Abstract
Recent studies have demonstrated that xylo-oligosaccharides (XOS), which are classified as emerging prebiotics, selectively enhance the growth of bifidobacteria in general and of Bifidobacterium animalis subsp. lactis strains in particular. To elucidate the metabolism of XOS in the well-documented and widely used probiotic strain B. animalis subsp. lactis BB-12, a combined proteomic and transcriptomic approach was applied, involving DNA microarrays, real-time quantitative PCR (qPCR), and two-dimensional difference gel electrophoresis (2D-DIGE) analyses of samples obtained from cultures grown on either XOS or glucose. The analyses show that 9 of the 10 genes that encode proteins predicted to play a role in XOS catabolism (i.e., XOS-degrading and -metabolizing enzymes, transport proteins, and a regulatory protein) were induced by XOS at the transcriptional level, and the proteins encoded by three of these (β-d-xylosidase, sugar-binding protein, and xylose isomerase) showed higher abundance on XOS. Based on the obtained results, a model for the catabolism of XOS in BB-12 is suggested, according to which the strain utilizes an ABC (ATP-binding cassette) transport system (probably for oligosaccharides) to bind XOS on the cell surface and transport them into the cell. XOS are then degraded intracellularly through the action of xylanases and xylosidases to d-xylose, which is subsequently metabolized by the d-fructose-6-P shunt. The findings obtained in this study may have implications for the design of a synbiotic application containing BB-12 and the XOS used in the present study.
Prebiotics are defined as food components that confer a health benefit on the host through modulation of the microbiota (17). Among different kinds of prebiotics, special focus has been given to nondigestible oligosaccharides (NDO), which are the most abundant nutrients in the lower gastrointestinal tract (GIT), the ecological niche of bifidobacteria. The majority of the members of the genus Bifidobacterium are capable of degrading NDO to monosaccharides, which in turn are converted into intermediates of the d-fructose-6-phosphate (F6P) shunt (also known as the bifid shunt), the central carbohydrate catabolic pathway characteristic of bifidobacteria.
Xylo-oligosaccharides (XOS) are NDO that have received increasing attention as potential prebiotic candidates (18). XOS are sugar oligomers composed of a β-1,4-linked xylopyranosyl backbone that are obtained by either chemical or, more commonly, enzymatic hydrolysis of xylan polysaccharides extracted from plant cell wall. The bifidogenic effect of XOS was demonstrated both by in vitro studies (22) and by small-scale in vivo human studies (2). Some intestinal bacterial strains are able to grow on XOS, yet numerous studies have demonstrated that the ability to utilize these oligosaccharides varies considerably among these bacteria (3, 25, 29). Moreover, a recent semicontinuous, anaerobic colon simulator study demonstrated that growth on XOS can also result in decreased levels of pathogenic strains, an increase in the levels of short-chain fatty acids, and a decrease in the concentrations of branched-chain amino acids, data that indicate a balanced microflora (11).
A few XOS-degrading enzymes have been identified and characterized in bifidobacteria. A β-d-xylosidase characterized from B. breve K-110 (26) was shown to hydrolyze xylan to xylose. In a study that tested the activity of arabino-XOS-degrading enzymes in B. adolescentis, B. infantis, and B. bifidum, all species demonstrated intracellular xylosidase and arabinosidase activities, whereas no xylanase activity was detected (32). An exo-oligoxylanase (RexA) was recombinantly expressed and characterized from B. adolescentis LMG10502. This enzyme acts at the reducing end and hydrolyzes birchwood xylan and oat spelt xylan (8).
B. animalis subsp. lactis BB-12 is a widely used commercial probiotic strain isolated in 1983 by Chr. Hansen A/S. The strain is included in a variety of food applications and dietary supplements and is ascribed various probiotic effects (12). In the present study, BB-12 was cultivated in a rich broth supplemented with either XOS (degree of polymerization [DP], 2 to 6) or glucose and the obtained samples were compared at both the transcriptional and the translational levels. To our knowledge, this is the first study comprising a comprehensive and complementary proteomic/transcriptomic approach in bifidobacteria. Based on the obtained results, a model was established for the fermentative catabolism of XOS by BB-12, with the aim to deepen the insight into the metabolic basis of the use of XOS as a supplement to probiotic preparations containing BB-12.
MATERIALS AND METHODS
Strain and growth medium.
Bifidobacterium animalis subsp. lactis strain BB-12 was obtained from Chr. Hansen A/S (BB-12 is a registered trademark of Chr. Hansen A/S, Hørsholm, Denmark). Glucose was purchased from Merck Chemicals (Darmstadt, Germany). XOS powder (DP, 2 to 6) was obtained from Shandong Longlive (Qindao, China) and contained (according to high-performance anion-exchange chromatography with pulsed amperometric detection [HPAEC-PAD]) 90% XOS (which comprised 3.7% xylose, 1.0% arabinose, 34.6% xylobiose, 36.6% xylotriose, 10.6% xylotetraose, 10.2% xylopentaose, and 3.3% xylohexaose) and 10% other mono- and disaccharides. The fermentation medium was MRS (4) made from its individual components, except for glucose, which was omitted. The pH of the medium was adjusted to 6.5 before sterilization (210 kPa, 121°C, 20 min). Carbohydrate solutions were filtered through a 0.22-μm-pore-size filter and added to the sterile broth to obtain a final carbohydrate concentration of 20 g/liter.
Growth experiments.
