Abstract
The incorporation of plant residues into soil not only represents an opportunity to limit soil organic matter depletion resulting from cultivation but also provides a valuable source of nutrients such as nitrogen. However, the consequences of plant residue addition on soil microbial communities involved in biochemical cycles other than the carbon cycle are poorly understood. In this study, we investigated the responses of one N-cycling microbial community, the nitrate reducers, to wheat, rape, and alfalfa residues for 11 months after incorporation into soil in a field experiment. A 20- to 27-fold increase in potential nitrate reduction activity was observed for residue-amended plots compared to the nonamended plots during the first week. This stimulating effect of residues on the activity of the nitrate-reducing community rapidly decreased but remained significant over 11 months. During this period, our results suggest that the potential nitrate reduction activity was regulated by both carbon availability and temperature. The presence of residues also had a significant effect on the abundance of nitrate reducers estimated by quantitative PCR of the narG and napA genes, encoding the membrane-bound and periplasmic nitrate reductases, respectively. In contrast, the incorporation of the plant residues into soil had little impact on the structure of the narG and napA nitrate-reducing community determined by PCR-restriction fragment length polymorphism (RFLP) fingerprinting. Overall, our results revealed that the addition of plant residues can lead to important long-term changes in the activity and size of a microbial community involved in N cycling but with limited effects of the type of plant residue itself.
Modern agricultural practices include a return of plant residues to soil, as this is considered sustainable to the environment. It is now recognized that the conversion of native land into cultivated systems leads to carbon losses, which can be up to 20 to 40% (17). Postharvest plant residues therefore represent an important source of carbon, helping to replenish soil organic matter that decomposes as a result of cultivation. Decomposing plant residues are also a source of nutrients, such as nitrogen, with reduced nitrate leaching compared to mineral fertilizers, which is beneficial for water quality (3). In addition, leaving the plant residue on the soil surface limits water losses by evaporation and prevents soil erosion by wind or water (15).
The biochemical composition of plant residues is one of the most important factors influencing their decomposition in soil (14, 28, 29, 51). Indeed, Manzoni et al. (28), using a data set of 2,800 observations, showed previously that the patterns of decomposition were regulated by the initial residue stoichiometry. Several other factors such as climatic conditions, soil type, or localization of the residue in the soil (incorporated or on the soil surface) were also reported previously to influence decomposition (2, 24, 29, 44). Microorganisms are the major decomposers of organic matter in soil, and therefore, the diversity and activity of the microbial community during plant residue decomposition has received much attention (6, 23, 26, 27, 35). It was shown previously that the biochemical composition of plant residues influences microbial respiration (8) and microbial community structure (7, 37). The recent development of carbon-labeling approaches has furthered our knowledge of the microorganisms that actively assimilate the carbon derived from various plant residues (10, 31). However, most of those studies focused on microorganisms involved in C mineralization, and in contrast, very little is known about the effect of plant residue decomposition on the microbial communities involved in biochemical cycles other than the carbon cycle. Thus, despite the influence of plant residues on nitrogen cycling (1, 4, 5, 16, 20), studies assessing the effect of the presence and composition of plant residues on the ecology of microbial communities involved in nitrogen cycling are rare (21, 32, 36).
The dissimilatory reduction of nitrate into nitrite is the first step in the processes of denitrification and the dissimilatory reduction of nitrate to ammonium (33, 41). The reduction of nitrate by denitrification leads to losses of nitrogen, which is often a limiting nutrient for plant growth in agriculture. Two types of dissimilatory nitrate reductases, differing in location, have been characterized: a membrane-bound nitrate reductase (Nar) and a periplasmic nitrate reductase (Nap) (9, 53). Nitrate reducers can harbor either Nar, Nap, or both (40, 47). Nitrate reducers are probably the most taxonomically diverse functional community within the nitrogen cycle, with members in most bacterial phyla and also archaea (42). Because of this high level of diversity of heterotrophs sharing the ability to produce energy from nitrate reduction, nitrate reducers are an excellent model system to investigate the response of the N-cycling community to plant residue addition.
The aim of this work was to determine how the incorporation of plant residues with contrasting biochemical compositions into soil affects the nitrate-reducing community. For this purpose, we monitored the dynamics of the potential activity, size, and structure of the nitrate-reducing community after the addition of wheat, rape, or alfalfa residues to soil in a field experiment. As the nature and availability of the substrate change during residue decomposition (38, 39, 48), the influence of the incorporation of different plant residues on the nitrate-reducing community was investigated at several sampling times for 11 months.
MATERIALS AND METHODS
Experimental field and sampling.
