damage the potato industry in cooler climates. However, R3bv2 will not become established if it cannot survive temperate winters. Previous experiments showed that in water at 4°C, R3bv2 does not survive as long as native U.S. strains, but R3bv2 remains viable longer than U.S. strains in potato tubers at 4°C. To further investigate the effects of temperature on this high-concern pathogen, we assessed the ability of R3bv2 and a native U.S. strain to survive typical temperate winter temperature cycles of 2 days at 5°C followed by 2 days at −10°C. We measured pathogen survival in infected tomato and geranium plants, in infected potato tubers, and in sterile water. The population sizes of both strains declined rapidly under these conditions in all three plant hosts and in sterile water, and no culturable R. solanacearum cells were detected after five to seven temperature cycles in plant tissue. The fluctuations played a critical role in loss of bacterial viability, since at a constant temperature of −20°C, both strains could survive in infected geranium tissue for at least 6 months. These results suggest that even when sheltered in infected plant tissue, R3bv2 is unlikely to survive the temperature fluctuations typical of a northern temperate winter.
To endure in an environment that does not provide consistent access to a living host, a pathogen must be able to persist during periods of suboptimal conditions (1). Understanding the limits of pathogen survival can lead to control methods for vulnerable regions, while climates that exceed these limits offer natural protection from establishment of an exotic pathogen (30).
Ralstonia solanacearum is a soilborne plant pathogen that causes bacterial wilt disease in over 200 plant species in warm-temperate and tropical climates worldwide (21). R. solanacearum can be transmitted by contaminated surface water and soil, latently infected plant cuttings, and discarded plant debris. This pathogen colonizes host plant vascular tissue after entering through naturally occurring root wounds. The bacterium multiplies rapidly in the xylem elements, inducing characteristic wilting before disseminating back into the environment to infect a new host or die (21).
One subgroup of the R. solanacearum species complex, now classified as phylotype II, sequevar 1, but historically and for regulatory purposes known as race 3, biovar 2 (R3bv2), causes brown rot of potato (2). Brown rot is a major source of potato crop losses in the tropical highlands worldwide, costing growers an estimated $950 million each year (2, 12). The potato is only one host of R3bv2, however, as the strain also infects tomato plants, eggplant, and many wild and horticultural plants (13, 25, 34, 47, 52). Infected geranium cuttings have been accidentally introduced to North America and Europe from the highland tropics, although the bacterium has not become established in North America (25, 29, 40, 41, 53). R3bv2 most likely originated in the Andes with potato plants, since isolates from around the world are essentially clonal (14, 22, 39, 48). Phylotype II, sequevar 7 (formerly race 1), strains that infect tomato, pepper, and tobacco plants are endemic to the warm temperate and subtropical zones of the southeastern United States, but these have never become established north of the mid-Atlantic states.
R3bv2 is a quarantine pest in North America and Europe. It is considered a threat because it can cause disease at cooler temperatures than tropical R. solanacearum strains and is widespread in the cool highland tropics (8, 9, 45, 46). If R3bv2 could overwinter in the harsher climate of the northern United States and Canada, it could threaten the $4 billion North American potato industry (http://www.agr.gc.ca/).
It has proven difficult to eradicate R3bv2 from northern Europe, where it appeared in the 1990s. Although the pathogen survives poorly in 4°C water or in field soil in the Netherlands (49, 50), R3bv2 can overwinter in reservoir hosts. One documented reservoir is the bittersweet nightshade, Solanum dulcamara, a common weed found near water in both Europe and North America (11). Although R3bv2 is still detected in waterways in northern Europe more than 15 years after its initial discovery, it has not caused significant disease-related losses, probably because the relatively cool summer temperatures are not optimal for wilt symptom development (4, 7, 12).
R. solanacearum remains viable for decades in pure water at room temperature in the laboratory and is also easily disseminated in irrigation water (10, 21, 35). However, in 4°C water, R3bv2 does not survive as long as some other R. solanacearum strains, including those endemic to the southern United States (31). Despite this poor cold survival in water, R3bv2 populations remain stable in potato tubers at 4°C, indicating that the pathogen is adapted to endure constant low temperatures when sheltered in host tissue (31). These data suggest that the cool climate epidemiology of R3bv2 strains involves interactions with host plants rather than direct physiological adaptations such as the increased membrane fluidity and RNA stability that contribute to cold tolerance in some food-borne mammalian pathogens (31, 33).
While previous studies found that R. solanacearum can survive months or years in soil in association with plant tissue, this trait has not previously been studied under the cold and fluctuating conditions typical of commercial potato-growing areas in North America (16-20, 37, 50). In addition, although plant pathogens commonly persist in decaying plant tissue, it was not known if sheltering inside dead hosts improves R3bv2 survival of suboptimal conditions. We therefore designed experiments to assess the survival of R3bv2 in infected plant tissue at stable subzero temperatures and during temperature cycles. We also studied the virulence of R3bv2 cells following long-term incubation inside geranium stems at subzero temperatures.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Two strains of R. solanacearum were used in this study. UW551 is an R3bv2 (phylotype II, sequevar 1) strain isolated in Wisconsin from a wilting geranium plant that originated in Kenya (53). K60, the type strain of R. solanacearum, is a phylotype II, sequevar 7, strain isolated from tomato plants in North Carolina and is historically known as race 1, biovar 1 (28). This study used spontaneous rifampin-resistant variants of both strains; these were previously shown to have wild-type virulence and fitness (42). Strains were streaked from −80°C glycerol or room temperature water stocks onto Casamino Acids-peptone-glucose (CPG) solid medium amended with rifampin at a final concentration of 50 mg/liter and incubated at 28°C for 48 h (23). Overnight cultures were grown in CPG broth amended with 50 mg/liter of rifampin and incubated at 28°C with shaking at 225 rpm for 20 h unless otherwise noted. Medium ingredients were obtained from Difco Laboratories (Detroit, MI), while all other chemicals and antibiotics were obtained from Sigma-Aldrich (St. Louis, MO).