Stationary-phase precultures in MRS medium (Oxoid, Basingstoke, United Kingdom) supplemented with 0.05% (wt/vol) cysteine chloride were harvested by centrifugation (3,200 × g for 10 min at 4°C) and washed in 5 ml 0.9 g/liter sodium chloride. The glucose- or XOS-containing broths and a control without added carbon source were inoculated with the washed precultures to an optical density at 600 nm (OD600) of 0.05. Here the inoculated media were divided in two (for duplicate measurements). Growth was monitored for 24 h by manual OD600 measurements; a linear correlation was established between OD and CFU/ml (1 × OD600 = 9.5 × 108 CFU/ml; correlation factor R2 = 0.97) from cultures grown on several carbon sources, including glucose and XOS, with OD values varying between 0.1 and 6.0. Growth experiments were carried out in 50-ml centrifuge tubes in a water bath at 37°C. These cultures were used for microarrays and quantitative PCR (qPCR) analyses. Identical setups of samples (in three biological replicates) were harvested for two-dimensional difference gel electrophoresis (2D-DIGE) and HPAEC-PAD analyses.
High-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD).
The analysis was undertaken on supernatants of BB-12 cultures grown on XOS collected after 0, 3, and 24 h of growth (samples for pH measurements were collected after 8 h). Due to technical problems the samples at 0 h could not be analyzed. Since there were limited growth and relatively low cell density at 3 h, the samples at this time point were used as an approximation to the XOS baseline values. Chromatography was performed using a Dionex AS3500 instrument (Dionex, Sunnyvale, CA) for separation of carbohydrates on a CarboPac PA-100 column (4 mm by 250 mm, p/N 43055), in combination with a CarboPac PA-100 guard column (4 mm by 50 mm). Chromatography was performed as previously described (6), with the following modifications: samples were diluted 1:20 in Milli-Q water, and the analyses were carried out by using a stepwise gradient at a flow rate of 0.8 ml/min and a total analysis time of 40 min. d-Xylose (Sigma), xylobiose, xylotriose, xylotetraose, xylopentaose, and xylohexaose (Megazyme, Bray, Ireland) were used as internal standards for quantification. All analyses were made in triplicate.
DNA microarray platform.
The design of the 65- to 75-mer oligonucleotides for the BB-12 whole-genome microarray platform was done as described previously (14) on a draft genome sequence of BB-12 (56 contigs). The platform specifications are available at the NCBI Gene Expression Omnibus (GEO) under platform accession no. GPL10040. The completed genome sequence of BB-12 can be found under GenBank accession no. CP001853.
Microarray production and analysis.
Isolation of total RNA, RNA quality control, microarray spotting, cDNA synthesis and labeling of total RNA, hybridization, washing and scanning of arrays, and preanalysis of arrays were done as described previously (14) with the following modifications: hybridization was performed with 60% instead of 40% (vol/vol) formamide due to the relatively high GC content of BB-12; microarray scanning was performed with “Auto-PMT” (Photo Multiplier Tube) set to a “saturation tolerance” of 0.1% instead of user-set PMT settings; genes were discarded if the standard deviation of the log2(ratio) of the replicate spots on each array was >0.5 rather than >0.8; and only genes where data were obtained from both the standard array and dye-swap array were included. The details are described at the NCBI Gene Expression Omnibus (GEO) under the series accession no. GSE20322.
Quantitative real-time PCR.
Primer sequences for genes analyzed by qPCR expression assays were designed using the Primer3 software (20). cDNA synthesis was carried out as described previously (14) with a total RNA input of 675 ng and performed in duplicates for each of the four biological replicates. The obtained cDNA samples were diluted 1:72.5 in nuclease-free water. qPCR was carried out on an ABI 7500 qPCR machine (Applied Biosystems, Foster City, CA) using SYBR green mastermix (Applied Biosystems) with 465 pmol/μl cDNA and 6 pmol/μl of each primer pair. PCR conditions used were as described previously (14). At the end of each reaction, the cycle threshold (CT) was manually set at the level that reflected the best kinetic PCR parameters and the 2−ΔΔCT method of relative quantification (9) was used to obtain expression values. The five genes (out of eight genes tested) that gave rise to the most consistent expression, as analyzed by the GeNorm software (28), were used for normalization of the results. The relative expression of each gene represented a mean of the values obtained for each of the four replicates, each being run in duplicate. A gene was considered to be regulated when a one-way analysis of variance (ANOVA) test gave a fold change value of >2 and a P value of <0.05.
Extraction of intracellular proteins.
Cells for proteome analyses were harvested after 8 h of growth by centrifugation (3,200 × g for 10 min at 4°C) at OD600s of 1.0 and 1.4 for the glucose and XOS cultures, respectively. Cell pellets were washed twice with 50 mM potassium phosphate buffer, pH 7.0, and kept frozen until disruption by passage through a French press (Duragauge P1603-136, Struers, Denmark) at 8,270 kPa. Cytosolic protein fractions were obtained as described previously (24), and the determination of protein concentration was performed using the Popov method (15).
Two-dimensional difference gel electrophoresis (2D-DIGE).