The experiment was conducted at the INRA experimental site at Epoisse, Burgundy, France (47.14°N, 5.06°E). The soil is a loamy clay soil with the following physicochemical properties: clay (<2 μm), 37.9%; fine silt (2 to 20 μm), 32.5%; silt (20 to 50 μm), 23.7%; fine sand (50 to 200 μm), 3.8%; sand (200 to 2,000 μm), 2.1%; pH 6.73; CaCO3, <1 g·kg−1; organic carbon, 14.7 g·kg−1; total nitrogen, 1.33 g·kg−1; and C/N ratio, 11.1. The experimental field was set up specifically to evaluate the influence of the crop residues on the structure and diversity of soil bacterial communities. Before the experiment, maize was grown on all plots, and soil characteristics over the entire experimental site were analyzed before the different treatments were set up. The field was shown to be homogeneous for the different soil parameters measured: soil texture, pH, CaCO3, and organic matter contents (O. Mathieu and J. Lévêque, personal communication). The field experiment consisted of four treatments: control soil, soil amended with wheat residues (Triticum aestivum [13 t ha−1 dry matter]), soil amended with rape residues (Brassica napus [17 t ha−1 dry matter]), and soil amended with alfalfa residues (Medicago sativa [4 t ha−1 dry matter]). The plots were amended with these residues only in July 2006, and the amount of residue incorporated for each crop corresponded to what was left behind in the soil after harvesting. The wheat, rape, and alfalfa residues had C/N ratios of 94, 51, and 27, respectively, as determined by total combustion in a 1500 CN elemental analyzer (Carlo Erba). Each treatment was laid out in three replicates consisting of three independent 3- by 8-m plots. At the beginning of the experiment on 4 July 2006, the residues were ground and incorporated into the first 15 cm of soil by tilling using a Rotavator. The control plots, i.e., without residues, were also tilled. Soil samples were collected from each of the four treatments 11 times between July 2006 and June 2007. At each sampling date, three soil subsamples were collected from the top soil layer (0 to 15 cm) over a 20- by 20-cm area for each of the 12 plots. These subsamples were pooled and mixed to obtain a single composite soil sample per plot, resulting in a total of 132 soil samples. Each soil sample was then sieved at 2 mm and stored at −20°C.
Measurement of potential nitrate reductase activity.
The potential nitrate reductase activity was determined by soil anaerobic incubation, with slight modifications of a protocol described previously by Kandeler (25). The method involved a determination of NO2−-N production after adding nitrate as a substrate and 2,4-dinitrophenol (DNP) as an uncoupler of oxidative phosphorylation that interfered with electron transfer but allowed nitrate reduction to continue. Optimal substrate and DNP inhibitor concentrations were determined in preliminary experiments. In detail, for each composite soil sample per plot, four soil aliquots of 0.2 g were incubated for 24 h at 28°C in a total volume of 1 ml containing 1 mM potassium nitrate. The optimum inhibitory concentration of DNP (0.9 mM) was then added to inhibit nitrite reduction. The soil mixture was extracted with 4 M KCl and centrifuged for 1 min at 13,000 × g. The nitrite concentration in the supernatant was determined before and after incubation of the soil samples with nitrate and DNP by a colorimetric reaction at 520 nm using a reagent composed of phosphoric acid (1%, vol/vol), N1-naphthyl ethylenediamine dichloride (2 g·liter−1), and sulfanilamide (40 g·liter−1).
Soil DNA extraction.
The DNA was extracted from three replicate samples from the treatments with wheat, rape, or alfalfa residues and from the control soil, according to a method described previously by Martin-Laurent et al. (30). Briefly, 1 g of each soil sample was mixed with a solution that contained 100 mM Tris (pH 8.0), 100 mM EDTA (pH 8.0), 100 mM NaCl, and 2% (wt/vol) sodium dodecyl sulfate. Two grams of 106-μm-diameter glass beads and eight 2-mm-diameter glass beads were added in a bead-beater tube. The samples were then homogenized for 30 s at 1,600 rpm in a mini-bead-beater cell disruptor (Mikrodismembrator; S. B. Braun Biotech International) and centrifuged at 7,000 × g for 5 min at 4°C after incubation for 30 min at 70°C. The collected supernatants were incubated for 10 min on ice with a 1/10 volume of 3 M potassium acetate (pH 5.5) and centrifuged at 14,000 × g for 5 min. After precipitation with 1 volume of ice-cold isopropanol, the nucleic acids were washed with 70% ethanol. For purification, aliquots (100 μl) of crude DNA extracts were loaded onto polyvinyl polypyrrolidone (PVPP) minicolumns (Bio-Rad, Marne la Coquette, France) and centrifuged at 1,000 × g for 2 min at 10°C. The eluate was collected and purified from residual impurities by using a Geneclean Turbo kit as recommended by the manufacturer (Q Biogene, Illkirch, France).
PCR-RFLP fingerprinting analysis of the nitrate-reducing community.