Survival in geranium tissue at constant subzero temperatures.
To measure the survival of R. solanacearum strains in stems of susceptible geranium plants, we petiole-wound inoculated 10-week-old geranium plants (cv. “Designer Dark Red”; Ball FloraPlant, West Chicago, IL) with a suspension of R. solanacearum cells as described previously (41) to give a final cell concentration of 2.5 × 103 CFU/plant for strain UW551 or 2.5 × 104 CFU/plant for strain K60. The plants were incubated in a growth chamber with a cycle of 14 h light at 24°C and 10 h dark at 19°C.
After about 21 days (corresponding to symptom development in over 80% of plants), symptomatic plants were unpotted and left to dry in a growth chamber with a cycle of 14 h light at 24°C and 10 h dark at 19°C for 14 days before being stripped of leaves. Each biological replicate was divided into three technical replicates, consisting of 10 plants each. Stem pieces extending approximately 1.5 cm above and 1.5 cm below the crown of each plant were harvested with a razor blade and sliced into 3-mm crosswise sections to ensure similar amounts of xylem tissue in all segments. Tissue around the crown was used, as it contains the most reliably high bacterial density (42). To enumerate initial and subsequent bacterial population sizes, a single crosswise ∼0.3-g tissue slice was ground in sterile water and serially dilution plated on CPG plates amended with 50 mg/liter rifampin and 100 mg/liter cycloheximide. Typical R. solanacearum colonies, which are cream colored with pink centers, slow-growing, irregular and mucoid, were counted after incubation at 28°C for 48 h, with a detection limit of 100 CFU.
Each separate technical replicate of pooled stem segments was placed into 50-ml conical screw-cap tubes with or without 9 times as much (wt/wt) nonsterile soil and incubated at −20°C, conditions representative of discarded host plants that are either tilled over or incorporated into an above-ground pile of refuse plants (cull pile). The soil was from a postharvest potato field at the University of Wisconsin West Madison Research Facility. The experiment contained three biological replicates, consisting of three technical replicates each for both soil and nonsoil samples. Viable bacterial population size in infected geranium tissue was measured at intervals by dilution plating as described above.
Survival following cycles of fluctuating temperature.
After initial preparation (outlined below for each host), infected plant samples were placed in a 5°C incubator. After 48 h, samples were shifted to a −10°C freezer. Following another 48 h, each replicate was sampled and placed again at 5°C to start the 96-h cycle again. Each fluctuating temperature cycle experiment in this study contained three biological replicates, with three technical replicates for each sampling, with the exception of the geranium assay, which consisted of six biological replicates, with three technical replicates each. Each biological replicate consisted of 30 plants, split into three technical replicates of 10 plants per replicate, with the exception of the potato tuber assay, which consisted of 25 potato tubers per each of the three biological replicates.
Geranium inoculation and growth was as described for the stable −20°C survival assay, except that stems were not dried. This was because preliminary experiments found that drying the stems did not affect R. solanacearum survival of cold stresses (data not shown). Stems were surface sterilized to reduce background contamination by fungi on dilution plates. Geranium stems were submerged in deionized water for 10 min, followed by treatment with 10% (vol/vol) bleach for 5 min, a 5-min deionized water rinse, treatment for 5 min in 10% (vol/vol) ethanol, and two final 5-min deionized water rinses. Stems were then dried, sliced, and placed in tubes without soil as described above. Samples were placed at 5°C to begin the fluctuating temperature cycles.
Potato minitubers (cv. “Russet Norkotah”; Sklarczyk Seed Farm, Johannesburg, MI) were surface sterilized as above and then inoculated with R. solanacearum as described previously (31). Briefly, each potato tuber was directly inoculated by pipetting 1.5 × 109 to 2.5 × 109 CFU into a needle wound and incubated in a sealed plastic box during temperature cycles. Tubers were destructively sampled after every cycle by using a no. 5 cork borer to excise the tissue around the point of inoculation. The tissue was then weighed, ground in water, and serially dilution plated as described above.
Unwounded tomato plants (wilt-susceptible cv. “Bonny Best”) were inoculated with R. solanacearum via a naturalistic soil soak method (43). Briefly, 12-day-old tomato plants were transplanted into 10-cm pots with 80 g soil. At 25 days old, a bacterial suspension was poured onto the potting mix to give a final concentration of 5.0 × 107 CFU/g soil and plants were incubated in a 28°C growth chamber and monitored daily. When wilt symptoms first appeared, approximately 4 cm of stem tissue spanning the cotyledons was removed and chopped into 5-mm segments. Segments from five or six plants per biological replicate were then pooled and initially sampled. Approximately 0.15 g of tissue was ground in sterile water and dilution plated as described above. Biological replicates were placed in 50-ml conical tubes before the fluctuating temperature cycles described above were begun.
