Abstract
Industrial penicillin production levels by the filamentous fungus Penicillium chrysogenum increased dramatically by classical strain improvement. High-yielding strains contain multiple copies of the penicillin biosynthetic gene cluster that encodes three key enzymes of the β-lactam biosynthetic pathway. We have analyzed the gene cluster dose effect on penicillin production using the high-yielding P. chrysogenum strain DS17690 that was cured from its native clusters. The amount of penicillin V produced increased with the penicillin biosynthetic gene cluster number but was saturated at high copy numbers. Likewise, transcript levels of the biosynthetic genes pcbAB [δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine synthetase], pcbC (isopenicillin N synthase), and penDE (acyltransferase) correlated with the cluster copy number. Remarkably, the protein level of acyltransferase, which localizes to peroxisomes, was saturated already at low cluster copy numbers. At higher copy numbers, intracellular levels of isopenicillin N increased, suggesting that the acyltransferase reaction presents a limiting step at a high gene dose. Since the number and appearance of the peroxisomes did not change significantly with the gene cluster copy number, we conclude that the acyltransferase activity is limiting for penicillin biosynthesis at high biosynthetic gene cluster copy numbers. These results suggest that at a high penicillin production level, productivity is limited by the peroxisomal acyltransferase import activity and/or the availability of coenzyme A (CoA)-activated side chains.
During the last 70 years, the increase in penicillin production by industrial strains of the filamentous fungus Penicillium chrysogenum has been based mainly on classical strain improvement (1, 14). The pathway of the biosynthesis of penicillin and the cellular localization of most of the critical enzymatic steps are known (12). Penicillin biosynthesis starts with the formation of the tripeptide δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine by the nonribosomal peptide synthetase δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine synthetase (ACVS; encoded by pcbAB [Pc21g21390]), whereupon the β-lactam ring is formed by isopenicillin N synthase (IPNS; encoded by pcbC [Pc21g21380]), resulting in the formation of isopenicillin N (IPN). These two enzymatic steps occur in the cytosol. Next, isopenicillin N enters the peroxisome, an alkaline compartment that contains acyltransferase (AT; encoded by penDE [Pc21g21370]), an enzyme that catalyzes side chain exchange using a coenzyme A (CoA)-activated organic acid (or fatty acids) as a substrate. The genes encoding these three key enzymes of the biosynthetic pathway are organized in a gene cluster, also termed the penicillin biosynthetic gene cluster (Fig. 1 A). Side chain activation is catalyzed by a group of enzymes termed CoA ligases. One of the enzymes involved in the activation of phenylacetic acid and phenoxyacetic acid in the biosynthesis of penicillins G and V is phenoxyacetic acid CoA ligase (PCL), which is encoded by the phl gene (Pc22g14900) (9, 14). The phl gene is located outside the biosynthetic gene cluster elsewhere in the genome. Together with acyltransferase, PCL is peroxisome localized (10).