The structure of the nitrate-reducing community was determined by using the narG and napA genes, which encode the membrane-bound and periplasmic nitrate reductases, respectively, as molecular markers. The DNA extracts from each replicate plot from the four treatments on four sampling dates (4 June 2006, 26 June 2006, 20 December 2006, and 15 March 2007, which resulted in a total of 48 DNA samples) were used for a restriction fragment length polymorphism (RFLP)-PCR fingerprinting analysis. The narG and napA genes were amplified by using primers narG1960F (5′-TAYGTSGGSCARGARAA-3′) and narG2650R (5′-TTYTCRTACCABGTBGC-3′), and primers napV67m (5′-AAYATGGCVGARATGCACCC-3′) and napV17m (5′-GRTTRAARCCCATSGTCCA-3′) (22), and previously described PCR conditions (22, 43). PCRs were carried out in a 50-μl mixture containing 1.5 mM MgCl2 buffer, 200 mM each deoxyribonucleoside triphosphate, 5 mM each primer, and 1.25 U of Taq polymerase (Qbiogene, France). At least three independent PCRs were performed, and the PCR products were then pooled for each replicate to minimize the effect of PCR bias. PCR products were purified by using the MiniElute gel extraction kit (Qiagen, France). Purified narG and napA PCR products were digested with the AluI restriction enzyme at 37°C for 4 h as previously described (22). The narG and napA RFLP fingerprints were obtained after separation by electrophoresis on a native 6% acrylamide-bisacrylamide (29:1) gel for 11 h at 5 mA. DNA staining was done by using Sybr green, and the resulting fluorescence was scanned with a phosphorimager (Storm 860; Molecular Dynamics, Sunnyvale, CA).
Estimation of the size of the nitrate-reducing community by qPCR.
A quantitative PCR (qPCR) assay was carried out with a 20-μl reaction mixture containing Sybr green PCR master mix (Absolute QPCR Sybr green; Rox ABgene, France), 1 μM each primer, 100 ng of T4 gene 32 (QBiogene, France), and 12.5 ng of DNA. Fragments of the narG and napA genes were amplified by using previously described primers and thermal cycling conditions (12). Thermal cycling, fluorescent data collection, and data analysis were carried out with an ABI Prism 7900HT sequence detection system according to the manufacturer's instructions (Applied Biosystems, France). Two independent quantitative PCR assays were performed for each gene and for the three DNA extracts from each treatment and each sampling date. Two to three no-template controls (NTCs) were run for each quantitative PCR assay. Serial dilutions of linearized plasmids containing the narG and napA genes from Pseudomonas aeruginosa PAO1 were used to generate standard curves, and the copy number of standard DNA genes was calculated as previously described (12). All narG and napA assays were run with DNA from P. aeruginosa PAO1 containing one copy of each of these two genes as an external standard. Assay specificity was verified by melting-curve analysis and gel electrophoresis. Tests for the potential presence of PCR inhibitors in DNA extracted from soil were performed by spiking soil DNA extracts with a known amount of standard DNA prior to qPCR. In all cases, no inhibition was detected.
Data analysis.
When data were not normally distributed, a log transformation was applied and a Student's t test was used to analyze the qPCR and activity data. For each type of residue, the percentage of the change from the control was calculated for the narG and napA gene copy numbers and for the nitrate reduction activity {[(Xresidue − Ycontrol)/Ycontrol] × 100}. Temporal variations between sampling dates were analyzed by using a Mann-Whitney test after arcsine transformation of the percentage data using the XLSTAT software (Addinsoft SARL, France). The narG and napA PCR-RFLP gels were analyzed by using One-DScan software, version 2.05 (Scanalytics Inc.). Data matrices (band presence and intensity) were then used to perform a principal-component analysis (PCA) with ADE-4 software. Pearson correlation analyses were performed by using the XLSTAT software (Addinsoft SARL, France) to test the relationships between nitrate reduction rates and temperature.
RESULTS
Dynamics of the activity of the nitrate-reducing community in relation to presence and composition of plant residues.
Leaving the plant residues in the field at the beginning of July 2006 immediately stimulated potential nitrate reduction activity, with rates ranging from 0.8 to 1.2 μg N-NO2− g−1 dry soil day−1 in the control plots and from 17.0 to 26.1 μg N-NO2− g−1 dry soil day−1 in the plant residue-amended soil during the first week (Fig. 1 A). This 20- to 25-fold stimulation of potential nitrate reduction activity in the plant residue-amended plot then rapidly decreased until fall 2006 (Fig. 1B). Significant differences in potential nitrate reduction between the amended and the control plots were still observed in winter 2006 and spring 2007 but with only 2- to 3-fold increases. The type of plant residue had a weak effect on potential nitrate reduction. Thus, no significant differences were observed between the wheat- and rape-amended plots for 11 months, while the alfalfa-amended plot first had higher and then had lower rates than the two other residue-amended plots (Fig. 1). The potential nitrate reduction activity in the plots with residues was highly correlated with temperature during the first 10 months (r2 of 0.59, 0.69, and 0.64 for wheat, rape, and alfalfa, respectively; P < 0.001) but not with rainfall.
FIG. 1.
(A) Dynamics of potential nitrate reduction rates in control plots and in plots amended with wheat, rape, or alfalfa residues. For each date, the same letters above the bars (means ± standard deviations; n = 9) indicate that treatments were not significantly different according to a Student's t test (P < 0.05). Rainfall (□) and temperature (×) data are indicated for each sampling date. (B) Percentage of changes from the control in the residue-amended plots. For the same residue, identical letters above the bars (means ± standard deviations; n = 9) indicate that the sampling times were not significantly different according to a Mann-Whitney test (P < 0.05).
Dynamics of the size of the nitrate-reducing community in response to plant residue incorporation.