Volumes corresponding to 50 μg protein from each of the samples (three biological replicates of cells grown on glucose or XOS), as well as an internal standard obtained by pooling aliquots containing 25 μg protein from each sample, were precipitated with chloroform-methanol (31). Precipitates were dissolved in a buffer containing 15 mM Tris, pH 8.5, and 8 M urea. Protein samples were labeled with fluorescent cyanine dyes as previously described (21), with the exception that 4 nmol dye (rather than 400 pmol) was used to label each of the samples. The labeled internal standard and the corresponding labeled samples for each gel were pooled, and the volume was made up to 350 μl by the addition of rehydration buffer (0.2% [vol/vol] carrier ampholytes, 0.4% [wt/vol] 3-[3-(cholamidopropyl)dimethylammonio]-1-propanesulfonate [CHAPS], and 50 mM dithiothreitol [DTT]). Isoelectric focusing was run using immobilized pH gradient strips (linear pH grade, 4 to 7; 18 cm; GE Healthcare, Piscataway, NJ). Polyacrylamide gel electrophoresis was performed using 10% (wt/vol) polyacrylamide gels on an EttanTM DALT-6 electrophoresis unit (GE Healthcare) overnight at 1 W/gel until the dye front reached the base of the gel. Fluorescence gel images were acquired by a Typhoon 9410 variable mode imager (GE Healthcare) at excitation and emission wavelengths of 488/520 nm (Cy2), 532/580 nm (Cy3), and 633/670 nm (Cy5) and a 100-μm resolution. Subsequent to fluorescence scanning, gels were stained using colloidal Coomassie brilliant blue (16).
Image analysis.
Analysis of the gel images was undertaken using Progenesis SameSpots software version 3.3 (Nonlinear Dynamics, Newcastle upon Tyne, United Kingdom). The gel images were aligned by automated calculation of alignment vectors after assigning 20 to 30 landmark vectors. The scanned gels were analyzed by intragel (methodological variance) and intergel (biological variance) analyses. A threshold of 1.5-fold for spot volume ratio change (in cases where proteins were identified from more than a single spot, the highest fold change values were considered) and an analysis of variance statistical significance test (P < 0.05) were chosen to identify differentially expressed protein spots, excluding spots giving rise to more than a single protein hit, as determined by mass spectrometry.
In-gel digestion and protein identification by mass spectrometry.
Differentially expressed protein spots from the 2D-DIGE analysis were manually excised and subjected to in-gel tryptic digestion as described previously (19) with minor modifications (33), excluding the reduction and alkylation steps (already performed in the equilibration step of the 2D-DIGE). Tryptic peptides were analyzed by an Ultraflex II matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Bruker-Daltonics, Bremen, Germany). Spectra were obtained as a summation of 10 individual spectra in positive reflector mode and externally calibrated using a tryptic digest of β-lactoglobulin (5 pmol/μl). Internal calibration was performed by using trypsin autolysis products and keratin contaminants that were subsequently removed from the peak list. The FlexAnalysis 3.0.96 and Biotools 3.1 software (Bruker-Daltonics) were used to analyze the recorded spectra. Protein identification was performed using the protein sequence database program Mascot (Matrix Science Ltd.) with the genome sequence of BB-12 (represented by the locus tag identifiers BIF_0XXXX that correspond to the GenBank accession no. CP001853) (5) as the taxonomy entry. The following search parameters were used in all Mascot database searches: carbamidomethylation (Cys) and methionine oxidation (+16 Da) as fixed and variable modifications, respectively, tolerance of one missed cleavage, and a maximum error tolerance of 80 ppm and 0.7 Da in the mass spectrometry (MS) and tandem MS data, respectively. No restrictions with respect to protein mass and pI were made. Protein/ion scores with P values of <0.05 were considered significant.
RESULTS
Comparison of growth on glucose and XOS.
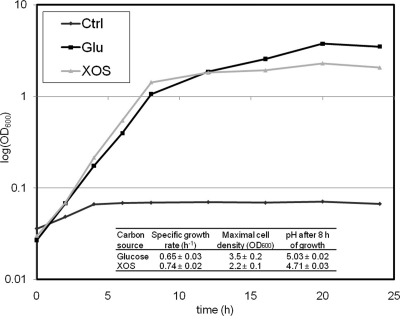
To test whether XOS can support the growth of B. animalis subsp. lactis BB-12, the strain was propagated in an MRS-reconstituted broth containing 20 g/liter of either glucose or XOS for 24 h (Fig. 1). The mean specific growth rates, as calculated from the exponential phase (2 to 6 h of growth), were slightly higher for the XOS-grown cultures. However, growth on XOS was markedly impaired after 6 to 7 h but less so for the glucose cultures. Accordingly, the mean turbidity value of the cultures measured after 24 h in the glucose cultures (3.5 ± 0.2) was markedly higher than that for XOS (2.2 ± 0.1). Similar experiments conducted with arabinoxylan or xylose resulted in very poor growth (data not shown). pH measurements after 8 h of growth showed that the decrease in pH from the initial 6.5 in the XOS culture (4.71 ± 0.03) was significantly greater than that in the glucose-containing cultures (5.03 ± 0.02). This is consistent with the faster growth, and thereby faster acidification, observed under growth on XOS. No significant differences were observed in the levels of the primary end products of the bifid shunt, acetate and lactate (data not shown).
FIG. 1.
Growth of B. animalis subsp. lactis BB-12 in a rich broth containing 2% (wt/vol) glucose or XOS, respectively, as well as a control culture where no carbon source was added. Growth studies were carried out in triplicate, and a representative data set is shown. Note that without an added carbon source there is only limited growth, which is presumably due to small amounts of other carbohydrates in the complex components of the MRS. Insert: mean values of specific growth rate, maximal cell density, and pH of culture supernatants after 8 h of growth.