For the samples suspended in sterile water, R. solanacearum cells from 24-h cultures were washed twice in sterile water and resuspended in sterile water at a density of 3.0 × 109 CFU/ml. Suspensions were starved at room temperature for 48 h and then separated into 1-ml aliquots in 1.7-ml microcentrifuge tubes. Initial and subsequent bacterial population sizes were determined by serially diluting an aliquot as described above. In this experiment, the detection limit was 10 cells because we plated 100 μl of the undiluted sample. The aliquots were then placed at 5°C to start the fluctuating temperature cycle described above.
We used enrichment culture to verify that no culturable R. solanacearum cells remained in samples that contained no bacteria detectable by dilution plating. Approximately 0.3 g geranium, 0.15 g tomato, or 0.4 g potato tuber tissue was incubated for 72 h with shaking at 28°C in 5 ml TTC supplemented with multiple antibiotics (CPG amended with 5 mg/liter crystal violet, 25 mg/liter bacitracin, 0.5 mg/liter penicillin G, 5 mg/liter chloramphenicol, 100 mg/liter cycloheximide, 50 mg/liter rifampin, and 100 mg/liter 2,3,5-triphenyl tetrazolium chloride). For water samples, enrichment culture was performed by pipetting 1 ml of the suspension into 4 ml of CPG amended with 50 mg/liter rifampin and 100 mg/liter cycloheximide and incubating the culture at 28°C with shaking at 225 rpm for 72 h. One hundred microliters of the resulting enrichment culture was then plated on CPG amended with 25 mg/liter rifampin, 100 mg/liter cycloheximide, and 5 g/liter sodium pyruvate to facilitate recovery of viable but not culturable (VBNC) cells (24). Colonies were counted after 48 h at 28°C.
Virulence assays.
We used a direct cut petiole inoculation method to measure the virulence of R. solanacearum strain UW551 cells that had been incubated for 6 to 7 months at −20°C inside geranium stem tissue (44). Briefly, the first true leaf of the 21-day-old tomato plants (wilt-susceptible cv. “Bonny Best”) was removed with a razor blade, and 2 μl of bacterial suspension containing 250 to 350 CFU was placed directly onto the cut petiole surface. The inoculum consisted of either UW551 streaked from a room temperature water stock or UW551 cells extracted from infected geranium tissue stored at −20°C, as described above. To prepare inoculum, infected geranium tissue was ground in sterile water in a 1:10 (wt/vol) dilution and the resulting suspension was directly pipetted onto the cut petiole. An aliquot was dilution plated to determine the inoculum concentration. Plants were incubated in a growth chamber at 28°C. Plants were blinded for treatment identity and rated daily on a disease index scale ranging from 0 to 4, where 0 indicates no disease, 1 indicates 1 to 25% of leaves wilted, 2 indicates 26 to 50% of leaves wilted, 3 indicates 51 to 75% of leaves wilted, and 4 indicates 76 to 100% of leaves wilted. Each experiment included 16 plants per treatment, and the assay was repeated three times.
Statistical analysis.
The statistical significance of all data was analyzed using the JMP 8.0.2 software program (SAS Institute, Cary, NC). Data on strain survival of fluctuating temperatures were tested for significance by comparing strains using analysis of covariance (ANCOVA) of one strain, using time as a continuous variable and biological replicates as a factor variable, and then plotting the predicted values from the fitted model against the actual values of the second strain. Data shown are the means of results from all biological replicates, presented as log (numbers of CFU/g + 1). The mean values for log (number of CFU/g + 1) are below the limit of detection in cases where some of the replicates for a time point gave detectable CFU and others did not. Differences in virulence on tomato plants between strains were tested using repeated-measures analysis of variance (ANOVA) to analyze disease severity as a function of time (44).
RESULTS
R. solanacearum R3bv2 survived well in infected geranium tissue at constant subzero temperatures.
Populations of R3bv2 strain UW551 in chopped geranium tissue declined very little over months of incubation at −20°C. Geranium tissue initially contained an average of 7.1 × 108 CFU/g; this declined to 4.6 × 107 to 9.5 × 107 CFU/g tissue after 182 days of incubation at −20°C. There was no difference in survival between samples incubated with soil versus samples incubated bare. UW551 in frozen geranium tissue remained detectable by dilution plating for up to 1 year after introduction to subzero temperatures, averaging 5.0 × 106 CFU/g tissue at 9 months and 2.6 × 105 CFU/g at 12 months. Similarly, cells of endemic U.S. strain K60 also remained detectable for months in infected geranium tissue, with populations of approximately 6.0 × 106 CFU/g tissue at 3 months and 7.5 × 104 CFU/g tissue at 4 months, decreasing from an initial population density of 1.8 × 108 CFU/g tissue (data not shown).
Virulence of R. solanacearum UW551 was reduced following survival at constant subzero temperatures.
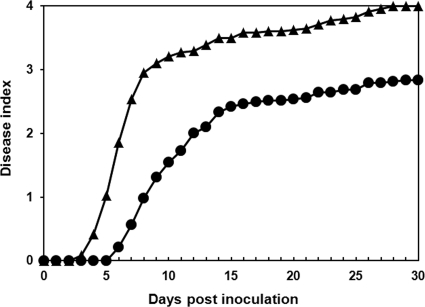
To determine if extended freezing affected bacterial interactions with hosts, we inoculated tomato plants with R. solanacearum strain UW551 cells extracted from infected geranium tissue stored at −20°C for 6 to 7 months, as described above. Post-in planta-freezing R. solanacearum cells behaved differently in culture from those that were grown in culture stocks. After 2 or more months of incubation in frozen geranium plants, UW551 cells no longer grew to a detectable level when inoculated directly into rich broth, although they remained easily detectable by dilution plating. Moreover, cells recovered from infected geranium tissue stored at −20°C were significantly less virulent on tomato plants than cells of the same strain inoculated directly from culture stocks (P < 0.001; repeated-measures ANOVA) (Fig. 1) About 20% of plants inoculated with recovered cells remained alive at the end of the 30-day assay; of these, some contained no detectable R. solanacearum cells, but others were latently infected and contained as much as 1 × 109 CFU/g tissue (data not shown).