The average gene copy number of narG was higher than that of napA, with densities of 1.6 × 107 to 28 × 107 and 3.4 × 106 to 45 × 106 copies per gram of dry soil, respectively (Fig. 2 and 3). Interestingly, the same temporal dynamics were observed for both genes with lower densities in fall 2006 and spring 2007. The incorporation of the plant residue into the soil resulted in an increase of the napA gene copy numbers on the first sampling date, while only the increase in the rape-amended plot was significant on the same date for narG. For the three following sampling dates (i.e., until 28 August), both the narG and napA gene copy numbers were higher in the residue-amended plots than in the control plots. After summer 2006, the presence of plant residues had a significant effect on the narG gene copy numbers only in March and June 2007. In contrast, the plant residue effect was stronger for the napA genes, with significant increases in the residue-amended plots at several sampling dates during winter 2006 and spring 2007. For both genes, a higher gene copy number was observed during the last months in the wheat and rape residue-amended plots than in the alfalfa residue-amended plots, but the differences were significant only for the napA gene (Fig. 2 and 3).
FIG. 2.
(A) Dynamics of the estimated size of the nitrate-reducing community using the narG gene as a molecular marker in control plots and in plots amended with wheat, rape, or alfalfa residues. For each date, the same letters above the bars (means ± standard deviations; n = 9) indicate that treatments were not significantly different according to a Student's t test (P < 0.05). (B) Percentage of changes from the control in the residue-amended plots. For the same residue, identical letters above the bars (means ± standard deviations; n = 9) indicate that the sampling times were not significantly different according to a Mann-Whitney test (P < 0.05).
FIG. 3.
(A) Dynamics of the estimated size of the nitrate-reducing community using the napA gene as a molecular marker in the control plots and in plots amended with wheat, rape, or alfalfa residues. For each date, identical letters above the bars (means ± standard deviations; n = 9) indicate that treatments were not significantly different according to a Student's t test (P < 0.05). (B) Percentage of changes from the control in the residue-amended plots. For the same residue, identical letters above the bars (means ± standard deviations; n = 9) indicate that the sampling times were not significantly different according to a Mann-Whitney test (P < 0.05).
Effect of the presence and composition of plant residues on the structure of the nitrate-reducing community.
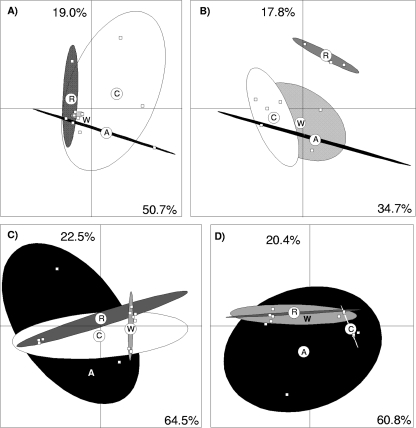
The structure of the nitrate-reducing community was analyzed at four sampling dates (Fig. 4 and 5), which were selected on the basis of the potential nitrate reduction activity data. The impact of gel-to-gel variations on gel analysis was avoided by running all samples from the same sampling date on the same gel. Twelve to twenty-seven bands were used for the analysis of the narG or napA RFLP gels (see Fig. S1 in the supplemental material). Principal-component analysis of the napA community did not reveal any significant differences between the different treatments, including the control plots, at all dates except 26 July. Thus, the napA community structure in the plots amended with rape residues separated on axis 2 from those from all the other plots (Fig. 5). Likewise, the addition of plant residues had little impact on the structure of the narG community, with a separate clustering of the plots amended with wheat residues on 15 March (Fig. 4D).
FIG. 4.
Principal-component analysis of the structure of the nitrate reducer community using the narG gene in the control plots (C) (white) and in plots amended with wheat (W) (light gray), rape (R) (dark gray), or alfalfa (A) (black) residues. (A) Data for 4 July 2006; (B) data for 26 July 2006; (C) data for 20 December 2006; (D) data for 15 March 2007. Statistical ellipses drawn over the plot replicates represent 90% confidence. The percentages of explained variations for the first two axes are indicated within the figures.
FIG. 5.
Principal-component analysis of the structure of the nitrate reducer community targeted using the napA gene in the control plots (C) (white) and in plots amended with wheat (W) (light gray), rape (R; dark gray), or alfalfa (A) (black) residues. (A) Data for 4 July 2006; (B) data for 26 July 2006; (C) data for 20 December 2006; (D) data for 15 March 2007. Statistical ellipses drawn over the plot replicates represent 90% confidence. The percentages of explained variations for the first two axes are indicated within the figures.
DISCUSSION
Temporal variation of nitrate reducer abundance, structure, and activity in response to decomposing plant residues.