HPAEC-PAD analysis of culture supernatants.
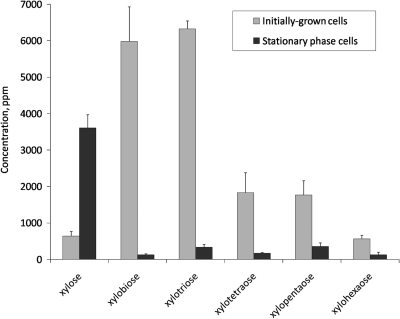
Supernatants of BB-12 cultures grown in the presence of XOS were collected at the stationary phase (24 h) and subjected to HPAEC-PAD analysis. The results (Fig. 2) showed that the concentration of all XOS (DP, 2 to 6) was markedly lower at the stationary phase than that in the initially grown cells (93% decrease in total). The concentration of the monomeric d-xylose was nearly 6-fold higher, yet this accumulation accounted for less than 20% of the decrease in the total XOS concentration. The results also show that the relative decrease in the concentration of XOS of different DP decreased with increasing chain length (decreases of 98, 95, 91, 80, and 77% for xylobiose, xylotriose, xylotetraose, xylopentaose, and xylohexaose, respectively). The level of arabinose in the above-mentioned culture supernatants was very low, and the absence of glucose, galactose, and lactose was verified by gas chromatography analyses (data not shown).
FIG. 2.
High-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) analysis of supernatants of B. animalis subsp. lactis BB-12 cultures propagated on XOS. Concentrations of different XOS species in the supernatants of BB-12 cultures grown on XOS collected from initially grown cells and stationary cells, respectively, are designated. The results represent mean values and standard errors for results from three biological replicates.
Microarray analysis.
The expression of genes from exponentially growing BB-12 cultured in the presence of either glucose or XOS was compared using whole-genome microarrays. Fourteen genes were found to be upregulated and 10 to be downregulated on XOS (Table 1; see Table S1 in the supplemental material). Of the genes induced upon growth on XOS, 11 are located successively in the genome of BB-12 and 9 of these genes, along with another gene located elsewhere in the genome, encode proteins likely to be involved in XOS catabolism. These proteins consist of XOS-degrading enzymes, transport-related proteins, d-xylose-metabolizing enzymes, and a transcriptional regulator, which may play a role in the regulation of the genes (Table 1). Six of the 10 repressed genes (relative to growth on XOS) code for carbohydrate-metabolizing or -binding proteins, e.g., maltose/maltodextrin-binding protein (BIF_00469) and sucrose phosphorylase (BIF_02090).
TABLE 1.
Proteins/genes predicted to play a role in the catabolism of XOS in B. animalis subsp. lactis BB-12a
| IDb | Description | Predicted function | Fold change upregulation on XOSc |
Additional informationd | ||
|---|---|---|---|---|---|---|
| DNA microarrays | qPCR | 2D-DIGE | ||||
| 501 | Xylose isomerase | Isomerization of d-xylose to d-xylulose | 32 | 19+++ | 13 | EC 5.3.1.5 COG2115, XylA (2 × 10−156) |
| 405 | β-Xylosidase | Cleavage of nonreducing end of β-1,4- xylo-oligosaccharides | 6.1 | 3.0++ | 1.8 | EC 3.2.1.37/EC 3.2.1.55 GH 43 (3 × 10−36); COG3507, XynB, β-xylosidase (4 × 10−34) |
| 92 | β-Xylosidase | Cleavage of nonreducing end of β-1,4- xylo-oligosaccharides | 31 | 10.3+++ | ND | EC 3.2.1.37/EC 3.2.1.55 GH 43 (1 × 10−84); COG3507, XynB, β-xylosidase (2 × 10−152) |
| 432 | Transcriptional repressor | Regulation of the transcription of the XOS gene cluster | 2.1 | 1.1 | ND | cd06267, PBP1_LacI_sugar_binding_like (8 × 10−66); cd01543, PBP1_XylR, ligand binding domain of DNA transcription repressor specific for xylose (1 × 10−15) |
| 212 | Sugar-binding protein | Binding of XOS at the cell surface | 29 | 21+++ | 6.2 | COG1653, UgpB, ABC-type sugar transport system periplasmic component (1 × 10−35); COG2182, MalE, maltose binding periplasmic proteins/domains (2 × 10−17) |
| 257 | Transporter | ABC-type permease, creating the pore in the membrane through which XOS are imported | 25 | 28+++ | ND | PRK10999, MalF, maltose transporter membrane protein (32%, 23%, 1 × 10−13); COG1175, UgpA, ABC-type sugar transport systems, permease components (7 × 10−58) |
| 258 | Transporter | ABC-type permease, creating the pore in the membrane through which XOS are imported | 18 | 14+++ | ND | COG3833, MalG, ABC-type maltose transport systems, permease component (32%, 25%, 7 × 10−37); TC 3.A.1.1.21e xylobiose porter BxlEFG(K) (31%, 22%, 2 × 10−34) |
| 633 | Endo-1,4-β-xylanase | Endohydrolysis of β-1,4-xylosidic linkages in XOS | 5.9 | 2.1+ | ND | EC 3.2.1.8 pfam04616, GH 43 (8 × 10−40); COG3507, XynB, β-xylosidase (3 × 10−46) |
| 928 | Endo-1,4-β-xylanase | Endohydrolysis of β-1,4-xylosidic linkages in XOS | 2.2 | 2.5+ | ND | EC 3.2.1.8 COG0657, Aes, esterase/lipase (6 × 10−16) |
| 829 | Xylulose kinase | Phosphorylation of xylulose to xylulose-5-P | 4.8 | 3.4+++ | ND | EC 2.7.1.17 TIGR01312, XylB, d-xylulose kinase (3 × 10−94) |
| 1629 | ABC transporter ATP- binding protein | Binding and hydrolysis of ATP to energize XOS transport via ABC-type transport systems | 1.2 | 0.82 | 1.8 | COG1129, MglA, ABC-type sugar transport systems, ATPase component (7 × 10−6) |
| 1681 | Sugar transport ATP-binding protein | Binding and hydrolysis of ATP to energize XOS transport via ABC-type transport systems | 1.1 | 1.1 | 1.6 | COG3839, MalK, ABC-type sugar transport systems, ATPase components (31%, 22%, 2 × 10−34) |
Fold-change upregulation values on XOS compared with glucose, as obtained using microarray (representing mean results of two analyses carried out using a dye swap), qPCR (mean of four values from four biological replicates, each run in duplicate), and 2D-DIGE analyses are listed. Information regarding function predictions is also given.