FIG. 1.
Ralstonia solanacearum R3bv2 had reduced virulence following long-term incubation in geranium tissue at −20°C. Young Bonny Best tomato plants were inoculated through a cut petiole with 250 to 350 CFU of R3bv2 strain UW551 either taken from water stock (closed triangles) or recovered from infected geranium tissue incubated at −20°C for 6 to 7 months (closed circles). Plants were rated daily on a disease index scale of 0 to 4, where 0 indicated healthy plants and 4 indicated 75 to 100% wilted plants. Each point represents the average disease index for four independent experiments, consisting of 16 plants per treatment; treatments differed by repeated-measures ANOVA (P < 0.001).
Survival in plant tissue was reduced by fluctuating temperatures.
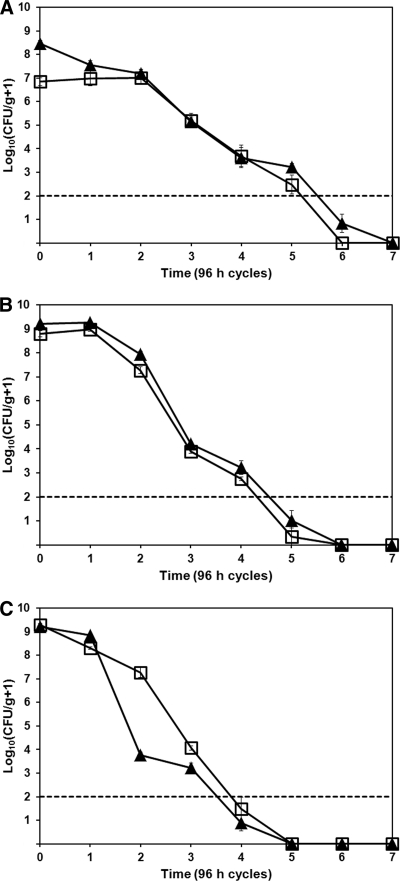
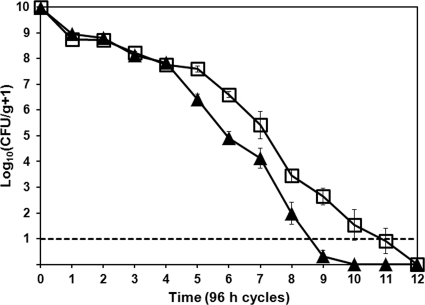
To determine the effect of more natural temperature conditions on bacterial survival, we measured population sizes of R. solanacearum strains UW551 and K60 from infected geranium, tomato, and potato tuber tissue subjected to temperatures that fluctuated around freezing. When exposed to a cycle of 2 days at −10°C followed by 2 days at 5°C, neither R3bv2 strain UW551 nor endemic southeastern U.S. strain K60 survived in geranium tissue longer than seven cycles (28 days) before falling below the limit of detection, which was 100 cells (Fig. 2 A). Under these conditions, which occur repeatedly during winters in the northern United States and Canada, there were no differences in survival between K60 and UW551 (R2 = 0.83). Populations of both strains remained high after the first two temperature cycles, averaging more than 1.0 × 107 CFU/g geranium tissue. Population sizes declined precipitously in subsequent cycles. After seven cycles (28 days), neither strain could be recovered by standard dilution plating on CPG, by enrichment culture in CPG broth, or by plating on CPG amended with sodium pyruvate, suggesting that geranium tissue did not protect either bacterial strain from freezing damage.
FIG. 2.
In infected host tissue, Ralstonia solanacearum populations declined rapidly following fluctuating temperature cycles of 48 h at 5°C and then 48 h at −10°C. Closed triangles represent the mean population sizes of R3bv2 strain UW551, and open squares represent endemic U.S. strain K60. Mean bacterial population sizes were measured by dilution plating: chopped stems of symptomatic R. solanacearum-inoculated geranium plants (A), chopped stems of wilting R. solanacearum-inoculated tomato plants (B), or cores of potato minitubers inoculated with ∼2 × 109 CFU of the relevant R. solanacearum strain (C). Infected plant material was sampled after each 4-day temperature cycle. The experiment was repeated six times, with three technical replicates per biological replicate (A) or three times with three technical replicates per biological replicate (B, C). The dashed line represents the detection limit. Bars represent the standard errors of the means.
We also measured the ability of R. solanacearum strains UW551 and K60 to survive temperature cycles in infected tissue of tomato plants, a natural host to both strains. Neither strain could survive more than six −10°C to 5°C temperature cycles (24 days) before falling below 100 cells/ml, the limit of detection (Fig. 2B). Again, there were no differences in survival between the two strains (R2 = 0.94), with populations of both strains declining rapidly after two cycles, as observed in the geranium experiments. A single freeze-thaw cycle did not reduce the population size of either strain, with both populations averaging around 1.0 × 109 CFU/g tomato tissue for both the initial and the first cycle samplings. After three cycles (12 days), pathogen populations plummeted to an average of 1.0 × 104 CFU/g tomato tissue, suggesting that multiple temperature fluctuation cycles reduced bacterial viability. After six cycles (24 days), neither strain was recoverable by either CPG broth enrichment culture or plating on CPG amended with sodium pyruvate.