The incorporation of the plant residues in the field resulted in an approximately 20-fold increase of the potential nitrate reduction activity within 1 day, which suggests that carbon was a strong limiting factor. Accordingly, Pascault et al. (39) previously showed a stimulation of soil microbial respiration in the amended plots in comparison to the control plots at the same experimental site. In a previous study, the addition of farmyard manure combined with mineral fertilizer resulted in potential nitrate reduction rates of more than 20 μg N-NO2− g−1 dry soil (13), which were in the same range as those observed for the residue-amended plots. The addition of crop residues to soil microcosms was shown previously to also promote denitrification (21, 32). Together with the nitrate concentration and oxygen partial pressure, the role of carbon as a major factor controlling respiratory nitrate reduction is well known (11, 50). Myrold and Tiedje (34) suggested previously that carbon availability in agricultural soil amended with alfalfa straw could be even more limiting for denitrification than nitrate concentration or oxygen partial pressure. In the residue-amended plots where carbon was not limiting, we found a very good correlation (P < 0.001) between potential nitrate reduction activity and temperature from 4 July 2006 until 4 May 2007, which also underlines the importance of temperature in regulating microbial activity (52). The Q10 for nitrate reduction over the temperature range of 2.3°C to 19.9°C could be up to 5, which is higher than the general range of 2 to 3 reported previously for microbial respiration (45). Higher Q10 values for anaerobic processes such as denitrification have been explained by an increase of the anaerobic soil volume resulting from higher microbial respiration (49). The temperature effect was less obvious when the last sampling date was taken into account, probably due to the fact that most of the carbon from the decomposing residues had already been used, thus resulting in limiting carbon availability. However, a weaker but still significant increase in the potential nitrate reductase activity was observed in June 2007, indicating a long-term stimulating effect of residue on N cycling, which lasted at least 11 months (Fig. 1). In contrast, Baggs et al. (5) previously observed a short-lived effect of crop residue incorporation on N2O fluxes, with most of the emissions occurring during the first 2 weeks and then returning to background levels after 30 to 40 days. Likewise, Aulakh et al. (4) reported that the effect of legume or cereal crop residues on denitrification was greater during the first week of decomposition. Since a significant residue effect was still observed after several months in our experiment, it is likely that the crop residue promoted nitrate reduction by supplying not only carbon but also nitrogen after mineralization and nitrification of the nitrogen present in the plants. However, this putative effect was probably more limited for the wheat than for the other residues, due to the high C/N ratio of the former (ratio of 94).
The stimulation of nitrate reducer activity resulting from the incorporation of plant residues into the soil was concomitant with an increase in the size of the nitrate reducer communities during the first 2 months (Fig. 2 and 3). This is consistent with data from a recent study by Miller et al. (32), which showed a similar increase in the abundance of denitrifiers in response to the higher availability of carbon substrate in soil resulting from decomposing residues. However, while we cannot rule out the possibility that the large increase in numbers of the nitrate reducers in response to residue incorporation during July 2006 might have contributed to the high initial stimulation of the potential nitrate reduction activity, in general, the size of the nitrate-reducing community was not an important driver of nitrate reductase activity over the 11 months of the experiment. As an example, the decrease of potential nitrate reduction during winter was not mirrored by a decrease of the narG or napA gene copy numbers (Fig. 2 and 3). This absence of a relationship between the size and activity of the nitrate reducer community is consistent with respiratory nitrate reduction being a facultative process, which therefore depends primarily on the presence of nitrate, oxygen limitation, and electron donor availability (50). However, differences in the sizes of the nitrate-reducing community might be reflected in the nitrate reduction activity when environmental conditions are not limiting for the expression of nitrate reductase. Accordingly, the concept of ecological regulation analysis has been extended to microbial ecology to understand to what extent biogeochemical fluxes are regulated by the abundance, diversity, or specific activity of the microorganisms performing the process (46). In contrast to potential nitrate reduction and abundance, the structure of the nitrate-reducing community was not significantly affected by the incorporation of plant residues, and most of the residue-amended plots overlapped the control plots at all sampling dates (Fig. 4 and 5), suggesting that plant residue decomposition stimulates the activity and abundance of nitrate reducers but has little impact on the composition of this community. This is consistent with a previous study which showed that the addition of carbon as artificial root exudates resulted in increased nitrate reductase activity but weak or no changes in the structure of the nitrate-reducing community (22).
Role of biochemical composition of plant residues in the nitrate-reducing community.