FIG. 1.
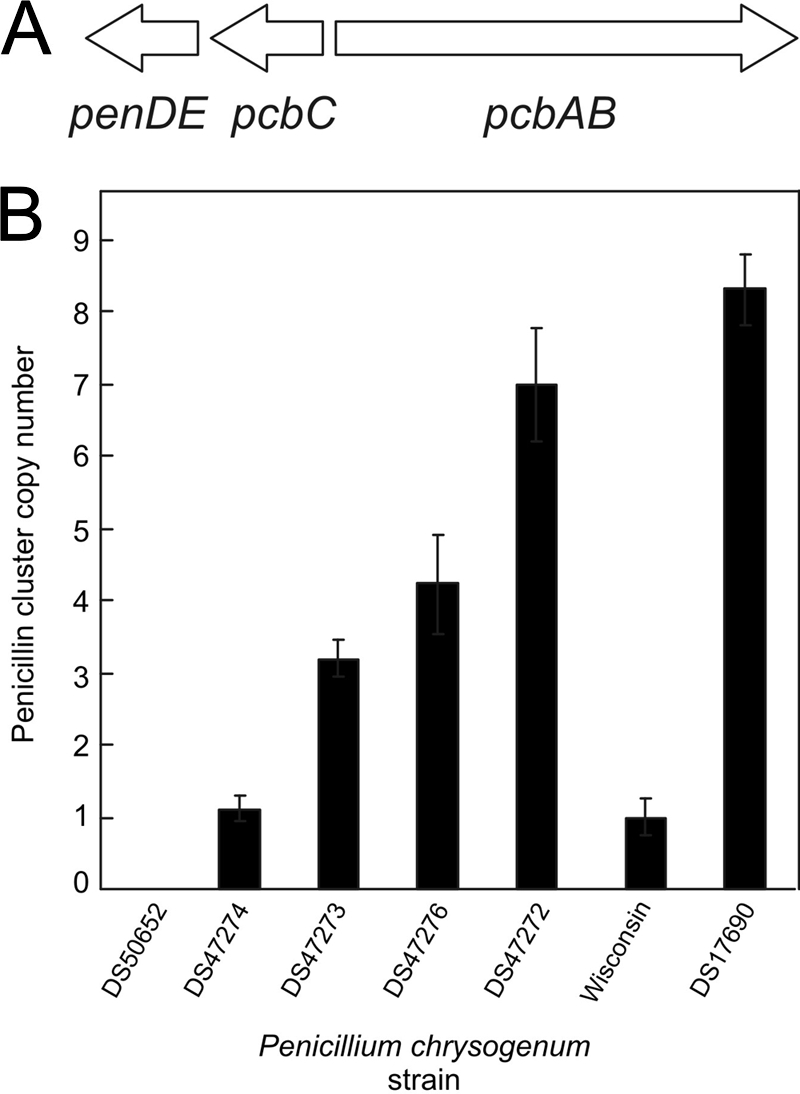
(A) Scheme of the penicillin biosynthetic gene cluster. (B) qPCR analysis of the penicillin cluster copy number in P. chrysogenum DS17690 strain and its derivatives. DS50652 is a cluster-free control strain, and Wisconsin54-1255, the ancestor of DS17690, contains a single penicillin cluster.
A remarkable phenomenon that occurred during classical strain improvement is the amplification of the penicillin biosynthetic gene cluster between tandem repeats (“amplicons”) (2, 13, 16), and this has been associated with increased levels of β-lactam production. The introduction of additional copies of the penicillin cluster genes into a single-copy strain like Wisconsin54-1255 also leads to increased penicillin titers (18). In addition, in a lineage of strains with increasing levels of penicillin production, the number of peroxisomes also increases (10, 11), although this does not seem to be a linear relationship. A recent transcriptomic analysis indicates that the genes encoding the main biosynthetic pathways yielding the three-amino-acid building blocks are upregulated in high-yielding strains relative to earlier production strains (19).
Recently, the complete genome sequence of P. chrysogenum has been elucidated (19). Also, a genetic toolbox for P. chrysogenum has been extended by the inactivation of the ku70 gene (17). Ku70 is involved in the nonhomologous end-joining (NHEJ) pathway that causes random integration, and its inactivation results in an increased gene targeting efficiency by homologous recombination. Therefore, now the phenotypes of gene deletions in industrial relevant strains can be analyzed. A disadvantage we noted in the use of these so-called “high-copy-number strains” is the genetic instability of the penicillin biosynthetic gene cluster-containing amplicons. During the transformation protocol, copies of the penicillin gene cluster amplicons are readily lost. This phenomenon has been exploited to generate a series of strains derived from a high-copy-number high-penicillin-yielding strain that differs in the number of penicillin biosynthetic gene cluster-containing amplicons (5). This now allowed for the first time a quantitative analysis of the dose effect of the penicillin biosynthetic gene cluster in a host that has been optimized for production by classical strain improvement. Thus, except for defined penicillin biosynthetic gene cluster amplification, this host bears all necessary modifications and mutations that render it an excellent vehicle for β-lactam production. Here we have analyzed this series of strains at the levels of DNA, RNA, and protein, the number of peroxisomes, and the main β-lactam biosynthetic pathway intermediates and end products. Our data suggest that the acyltransferase activity becomes limiting at high copy numbers. The implications of these results for high-level penicillin production are discussed.
MATERIALS AND METHODS
Fungal strains, media, and culture conditions.