The potato is the most economically important host of R3bv2 strains like UW551, and this pathogen is often disseminated in latently infected potato tubers. Strain K60 can also infect potato plants. We therefore compared the abilities of UW551 and K60 to survive freeze-thaw cycles in infected potato tubers. In this host, neither strain could survive more than five −10°C-to-5°C cycles (20 days) before falling below detectability (Fig. 2C). There was little difference between the survival rates of UW551 and K60 over the course of five cycles (R2 = 0.94), although UW551 populations underwent a larger decline following cycle 2, dropping to an average of 7.7 × 103 from 7.8 × 108 CFU/g tissue. K60 population sizes dropped to 2.2 × 105 from 2.1 × 108 CFU/g tissue over the same interval.
Fluctuating temperatures also reduced R. solanacearum survival in water.
To separate the contribution of temperature changes from any protective or antagonistic effect of surrounding host plant tissue, we measured the survival of strains UW551 and K60 following freeze-thaw cycles in sterile water. Both strains survived cycles of −10°C to 5°C in water better than they did in any plant tissue tested. In sterile water, UW551 survived until the 10th cycle (40 days) before falling below the detection limit, which was 10 cells, while K60 survived until the 12th cycle (48 days) (Fig. 3). R. solanacearum cells in water did not undergo the rapid population decline observed in every plant survival assay following the second temperature cycle. After four cycles, populations of both UW551 and K60 remained large, at 9.5 × 107 and 8.4 × 107 CFU/ml, respectively. While UW551 populations declined quickly after four temperature cycles, the K60 population size declined more gradually. In the absence of host tissue, K60 survived fluctuating temperatures better than UW551. Although K60 survived longer than UW551, these differences were not found to be significant when strain survival over the entire time course was considered (R2 = 0.90).
FIG. 3.
In sterile water, Ralstonia solanacearum populations declined following fluctuating temperature cycles of 48 h at 5°C and then 48 h at −10°C. Closed triangles represent the mean population sizes of R3bv2 strain UW551, and open squares represent endemic U.S. strain K60. Bacterial population size was determined after every cycle by dilution plating. The experiment was repeated three times, with three technical replicates per biological replicate. The dashed line represents the detection limit. Bars represent the standard errors of the means.
DISCUSSION
Previous studies suggested that R. solanacearum cells might persist best in the environment when sheltered in plant debris (31). Unexpectedly, we found that this pathogen was less likely to survive cyclic cold stress when associated with a host than when in pure water. The reason for this reduced survival may be that when R. solanacearum cells thaw inside plant tissue where nutrients are available, the bacteria undergo a respiratory burst that increases their vulnerability to damage inflicted by a subsequent freeze. A freeze-thaw cycle is known to induce a respiratory surge in soilborne bacteria, likely stimulated by nutrients released from lysing dead cells (36, 38, 51). This creates a period of prosperity for the remaining microbes (38), but this temporary bounty may actually be detrimental to long-term survival. In contrast, cells in water may be less vulnerable because starvation-induced stress proteins can protect bacteria from temperature damage (26, 27). We previously found that starved cells of all R. solanacearum strains tested were more resistant to cold stress than cells that were actively multiplying (31). This supports the idea that an R. solanacearum population with access to more nutrients in the form of host tissue will begin growing more swiftly after a thaw but will be more vulnerable to dying from another freeze than bacteria in a lower-nutrient-level environment.
The living plant's own defenses may also indirectly increase bacterial freezing tolerance. R. solanacearum cells growing in plants experience stress from host reactive oxygen species (ROS) and antimicrobial compounds (5, 6, 15). The diverse stress-protective mechanisms expressed by this pathogen during bacterial wilt pathogenesis may also protect it from an initial cycle of freezing stress. However, when the surviving bacteria begin to grow again after the first thaw, these protective mechanisms may no longer be active, because the dead plant no longer induces them. This mechanism could also explain why the second temperature cycle was far more detrimental to bacterial survival than the first. This cycle of nutrient-facilitated growth, death by freezing damage, and renewed growth of the survivors then continues as the temperature fluctuates until the entire pathogen population is nonviable.
Following cold stress, R. solanacearum can become viable but not culturable (VBNC) because of reduced tolerance for reactive oxidase species (49). We previously found that during prolonged storage in water at 4°C, R. solanacearum populations gradually diverged into (i) cells that formed colonies on conventional medium, (ii) cells that formed colonies only on medium treated with catalase to reduce ROS, and (iii) cells that did not grow on either medium but had intact membranes in a live/dead stain (31). To determine if bacteria were moving into a VBNC state following the rapid temperature cycling in these experiments, we plated samples on medium containing sodium pyruvate, which, like catalase, increases the recovery of R. solanacearum cells damaged by ROS stress (24). However, this did not improve recovery of cells from any tissue or water culture samples, suggesting that these populations did not contain significant numbers of VBNC cells. Live/dead staining of samples would confirm this result.