Since the regular agricultural practices were followed strictly in our study, not only the types of residue but also the amounts of residue differed between the amended plots. As a consequence, the effects of the type and amounts of the residues were tightly linked in our study. Nevertheless, the fact that the stimulation by alfalfa observed during the first month after incorporation was similar to or even higher than that observed for the wheat- and rape-amended plots, despite the much smaller amount of dry matter incorporated into the alfalfa plots, indicates a significant role of residue biochemistry in regulating nitrate reduction rates. Differences in residue biochemistry were shown not only by an analysis of their C/N ratios but also by assessing residue quality using near-infrared spectroscopy during the entire experiment (39). At later stages of decomposition, it was not possible to discriminate between the effects of the quality and amount of the plant residues. Indeed, the smaller effect of alfalfa residues than those of the wheat or rape residues at several sampling dates after summer 2006 could be due either to the initial smaller amount of alfalfa residues incorporated into the soil or to their biochemical composition, since crop residues with a low C/N ratio commonly decompose faster than residues with a high C/N ratio (18). Similarly, Pascault et al. (39) reported lower microbial respiration in response to alfalfa residues than in response to wheat or rape residues at later stages of decomposition. The size of the nitrate-reducing community was also affected by the type of residues. A trend toward higher narG gene copy numbers was observed at the beginning of the experiment (7 and 26 July) in the rape-amended plots than in the other plots. The same trend was observed for napA, with a significant difference on 26 July (Fig. 3). Interestingly, we also found that the structure of the napA community in the rape-amended plots was significantly different from that of the other plots on the same sampling date (Fig. 5), which indicates that the rape residues had a short-term effect on the diversity and abundance of the nitrate-reducing bacteria but not on their activity. These results also suggest that the napA community was more affected by the type and amount of plant residues than the narG community. This difference between the effects of residues on the napA and narG genes can be explained by the fact that the narG and napA communities do not fully overlap, since nitrate reducers can possess either Nar, Nap, or both (47). In contrast to our results, no significant difference in denitrifier abundance between soils amended with soybean, red clover, and barley residues was observed previously by Henderson et al. (21). Overall, no strong effect of the type of residue on the structure of the nitrate-reducing community was apparent with either narG or napA as a molecular marker. Likewise, other authors reported the absence of a general relationship between residue biochemistry and microbial community structure in soil (7). On the other hand, Pascault et al. (39) found that in the same experimental field, the composition of the microbial community present on the residues was related to residue biochemistry, indicating that the influence of residues on microbial community composition was limited to the vicinity of the decomposing material. Accordingly, microbial gradients induced by plant roots decomposing in the soil were identified (19).
Conclusion.
Our results indicate that not only the activity but also other components of N-cycling communities, such as their abundance, can be significantly modified by incorporating plant residues into soil. The stimulating effect of plant residues on the abundance and activity of the nitrate-reducing community was shown to last throughout the 11 months. Our results also suggest that the management of the type of residue to be incorporated into the soil had a limited influence on nitrate reducers. Further studies will be required to verify whether our findings apply to other N-cycling communities.
Supplementary Material
Acknowledgments
We thank the Genotyping Service (SSG) in Dijon, France. We are grateful to V. Nowak, F. Bastian, S. Dequiedt, and C. Faivre for technical assistance.
This ECOGER project was supported by an ECCO grant from the French National Research Agency (ANR).
Footnotes
Published ahead of print on 10 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ambus, P., and E. S. Jensen. 1997. Nitrogen mineralisation and denitrification as influenced by crop residue particle size. Plant Soil 197:261-270. [Google Scholar]
- 2.Angers, D. A., and S. Recous. 1997. Decomposition of wheat straw and rye residues as affected by particle size. Plant Soil 189:197-203. [Google Scholar]
- 3.Aulakh, M., T. Khera, K. Singh, B. Singh, and J. W. Doran. 2000. Yields and nitrogen dynamics in a rice-wheat system using green manure and inorganic fertilizer. Soil Sci. Soc. Am. J. 64:1867-1876. [Google Scholar]
- 4.Aulakh, M. S., J. W. Doran, D. T. Walters, A. R. Mosier, and D. D. Francis. 1991. Crop residue type and placement effects on denitrification and mineralization. Soil Sci. Soc. Am. J. 55:1020-1025. [Google Scholar]
- 5.Baggs, E. M., R. M. Rees, K. A. Smith, and A. J. A. Vinten. 2000. Nitrous oxide emission from soils after incorporating crop residues. Soil Use Manage. 16:82-87. [Google Scholar]