Gene/protein number (the “BIF_” prefix was removed).
+, P < 0.05; ++, P < 0.01; +++, P < 0.001 (one-way ANOVA test). For complete data on the 2D-DIGE analysis see Table S2 in the supplemental material. ND, proteins not detected to be upregulated.
Sequence-based similarities to conserved domains, as predicted using BLASTP (1) (percent amino acid identity, percent similarity, and expected values, respectively, are indicated in parentheses).
The TC number corresponds to the Transporter Classification (TC) system and was predicted using the Transporter Classification Database (TCDB; http://www.tcdb.org/) (23).
Real-time qPCR analysis.
To verify the results obtained in the microarray analyses, 12 putative XOS-related genes were subjected to qPCR analysis (Table 1). Except for the transcriptional regulator BIF_00432, the induction of all 10 genes upregulated in the microarray analysis was confirmed by qPCR analysis. The correlation obtained between the upregulation in the microarrays and qPCR analyses was good, yet the fold change values were higher according to the microarray analysis for seven of the nine genes found to be differentially expressed in both analyses (Table 1). No upregulation at the transcript level was detected, however, for the gene coding for ATP-binding protein BIF_01681 that showed increased abundance at the protein level in cultures grown on XOS (Table 1).
Comparative proteome analysis using 2D-DIGE.
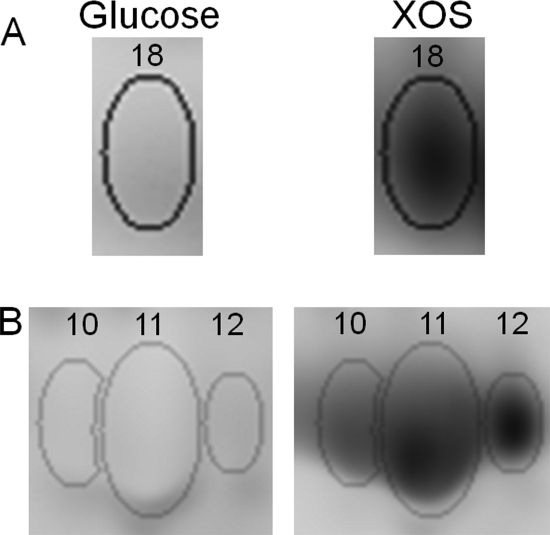
A total of 28 differentially abundant protein spots were identified in total (see Fig. S2 in the supplemental material). The proteins in all of these spots were identified by MALDI-TOF MS/MS, amounting to 25 unique proteins (see Table S2 in the supplemental material). Among these, eight showed increased spot intensity whereas seven exhibited decreased abundance in the XOS cultures compared to the glucose-grown cultures. Three proteins were regulated more than 5-fold: xylose isomerase (BIF_00501) and sugar-binding protein (BIF_00212) were upregulated on XOS (Fig. 3), whereas the abundance of enolase (BIF_01197) was decreased. Ten additional proteins were detected in seven differentially abundant spots (six of which were increased) whose analyses gave rise to more than a single protein identification, rendering the determination of the level of regulation impossible. Carbohydrate-metabolizing proteins, including XOS-metabolizing proteins and enzymes of the bifid shunt, accounted for the majority of the proteins identified from the eight protein spots whose abundance increased in the XOS samples.
FIG. 3.
Comparison of the protein abundance in B. animalis subsp. lactis BB-12 grown on XOS or glucose (as observed by spot patterns on a gel image). Images (from a representative 2D-DIGE gel) were obtained by differential labeling of protein samples with fluorescent dyes. Spots A (spot 18; see Fig. S2 and Table S2 in the supplemental material) and B (spots 10 to 12) correspond to sugar-binding protein (BIF_00212) and xylose isomerase (BIF_00501), respectively.
With respect to XOS metabolism, four of the eight overexpressed proteins are predicted to be related to the utilization of these oligosaccharides. The genes coding for three of these proteins—sugar-binding protein (BIF_00212; Fig. 3A), xylose isomerase (BIF_00501; Fig. 3B), and β-xylosidase (BIF_00405)—were upregulated also at the transcriptional level according to both microarrays and qPCR analysis during growth on XOS (Table 1). The abundance of the ATP-binding protein BIF_01681 was increased when grown in the presence of XOS, yet this induction was not observed in qPCR. Another ATP-binding protein, BIF_01629, was identified in a differentially abundant spot also containing another protein. These two proteins may be involved in an ABC transport system that transports XOS across the cell membrane.