Prolonged but constant low-temperature stress appeared to broadly weaken R. solanacearum. UW551 cells subjected to extended periods at −20°C inside geranium tissue could form colonies on plates, but they were debilitated. They grew poorly or not at all in rich broth and did not recover even after repeated subculturing. Of greater potential epidemiological importance, they were also significantly reduced in bacterial wilt virulence. Because relatively few viable cells were present in the frozen geranium tissue, we inoculated tomato plants through a cut petiole, a method that requires only a few hundred pathogen cells. Petiole inoculation, which strongly favors the pathogen because it bypasses the natural infection route into the tomato, usually results in rapid disease progress and typically kills the plant in less than 2 weeks (44). However, plants inoculated with cells recovered from frozen geranium stems wilted slowly or never developed symptoms. This reduction in virulence suggests that R3bv2 cells that survived prolonged exposure to cold or to temperature fluctuations in the field might be significantly impaired in their ability to cause wilt disease. Additional physiological studies are needed to understand the mechanisms underlying this unexpected result.
Potato brown rot is widespread and persistent in many tropical highland regions around the world, demonstrating that R3bv2 survives in that environment (12). Temperatures in the Andean highlands where R3bv2 originated are cool but moderate, averaging 11°C year-round, with a mean minimum temperature of 0.1°C and a mean maximum temperature of 21°C at 3,400 m above sea level (http://www.worldweather.org/). Mean low winter temperatures in the United Kingdom and the Netherlands, where R3bv2 has persisted in waterways and weeds, average 1.0°C and 0.9°C, respectively. However, the winters in the northern United States and Canada are significantly colder than those in northern Europe. Average low temperatures in the upper Midwest are −12°C during the coldest months, with a soil frost depth typically exceeding 75 cm. Subfreezing temperatures alternating with temperatures above freezing commonly occur in 6 months of the year (http://www.esrl.noaa.gov/psd/data/usclimate/states.fast.html). These conditions are stressful for many bacteria; Salmonella enterica serovar Typhimurium and Escherichia coli cannot survive the freeze-thaw cycles of a typical northern U.S. winter (32). In our experiments, R3bv2 strains did not survive successive temperature fluctuations around freezing, suggesting that R3bv2 has no special adaptations for surviving repeated freeze-thaw cycles. Further, an endemic North American sequevar 7 R. solanacearum strain also could not survive repeated moderate freeze-thaw cycles. This may explain why these strains have never become established north of the mid-Atlantic states, even though they are common in the southeast and have probably been disseminated repeatedly in soil, vegetable seedlings, tubers, and cuttings.
Taken together, the results presented here suggest that R3bv2 is not likely to become established in commercial potato-growing regions of North America because it is not adapted to survive frequent near-freezing temperature fluctuations even when sheltered in host plant tissue. Additional research under more-realistic conditions would permit exploration of other potentially important survival parameters, such as desiccation and microbial competition (3, 49). Specifically, it would be desirable to quantify the ability of R3bv2 to survive a northern winter in potato or geranium cull piles or at various depths in natural soil. However, regulations governing research with Select Agent pathogens preclude experiments for measuring R3bv2 survival in the field.
Acknowledgments
This research was supported by USDA CSREES Plant Biosecurity project 2006-04560, by the USDA-ARS Floral and Nursery Crops Research Initiative, and by the University of Wisconsin College of Agricultural and Life Sciences.
We thank AgDia, Inc. (Elkhart, IN), and Ball FloraPlant (West Chicago, IL) for donated research materials and Michael Klopmeyer and Timothy Denny for useful discussions.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Agrios, G. N. 2005. Plant Pathology, 5th ed. Elsevier Academic Press, Amsterdam, Netherlands.
- 2.Allen, C., A. Kelman, and E. R. French. 2001. Brown rot of potatoes, p. 11-13. In W. R. Stevenson, R. Loria, G. D. Franc, and D. P. Weingartner (ed.), Compendium of potato diseases, 2nd ed. APS Press, St. Paul, MN.
- 3.Alvarez, B., M. Lopez, and E. Biosca. 2007. Influence of native microbiota on survival of Ralstonia solanacearum phylotype II in river water microcosms. Appl. Environ. Microbiol. 73:7210-7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez, B., M. Lopez, and E. Biosca. 2008. Survival strategies and pathogenicity of Ralstonia solanacearum phylotype II subjected to prolonged starvation in environmental water microcosms. Microbiology 154:3590-3598. [DOI] [PubMed] [Google Scholar]
- 5.Brown, D., and C. Allen. 2004. Ralstonia solanacearum genes induced during growth in tomato: an inside view of bacterial wilt. Mol. Microbiol. 53:1641-1660. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. G., J. K. Swanson, and C. Allen. 2007. Two host-induced Ralstonia solanacearum genes, acrA and dinF, encode multidrug efflux pumps and contribute to bacterial wilt virulence. Appl. Environ. Microbiol. 73:2777-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caruso, P., J. L. Palomo, E. Bertolini, B. Alvarez, M. M. López, and E. G. Biosca. 2005. Seasonal variation of Ralstonia solanacearum biovar 2 populations in a Spanish river: recovery of stressed cells at low temperatures. Appl. Environ. Microbiol. 71:140-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciampi, L., and L. Sequeira. 1980. Influence of temperature on virulence of race 3 strains of Pseudomonas solanacearum. Am. Potato J. 57:307-317. [Google Scholar]
- 9.Ciampi, L., L. Sequeira, and E. R. French. 1980. Latent infection of potato tubers by Pseudomonas solanacearum. Am. Potato J. 57:377-386. [Google Scholar]
- 10.Denny, T. P. 2006. Plant pathogenic Ralstonia species, p. 573-644. In S. S. Gnanamanickam (ed.), Plant-associated bacteria. Springer Publishing, Dordrecht, Netherlands.