- 6.Bastian, F., L. Bouziri, B. Nicolardot, and L. Ranjard. 2009. Impact of wheat straw decomposition on successional patterns of soil microbial community structure. Soil Biol. Biochem. 2009:262-275. [Google Scholar]
- 7.Baumann, K., P. Marschner, R. J. Smernik, and J. A. Baldock. 2009. Residue chemistry and microbial community structure during decomposition of eucalypt, wheat and vetch residues. Soil Biol. Biochem. 41:1966-1975. [Google Scholar]
- 8.Bending, G. D., M. K. Turner, and I. G. Burns. 1998. Fate of nitrogen from crop residues as affected by biochemical quality and the microbial biomass. Soil Biol. Biochem. 30:2055-2065. [Google Scholar]
- 9.Berks, B. C., S. J. Ferguson, J. W. B. Moir, and D. J. D. Richardson. 1995. Enzymes and associated electron transports systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1232:97-173. [DOI] [PubMed] [Google Scholar]
- 10.Bernard, L., C. Mougel, P.-A. Maron, V. Nowak, J. Lévêque, C. Hénault, F. Z. Haichar, O. Berge, C. Marol, J. Balesdent, F. Gibiat, P. Lemanceau, and L. Ranjard. 2007. Dynamics and identification of soil microbial populations actively assimilating carbon from 13C-labelled wheat residue as estimated by DNA- and RNA-SIP techniques. Environ. Microbiol. 9:752-764. [DOI] [PubMed] [Google Scholar]
- 11.Brettar, I., J. Sanchez-Perez, and M. Tréolières. 2002. Nitrate elimination by denitrification in hardwood forest soils of the Upper Rhine floodplain—correlation with redox potential and organic matter. Hydrobiologia 469:11-21. [Google Scholar]
- 12.Bru, D., A. Sarr, and L. Philippot. 2007. Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl. Environ. Microbiol. 73:5971-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chèneby, D., A. Brauman, B. Rabary, and L. Philippot. 2009. Differential responses of nitrate reducer community size, structure, and activity to tillage systems. Appl. Environ. Microbiol. 75:3180-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheshire, M. V., and S. J. Chapman. 1996. Influence of the N and P status of plant material and of added N and P on the mineralisation of C from 14C-labelled ryegrass in soil. Biol. Fertil. Soils 21:166-170. [Google Scholar]
- 15.Cook, R. J. 2006. Towards cropping systems that enhance productivity and sustainability. Proc. Natl. Acad. Sci. U. S. A. 103:18389-18394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corbeels, M., G. Hofman, and O. VanCleemput. 2000. Nitrogen cycling associated with the decomposition of sunflower stalks and wheat straw in a vertisol. Plant Soil 218:71-82. [Google Scholar]
- 17.Davidson, E. A., and I. L. Ackerman. 1993. Changes in soil carbon inventories following cultivation of previously untilled soils. Biogeochemistry 20:161-193. [Google Scholar]
- 18.Eiland, F., M. Klamer, A. M. Lind, M. Leth, and E. Bååth. 2001. Influence of initial C/N ratio on chemical and microbial composition during long term composting of straw. Microb. Ecol. 41:272-280. [DOI] [PubMed] [Google Scholar]
- 19.Gaillard, V., C. Chenu, S. Recous, and G. Richard. 1990. Carbon, nitrogen and microbial gradients induced by plant residues decomposing in soil. Eur. J. Soil Sci. 50:567-578. [Google Scholar]
- 20.Garcia-Ruiz, R., and E. M. Baggs. 2007. N2O emission from soil following combined application of fertiliser-N and ground weed residues. Plant Soil 299:263-274. [Google Scholar]
- 21.Henderson, S. L., C. E. Dandie, C. L. Patten, B. J. Zebarth, D. L. Burton, J. T. Trevors, and C. Goyer. 2010. Changes in denitrifier abundance, denitrification mRNA levels, nitrous oxide emissions and denitrification in anoxic soil microcosms amended with glucose and plant residues. Appl. Environ. Microbiol. 76:2155-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henry, S., S. Texier, S. Hallet, D. Bru, C. Dambreville, D. Chèneby, F. Bizouard, J. C. Germon, and L. Philippot. 2008. Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environ. Microbiol. 10:3082-3092. [DOI] [PubMed] [Google Scholar]
- 23.Hu, S. J., A. H. C. van Bruggen, and N. J. Grünwald. 1999. Dynamics of bacterial populations in relation to carbon availability in a residue-amended soil. Appl. Soil Ecol. 13:21-30. [Google Scholar]
- 24.Jensen, L. S., T. Mueller, J. Magid, and N. E. Nielsen. 1997. Temporal variation of C and N mineralization, microbial biomass and extractable organic pools after oilseed rape straw incorporation in the field. Soil Biol. Biochem. 29:1043-1055. [Google Scholar]
- 25.Kandeler, E. 1996. Nitrate reductase activity, p. 176-179. In F. Schinner, R. Öhlinger, E. Kandeler, and R. Margesin (ed.), Methods in soil biology. Springer, Berlin, Germany.
- 26.Ladd, J. N., M. Amato, Y. Li-Kai, and J. E. Schultz. 1994. Differential effects of rotation, plant residue and nitrogen fertilizer on microbial biomass and organic matter in an Australian alfisol. Soil Biol. Biochem. 26:821-831. [Google Scholar]
- 27.Lundquist, E. J., L. E. Jackson, K. M. Scow, and C. Hsu. 1999. Changes in microbial biomass and community composition, and soil carbon and nitrogen pools after incorporation of rye into three California agricultural soils. Soil Biol. Biochem. 31:221-236. [Google Scholar]
- 28.Manzoni, S., R. B. Jackson, J. A. Trofymow, and A. Porporato. 2008. The global stoichiometry of litter nitrogen mineralization. Science 321:684-686. [DOI] [PubMed] [Google Scholar]