DISCUSSION
The induction of a few putative XOS-related proteins at the transcript and/or protein level provided the basis for the establishment of a model for XOS catabolism in BB-12. The bacterium attained a slightly higher growth rate but a lower growth yield on XOS than on glucose. Earlier studies showed that BB-12 is capable of utilizing XOS as a carbon source but cannot ferment xylan, arabinoxylan (3), or xylose. Vernazza et al. (30) reported a markedly lower growth rate of BB-12 on XOS than on glucose, yet comparison of growth rates and cell yields are potentially problematic due to differences in the growth medium and in the XOS used between the current study and the study carried out by Vernazza and colleagues. Cessation of growth was observed in the XOS-cultivated cultures after 6 to 7 h, which is probably related to the lower pH compared with growth on glucose (4.71 and 5.03, respectively).
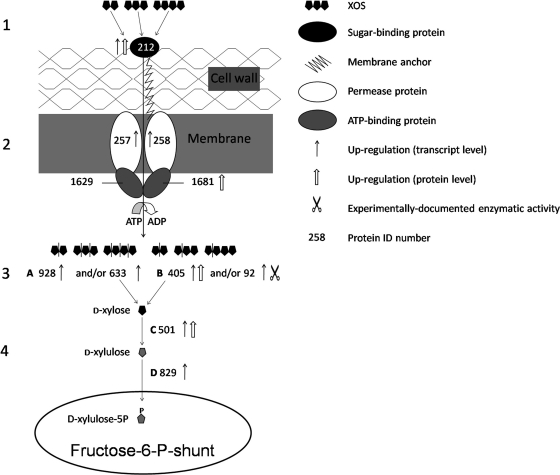
The capability of BB-12 to utilize XOS is predicted, according to the suggested model (Fig. 4), to be facilitated by a multistep mechanism consisting of the following steps: (i) binding of XOS at the cell surface, (ii) transport across the membrane by ABC-type oligosaccharide transport system(s), (iii) intracellular degradation of XOS to d-xylose by XOS-degrading enzymes, and (iv) two-step conversion of d-xylose to d-xylulose 5-phosphate (X5P).
FIG. 4.
Proposed model for the catabolism of XOS in B. animalis subsp. lactis BB-12 comprising the following steps. 1, binding of XOS at the cell surface by a sugar-binding protein. 2, transport of XOS by an ABC transport system. 3, degradation of XOS to d-xylose (by a combined action of an endo-1,4-β-xylanase and a β-xylosidase). 4, conversion of d-xylose to xylulose-5-P, a key metabolite of the fructose-6-P shunt. A, endo-1,4-β-xylanase; B, β-xylosidase; C, xylose isomerase; D, xylulose kinase. The numbers indicate the respective genes in the genome of BB-12, omitting the “BIF_0.”
Binding of XOS at the cell surface is carried out, according to the suggested model, by the sugar-binding protein BIF_00212, which is upregulated on XOS at the transcriptional level. In the present study, the abundance of this protein was increased upon growth on XOS, and similar observations were recorded in extracellular proteome analysis (O. Gilad, S. Jacobsen, B. Stuer-Lauridsen, and B. Svensson, unpublished data).
Transport of XOS across the cell membrane is predicted to be facilitated by an ABC-type sugar transport system(s) capable of importing a variety of oligosaccharides, as was postulated (at the genus level) by Palframan et al. (13). The fact that nearly all of the XOS were consumed after 24 h of growth (Fig. 2) implies that the oligosaccharides were taken up by the cells. The proteins predicted to take part in XOS transport are the transporter proteins BIF_00257 and BIF_00258. The transcription of the genes encoding these permeases was markedly upregulated on XOS (Table 1). In addition to this, preliminary data show that both proteins were identified in the membrane fraction obtained from BB-12 cultures cultivated on XOS (O. Gilad, A. Margolles, S. Jacobsen, B. Stuer-Lauridsen, K. Hjernø, O. N. Jensen and B. Svensson, unpublished data). Sequence similarity searches of BIF_00257 and BIF_00258 (Table 1) demonstrate that these proteins resemble oligosaccharide transporters. The ATP-binding proteins that energize transport of XOS through the ABC transport system may be BIF_01629 and BIF_01681; the latter is differentially expressed in cultures grown on XOS.
The degradation of XOS transported across the membrane is suggested to be facilitated by the action of endo-1,4-β-xylanases (BIF_00633 and BIF_00928) and/or β-xylosidases (BIF_00092 and BIF_00405). Endo-1,4-β-xylanase cleaves XOS randomly, while β-xylosidase degrades the xylo-oligomeric chain at the nonreducing end, releasing d-xylose. The genes coding for these four enzymes were induced upon growth on XOS according to both DNA microarray and qPCR analyses. According to the glycoside hydrolase classification system (http://www.cazy.org/Glycoside-Hydrolases.html) (7), BIF_00633, BIF_00092, and BIF_00405 are predicted to belong to glycoside hydrolase family 43 (GH43), which comprises both endo-1,4-β-xylanases and β-xylosidases. Preliminary data show that recombinant BIF_00092 degrades β-d-(1,4)-xylo-oligosaccharides with DP of 2 to 5 (xylohexaose was not tested; A. H. Viborg, S. Jacobsen, K. I. Sørensen, O. Gilad and B. Svensson, unpublished data). The upregulation of BIF_00405 on XOS according to 2D-DIGE analysis provides additional experimental evidence supporting the suggested role of this xylosidase in XOS degradation.