- 11.Elphinstone, J. 1996. Survival and possibilities for extinction of Pseudomonas solanacearum (Smith) in cool climates. Potato Res. 39:403-410. [Google Scholar]
- 12.Elphinstone, J. G. 2005. The current bacterial wilt situation: a global overview, p. 9-28. In C. Allen, P. Prior, and A. C. Hayward (ed.), Bacterial wilt: the disease and the Ralstonia solanacearum species complex. American Phytopathological Society Press, St. Paul, MN.
- 13.Elphinstone, J. G., H. M. Stanford, and D. E. Stead. 1998. Survival and transmission of Ralstonia solanacearum in aquatic plants Solanum dulcamara and associated surface water in England. OEPP Bull. 28:93-94. [Google Scholar]
- 14.Fegan, M., and P. Prior. 2005. How complex is the “Ralstonia solanacearum species complex”?, p. 449-461. In C. Allen, P. Prior, and A. C. Hayward (ed.), Bacterial wilt disease and the Ralstonia solanacearum species complex. APS Press, St. Paul, MN.
- 15.Flores-Cruz, Z., and C. Allen. 2009. Ralstonia solanacearum encounters an oxidative environment during tomato infection. Mol. Plant Microbe Interact. 22:773-782. [DOI] [PubMed] [Google Scholar]
- 16.Graham, J., D. A. Jones, and A. B. Lloyd. 1979. Survival of Pseudomonas solanacearum race 3 in plant debris and in latently infected potato tubers. Phytopathology 69:1100-1103. [Google Scholar]
- 17.Graham, J., and A. B. Lloyd. 1979. Survival of potato strain (race 3) of Pseudomonas solanacearum in the deeper soil layers. Aust. J. Agric. Res. 30:489-496. [Google Scholar]
- 18.Granada, G. A., and L. Sequeira. 1983. Survival of Pseudomonas solanacearum at low temperature. Fitopatologia 18:22-24. [Google Scholar]
- 19.Granada, G. A., and L. Sequeira. 1983. Survival of Pseudomonas solanacearum in soil, rhizosphere, and plant roots. Can. J. Microbiol. 29:433-440. [Google Scholar]
- 20.Harrison, D. E. 1961. Bacterial wilt of potatoes I. Field symptoms of the disease and studies on the causal organism, Pseudomonas solanacearum var. asiaticum. Aust. J. Agric. Res. 12:854-871. [Google Scholar]
- 21.Hayward, A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29:65-87. [DOI] [PubMed] [Google Scholar]
- 22.Hayward, A. C. 1994. Systematics and phylogeny of Pseudomonas solanacearum and related bacteria, p. 123-135. In A. C. Hayward and G. L. Hartman (ed.), bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford, United Kingdom.
- 23.Hendrick, C. A., and L. Sequeira. 1984. Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl. Environ. Microbiol. 48:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imazaki, I., and K. Nakaho. 2009. Pyruvate-amended modified SMSA medium: improved sensitivity for detection of Ralstonia solanacearum. J. Gen. Plant Pathol. 76:52-61. [Google Scholar]
- 25.Janse, J. D., H. E. van den Beld, J. Elphinstone, S. Simpkins, N. A. A. Tjou-Tam-Sin, and J. van Vaerenbergh. 2004. Introduction to Europe of Ralstonia solanacearum biovar 2, race 3 in Pelargonium zonale cuttings. J. Plant Pathol. 86:147-155. [Google Scholar]
- 26.Jenkins, D. E., J. E. Schultz, and A. Matin. 1988. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J. Bacteriol. 170:3910-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jouper-Jaan, A., A. E. Goodman, and S. Kjelleberg. 1992. Bacteria starved for prolonged periods develop increased protection against lethal temperatures. FEMS Microbiol. Lett. 101:229-236. [Google Scholar]
- 28.Kelman, A. 1954. The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance in tetrazolium medium. Phytopathology 44:693-695. [Google Scholar]
- 29.Kim, S. H., T. N. Olson, N. W. Schaad, and G. W. Moorman. 2003. Ralstonia solanacearum race 3, biovar 2, the causal agent of brown rot of potato, identified in geranium in Pennsylvania, Delaware, and Connecticut. Plant Dis. 87:450. [DOI] [PubMed] [Google Scholar]
- 30.Maloy, O. C. 1993. Plant disease control: principles and practice. John Wiley & Sons, Inc., New York, NY.
- 31.Milling, A., F. Meng, T. P. Denny, and C. Allen. 2009. Interactions with hosts at cool temperatures, not cold tolerance, explain the unique epidemiology of Ralstonia solanacearum race 3 biovar 2. Phytopathology 99:1127-1134. [DOI] [PubMed] [Google Scholar]
- 32.Natvig, E., S. Ingham, B. Ingham, L. Cooperband, and T. Roper. 2002. Salmonella enterica serovar Typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soils with incorporated bovine manure. Appl. Environ. Microbiol. 68:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phadtare, S. 2004. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6:125-136. [PubMed] [Google Scholar]
- 34.Pradhanang, P. M., J. G. Elphinstone, and R. T. V. Fox. 2000. Identification of crop and weed hosts of Ralstonia solanacearum biovar 2 in the hill of Nepal. Plant Pathol. 49:403-413. [Google Scholar]
- 35.Schaad, N. W., J. B. Jones, and W. Chun. 2001. Laboratory guide for the identification of plant pathogenic bacteria, 3rd ed. APS Press, St. Paul, MN.