- 29.Martens, D. A. 2000. Plant residues biochemistry regulates soil carbon cycling and carbon sequestration. Soil Biol. Biochem. 32:361-369. [Google Scholar]
- 30.Martin-Laurent, F., L. Philippot, S. Hallet, R. Chaussod, J. C. Germon, G. Soulas, and G. Catroux. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMahon, S. K., M. A. Williams, P. J. Bottomley, and D. D. Myrold. 2005. Dynamics of microbial communities during decomposition of carbon-13 labeled ryegrass fractions in soil. Soil Sci. Soc. Am. J. 69:1238-1247. [Google Scholar]
- 32.Miller, M. N., B. J. Zebarth, C. E. Dandie, D. L. Burton, C. Goyer, and J. T. Trevors. 2008. Crop residue influence on denitrification, N2O emissions and denitrifier community abundance in soil. Soil Biol. Biochem. 40:2553-2562. [Google Scholar]
- 33.Moreno-Vivián, C., P. Cabello, M. Martinez-Luque, R. Blasco, and F. Castillo. 1999. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J. Bacteriol. 181:6573-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myrold, D. D., and J. M. Tiedje. 1985. Establishment of denitrification capacity in soil: effects of carbon, nitrate and moisture. Soil Biol. Biochem. 6:819-822. [Google Scholar]
- 35.Neely, C. L., M. H. Beare, W. L. Hargrove, and D. C. Coleman. 1991. Relationships between fungal and bacterial substrate-induced respiration, biomass and plant residue decomposition. Soil Biol. Biochem. 23:947-954. [Google Scholar]
- 36.Nelson, D. R., and P. M. Mele. 2006. The impact of crop residue amendments and lime on microbial community structure and nitrogen-fixing bacteria in the wheat rhizosphere. Aust. J. Soil Res. 44:319-329. [Google Scholar]
- 37.Nicolardot, B., L. Bouziri, F. Bastian, and L. Ranjard. 2007. A microcosm experiment to evaluate the influence of the location and quality of plant residues on residue decomposition and genetic structure of soil microbial communities. Soil Biol. Biochem. 39:1631-1644. [Google Scholar]
- 38.Palm, C. A., C. N. Gachengo, R. J. Delve, G. Cadisch, and K. E. Giller. 2001. Organic inputs for soil fertility management in tropical agroecosystems: application of an organic resource database. Agric. Ecosyst. Environ. 83:27-42. [Google Scholar]
- 39.Pascault, N., L. Cécillon, O. Mathieu, C. Hénault, A. Sarr, J. Leveque, P. Farcy, L. Ranjard, and P.-A. Maron. 1 July 2010, posting date. In situ dynamics of microbial communities during decomposition of wheat, rape, and alfalfa residues. Microb. Ecol. [Epub ahead of print.] doi: 10.1007/s00248-010-9705-7. [DOI] [PubMed]
- 40.Philippot, L. 2002. Denitrifying genes in bacterial and archaeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 41.Philippot, L. 1999. Dissimilatory nitrate reductases in bacteria. Biochim. Biophys. Acta 1446:1-23. [DOI] [PubMed] [Google Scholar]
- 42.Philippot, L. 2005. Tracking nitrate reducers and denitrifiers in the environment. Biochem. Soc. Trans. 33:204-208. [DOI] [PubMed] [Google Scholar]
- 43.Philippot, L., S. Piutti, F. Martin-Laurent, S. Hallet, and J. C. Germon. 2002. Molecular analysis of the nitrate-reducing community from unplanted and maize-planted soil. Appl. Environ. Microbiol. 68:6121-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poll, C., S. Marhan, J. Ingwersen, and E. Kandeler. 2008. Dynamics of litter carbon turnover and microbial abundance in a rye detritusphere. Soil Biol. Biochem. 40:1306-1321. [Google Scholar]
- 45.Raich, J., and W. Schlesinger. 1992. The global carbon dioxide flux in soil respiration and its relationship to vegetation and climate. Tellus 44B:81-89. [Google Scholar]
- 46.Röling, W. F. M. 2007. Do microbial numbers count? Quantifying the regulation of biogeochemical fluxes by population size and cellular activity. FEMS Microbiol. Ecol. 62:202-210. [DOI] [PubMed] [Google Scholar]
- 47.Roussel-Delif, L., S. Tarnawski, J. Hamelin, L. Philippot, M. Aragno, and N. Fromin. 2005. Frequency and diversity of nitrate reductase genes among nitrate-dissimilating Pseudomonas in the rhizosphere of perennial grasses grown in field conditions. Microb. Ecol. 49:63-72. [DOI] [PubMed] [Google Scholar]
- 48.Shepherd, K. D., B. Vanlauwe, C. N. Gachengo, and C. A. Palm. 2005. Decomposition and mineralization of organic residues predicted using near infrared spectroscopy. Plant Soil 277:315-333. [Google Scholar]
- 49.Smith, K. A., T. Ball, F. Conen, K. E. Dobbie, J. Massheder, and A. Rey. 2003. Exchange of greenhouse gases between soil and atmosphere: interactions of soil physical factors and biological processes. Eur. J. Soil Sci. 54:779-791. [Google Scholar]
- 50.Tiedje, J. M. 1988. Ecology of denitrification and dissimilatory nitrate reduction to ammonium, p. 179-244. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. John Wiley & Sons, Inc., New York, NY.
- 51.Trinsoutrot, I., S. Recous, B. Bentz, M. Lineres, D. Chèneby, and B. Nicolardot. 2000. Biochemical quality of crop residues and carbon and nitrogen mineralization kinetics under nonlimiting nitrogen conditions. Soil Sci. Soc. Am. J. 64:918-926. [Google Scholar]
- 52.Winkler, J., R. Cherry, and W. Schlesinger. 1996. The Q10 relationship of microbial respiration in a temperate forest soil. Soil Biol. Biochem. 28:1067-1072. [Google Scholar]
- 53.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.