The final step in the model for XOS catabolism comprises conversion of d-xylose (formed by the action of the XOS-degrading enzymes) to X5P, as described for Lactobacillus pentosus MD353 (10). d-Xylose is isomerized by xylose isomerase to d-xylulose, which is phosphorylated by xylulose kinase to X5P (Fig. 4). Xylose isomerase was highly upregulated at both the protein and transcript levels (fold change, 32, 19, and 13 for the microarray, qPCR, and 2D-DIGE analyses, respectively) in the XOS cultures.
The proposed model is currently being subjected to verification by comparative proteome analysis of the membrane proteins of the bacterium, as well as by characterization of enzymes predicted to participate in XOS catabolism. The activity and importance of the proteins predicted to play a role in growth on XOS can be verified by knockout studies.
A similar model for XOS utilization was suggested for B. adolescentis LMG10502 (8), but a somewhat alternative model concerning B. longum biotype longum was introduced by van den Broek and coworkers (27). According to the latter model, XOS are degraded extracellularly and imported into the cell through a concerted binding-cleavage-transport mechanism that involves cleavage by extracellular membrane-anchored endo- and exoxylanases (27).
To conclude, the present study shows that BB-12 possesses the metabolic capacity to utilize XOS as a primary carbon source, a trait described in detail in the proposed transcriptomics/proteomics-based model for XOS catabolism. This model is expected to broaden the insight into the way by which XOS, which are gaining increasing industrial and scientific attention as emerging prebiotics, are fermented by bifidobacteria. In addition, since XOS was shown to be specifically efficient in promoting the in vitro growth of strains belonging to the B. animalis subsp. lactis taxon (18), e.g., BB-12, a synbiotic preparation containing a combination of the two may be taken into consideration.
Supplementary Material
Acknowledgments
We thank Maiken Lund Jensen, Karen Fuglede Appel, and Dorte Myling-Petersen (Chr. Hansen A/S) and Birgit Andersen, Morten Ejby Hansen, and Alexander Holm Viborg (Department of Systems Biology, DTU) for technical assistance. Marianne Richelieu (Chr. Hansen A/S) is gratefully acknowledged for running the HPAEC-PAD analysis. We extend our appreciation to Abelardo Margolles (Instituto de Productos Lácteos de Asturias [CSIC], Spain), Mads Bennedsen, and Ulf Houlberg (Chr. Hansen A/S) for stimulating discussions and Anette Wind (Chr. Hansen A/S) for coordinating the project.
The current study is a part of an industrial Ph.D. project, carried out by Ofir Gilad, that is partly financed by the Danish Ministry of Science, Technology and Innovation. We thank the Danish Research Council for Natural Science and the Centre for Advanced Food Studies for financial support to the DIGE platform and the mass-spectrometry instrumentation.
Footnotes
Published ahead of print on 17 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung, Y., C. Hsu, C. Ko, and Y. Chan. 2007. Dietary intake of xylooligosaccharides improves the intestinal microbiota, fecal moisture, and pH value in the elderly. Nutr. Res. 27:756-761. [Google Scholar]
- 3.Crittenden, R., S. Karppinen, S. Ojanen, M. Tenkanen, R. Fagerstrom, J. Matto, M. Saarela, T. Mattila-Sandholm, and K. Poutanen. 2002. In vitro fermentation of cereal dietary fibre carbohydrates by probiotic and intestinal bacteria. J. Sci. Food Agric. 82:781-789. [Google Scholar]
- 4.de Man, J., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 5.Garrigues, C., E. Johansen, and M. B. Pedersen. 2010. Complete genome sequence of Bifidobacterium animalis subsp. lactis BB-12, a widely consumed probiotic strain. J. Bacteriol. 192:2467-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gullón, P., P. Moura, M. P. Esteves, F. M. Girio, H. Domínguez, and J. C. Parajó. 2008. Assessment on the fermentability of xylooligosaccharides from rice husks by probiotic bacteria. J. Agric. Food Chem. 56:7482-7487. [DOI] [PubMed] [Google Scholar]
- 7.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagaert, S., S. Van Campenhout, A. Pollet, T. M. Bourgois, J. A. Delcour, C. M. Courtin, and G. Volckaert. 2007. Recombinant expression and characterization of a reducing-end xylose-releasing exo-oligoxylanase from Bifidobacterium adolescentis. Appl. Environ. Microbiol. 73:5374-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 10.Lokman, B. C., M. Heerikhuisen, R. J. Leer, A. van den Broek, Y. Borsboom, S. Chaillou, P. W. Postma, and P. H. Pouwels. 1997. Regulation of expression of the Lactobacillus pentosus xylAB operon. J. Bacteriol. 179:5391-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mäkeläinen, H., S. Forssten, M. Saarinen, J. Stowell, N. Rautonen, and A. C. Ouwehand. 2009. Xylo-oligosaccharides enhance the growth of bifidobacteria and Bifidobacterium lactis in a simulated colon model. Benef. Microbes 1:81-91. [DOI] [PubMed] [Google Scholar]
- 12.Ouwehand, A. C., S. Salminen, and E. Isolauri. 2002. Probiotics: an overview of beneficial effects. Antonie Van Leeuwenhoek 82:279-289. [PubMed] [Google Scholar]
- 13.Palframan, R. J., G. R. Gibson, and R. A. Rastall. 2003. Carbohydrate preferences of Bifidobacterium species isolated from the human gut. Curr. Issues Intest. Microbiol. 4:71-75. [PubMed] [Google Scholar]
- 14.Pedersen, M. B., C. Garrigues, K. Tuphile, C. Brun, K. Vido, M. Bennedsen, H. Møllgaard, P. Gaudu, and A. Gruss. 2008. Impact of aeration and heme-activated respiration on Lactococcus lactis gene expression: identification of a heme-responsive operon. J. Bacteriol. 190:4903-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popov, N., M. Schmitt, S. Schulzeck, and H. Matthies. 1975. Eine störungsfreie Mikromethode zur bestimmung des Proteingehaltes in Gewebehomogenaten. Acta Biol. Med. Ger. 34:1441-1446. [PubMed] [Google Scholar]
- 16.Rabilloud, T., and S. Charmont. 2000. Detection of proteins on two-dimensional electrophoresis., p. 109-110. In T. Rabilloud (ed.), Proteome research: two-dimensional electrophoresis and identification methods. Springer Verlag, Berlin, Heidelberg, Germany.