- 36.Schimel, J., and C. Mikan. 2004. Changing microbial substrate use in Arctic tundra soils through a freeze-thaw cycle. Soil Biol. Biochem. 37:1411-1418. [Google Scholar]
- 37.Shamsudin, N., A. B. Lloyd, and J. Graham. 1978. Survival of the potato strain of Pseudomonas solanacearum in soil. J. Aust. Inst. Agric. Sci. 44:212-215. [Google Scholar]
- 38.Skogland, T., S. Lomeland, and J. Goksøyr. 1988. Respiratory burst after freezing and thawing of soil: experiments with soil bacteria. Soil Biol. Biochem. 20:851-856. [Google Scholar]
- 39.Smith, J., L. Offord, M. Holderness, and G. S. Saddler. 1995. Genetic diversity of Burkholderia solanacearum (synonym Pseudomonas solanacearum) race 3 in Kenya. Appl. Environ. Microbiol. 61:4263-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strider, D. L., R. K. Jones, and R. A. Haygood. 1981. Southern wilt of geranium caused by Pseudomonas solanacearum. Plant Dis. 65:52-53. [Google Scholar]
- 41.Swanson, J., L. Montes, L. Mejia, and C. Allen. 2007. Detection of latent infections of Ralstonia solanacearum race 3 biovar 2 in geranium. Plant Dis. 91:828-834. [DOI] [PubMed] [Google Scholar]
- 42.Swanson, J., J. Yao, J. Tans-Kersten, and C. Allen. 2005. Behavior of Ralstonia solanacearum race 3 biovar 2 during latent and active infection of geranium. Phytopathology 95:136-146. [DOI] [PubMed] [Google Scholar]
- 43.Tans-Kersten, J., Y. Guan, and C. Allen. 1998. Ralstonia solanacearum pectin methylesterase is required for growth on methylated pectin but not for bacterial wilt virulence. Appl. Environ. Microbiol. 64:4918-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tans-Kersten, J., H. Huang, and C. Allen. 2001. Ralstonia solancearum needs motility for invasive virulence on tomato. J. Bacteriol. 183:3597-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thurston, H. D., and J. C. Lozano. 1968. Resistance to bacterial wilt of potatoes in Columbian clones of Solanum phureja. Am. Potato J. 40:51-55. [Google Scholar]
- 46.Tung, P. X., E. T. Rasco, P. VanderZaag, and P. Schmiediche. 1990. Resistance to Pseudomonas solanacearum in the potato: II. Aspects of host-pathogen-environment interaction. Euphytica 45:211-215. [Google Scholar]
- 47.Tusiime, G., E. Adipala, F. Opio, and A. S. Bhagsari. 1998. Weeds as latent hosts of Ralstonia solanacearum in highland Uganda: implications to development of an integrated control package for bacterial wilt. .In P. Prior, C. Allen, and J. Elphinstone (ed.), Bacterial wilt disease: molecular and ecological aspects. Springer Verlag, Berlin, Germany.
- 48.Van der Wolf, J. M., P. J. M. Bonants, J. J. Smith, M. M. Hagenaar, E. Nijhuis, J. R. C. M. V. Beckhoven, G. S. Saddler, A. Trigalet, and R. Feuillade. 1998. Genetic diversity of Ralstonia solanacearum race 3 in Western Europe determined by AFLP, RC-PFGE, and Rep-PCR., p. 44-49. In P. Prior, C. Allen, and J. G. Elphinstone (ed.), Bacterial wilt disease: molecular and ecological aspects. Springer Verlag, Berlin, Germany.
- 49.van Elsas, J. D., P. Kastelein, P. M. de Vries, and L. S. van Overbeek. 2001. Effects of ecological factors on the survival and physiology of Ralstonia solanacearum bv. 2 in irrigation water. Can. J. Microbiol. 47:842-854. [DOI] [PubMed] [Google Scholar]
- 50.van Elsas, J. D., P. Kastelein, P. van Bekkum, J. M. van der Wolf, P. M. de Vries, and L. S. van Overbeek. 2000. Survival of Ralstonia solanacearum biovar 2, the causative agent of potato brown rot, in field and microcosm soils in temperate climates. Phytopathology 90:1358-1366. [DOI] [PubMed] [Google Scholar]
- 51.Walker, V., G. Palmer, and G. Voordouw. 2006. Freeze-thaw tolerance and clues to the winter survival of a soil community. Appl. Environ. Microbiol. 72:1784-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wenneker, M., M. S. W. Verdel, R. M. W. Groeneveld, C. Kempenaar, A. R. van Beuningen, and J. D. Janse. 1999. Ralstonia (Pseudomonas) solanacearum race 3 (biovar 2) in surface water and natural weed hosts: first report on stinging nettle (Urtica dioica). Eur. J. Plant Pathol. 105:307-315. [Google Scholar]
- 53.Williamson, L., N. Kazuhiro, B. Hudelson, and C. Allen. 2002. Ralstonia solanacearum race 3, biovar 2 strains isolated from geranium are pathogenic on potato. Plant Dis. 86:987-991. [DOI] [PubMed] [Google Scholar]