- 17.Reid, G., M. E. Sanders, H. R. Gaskins, G. R. Gibson, A. Mercenier, R. Rastall, M. Roberfroid, I. Rowland, C. Cherbut, and T. R. Klaenhammer. 2003. New scientific paradigms for probiotics and prebiotics. J. Clin. Gastroenterol. 37:105-118. [DOI] [PubMed] [Google Scholar]
- 18.Roberfroid, M. 2007. Prebiotics: the concept revisited. J. Nutr. 137:830S-837S. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeld, J., J. Capdevielle, J. Guillemot, and P. I. Ferrara. 1992. In-gel digestion of proteins for internal sequence-analysis after 1-dimensional or 2-dimensional gel-electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 20.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz, L., Y. Coute, B. Sánchez, C. G. de los Reyes-Gavilán, J. C. Sanchez, and A. Margolles. 2009. The cell-envelope proteome of Bifidobacterium longum in an in vitro bile environment. Microbiology 155:957-967. [DOI] [PubMed] [Google Scholar]
- 22.Rycroft, C. E., M. R. Jones, G. R. Gibson, and R. A. Rastall. 2001. A comparative in vitro evaluation of the fermentation properties of prebiotic oligosaccharides. J. Appl. Microbiol. 91:878-887. [DOI] [PubMed] [Google Scholar]
- 23.Saier, M. H., Jr., M. R. Yen, K. Noto, D. G. Tamang, and C. Elkan. 2009. The Transporter Classification Database: recent advances. Nucleic Acids Res. 37:D274-D278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sánchez, B., M. C. Champomier-Vergès, P. Anglade, F. Baraige, C. G. de Los Reyes-Gavilán, A. Margolles, and M. Zagorec. 2005. Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187:5799-5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos, A., M. San Mauro, and D. M. Diaz. 2006. Prebiotics and their long-term influence on the microbial populations of the mouse bowel. Food Microbiol. 23:498-503. [DOI] [PubMed] [Google Scholar]
- 26.Shin, H. Y., J. H. Lee, J. Y. Lee, Y. O. Han, M. J. Han, and D. H. Kim. 2003. Purification and characterization of ginsenoside Ra-hydrolyzing β-D-xylosidase from Bifidobacterium breve K-110, a human intestinal anaerobic bacterium. Biol. Pharm. Bull. 26:1170-1173. [DOI] [PubMed] [Google Scholar]
- 27.van den Broek, L. A., S. W. Hinz, G. Beldman, J. P. Vincken, and A. G. Voragen. 2008. Bifidobacterium carbohydrases—their role in breakdown and synthesis of (potential) prebiotics. Mol. Nutr. Food Res. 52:146-163. [DOI] [PubMed] [Google Scholar]
- 28.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1-research0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Laere, K. M., R. Hartemink, M. Bosveld, H. A. Schols, and A. G. Voragen. 2000. Fermentation of plant cell wall derived polysaccharides and their corresponding oligosaccharides by intestinal bacteria. J. Agric. Food Chem. 48:1644-1652. [DOI] [PubMed] [Google Scholar]
- 30.Vernazza, C. L., G. R. Gibson, and R. A. Rastall. 2006. Carbohydrate preference, acid tolerance and bile tolerance in five strains of Bifidobacterium. J. Appl. Microbiol. 100:846-853. [DOI] [PubMed] [Google Scholar]
- 31.Wessel, D., and U. Flügge. 1984. A method for the quantitative recovery of protein in dilute-solution in the presence of detergents and lipids. Anal. Biochem. 138:141-143. [DOI] [PubMed] [Google Scholar]
- 32.Zeng, H., Y. Xue, T. Peng, and W. Shao. 2007. Properties of xylanolytic enzyme system in bifidobacteria and their effects on the utilization of xylooligosaccharides. Food Chem. 101:1172-1177. [Google Scholar]
- 33.Zhang, X., L. Shi, S. Shu, Y. Wang, K. Zhao, N. Xu, S. Liu, and P. Roepstorff. 2007. An improved method of sample preparation on AnchorChip targets for MALDI-MS and MS/MS and its application in the liver proteome project. Proteomics 7:2340-2349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.