Penicillium chrysogenum strain DS17690 and its derivatives and P. chrysogenum Wisconsin54-1255, the ancestor of DS17690, were kindly supplied by DSM Anti-Infectives. Spores were inoculated in YGG medium containing the following (in g/liter): KCl, 10.0; glucose, 20.0; yeast nitrogen base (YNB), 6.66; citric acid, 1.5; K2HPO4, 6.0; and yeast extract, 2.0. After inoculation, spores were grown for 24 h in a shaking incubator at 220 rpm and 25°C. On day 0, the mycelium was diluted 7 times in penicillin V production medium (PPM) containing the following (in g/liter): glucose, 5.0; lactose, 75; urea, 4.0; Na2SO4, 4.0; CH3COONH4, 5.0; K2HPO4, 2.12; KH2PO4, 5.1; and phenoxyacetic acid, 2.5. After dilution, the mycelium was supplemented with a trace element solution (pH 6.3 ± 0.1) and grown for 5 days in a shaking incubator at 220 rpm and 25°C.
gDNA extraction, total RNA extraction, and cDNA amplification.
Genomic DNA (gDNA) was isolated after 96 h of inoculation in penicillin-producing medium using a modified yeast genomic DNA isolation protocol (3) in which the fungal mycelium is broken in a FastPrep FP120 system (Qbiogene). Isolated genomic DNA was measured using NanoDrop ND-1000 (Thermo Scientific) and set at a concentration of 40 ng per quantitative PCR (qPCR) mixtures of 25 μl. Total RNA of transformants was isolated after 5 days of growth in a penicillin-producing medium using Trizol (Invitrogen), with additional DNase treatment by the Turbo DNA-free kit (Ambion). Total RNA was measured with NanoDrop ND-1000 and set at a concentration of 500 ng per cDNA reaction. cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad) in a 10-μl end volume.
qPCR analysis.
The primers for analyzing the expression of the genes in the penicillin biosynthetic gene cluster (pcbAB [Pc21g21390], pcbC [Pc21g21380], and penDE [Pc21g21370]) and the phenoxyacetic acid CoA ligase (phl [Pc22g14900]) are indicated in Table 1. Primers were designed around an intron in the case of the γ-actin (Pc20g11630), penDE, and phl genes in order to be able to separate amplification on gDNA and cDNA. For expression analyses, the γ-actin gene was used as a control for normalization. A negative reverse transcriptase (RT) control was used to determine the gDNA contamination in isolated total RNA. To analyze the number of penicillin clusters in the strains, the γ-actin gene and an intergenic target (between Pc20g07090 and Pc20g07100) were used as reference templates. The primers for pcbAB and pcbC were used to assess the cluster copy number with gDNA. P. chrysogenum Wisconsin54-1255 and strain DS50652 were used as controls containing a single cluster and no penicillin gene clusters, respectively.
TABLE 1.
Primers used for qPCR
| Target | Primer sequence (5′-3′) |
|
|---|---|---|
| Forward | Reverse | |
| γ-actin cDNA | CTGGCGGTATCCACGTCACC | AGGCCAGAATGGATCCACCG |
| γ-actin gDNA | TTCTTGGCCTCGAGTCTGGCGG | GTGATCTCCTTCTGCATACGGTCG |
| IGR (Pc20g07090) | GTTCCTATAGGACGTAGCTCCGC | AAATCAGCTCTACTAGCGATCCGC |
| pcbAB | CACTTGACGTTGCGCACCGGTC | CTGGTGGGTGAGAACCTGACAG |
| penDE | CATCCTCTGTCAAGGCACTCC | CCATCTTTCCTCGATCACGC |
| pcbC | AGGGTTACCTCGATATCGAGGCG | GTCGCCGTACGAGATTGGCCG |
| phl | CTGGGTATGGAGACAGCTGCCG | CGTGCCTCGACTCCAGGGAGC |
The expression levels and gene copy numbers were analyzed, in triplicate, with a MiniOpticon system (Bio-Rad) using the Bio-Rad CFX manager software, with which the threshold cycle (CT) values were determined automatically by regression. The SensiMix SYBR mix (Bioline) was used as a master mix for qPCR with 0.4-μM primers. The following thermocycler conditions were used: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Subsequently, a melting curve was generated to determine the specificity of the qPCRs. The efficiency of the primers used for the copy number determination was assessed through the use of four dilutions of gDNA. The γ-actin reference gene, the intergenic region, and the pcbAB and pcbC genes showed efficiencies of 100.17% (R2 = 1.000), 94.34% (R2 = 0.997), 96.78% (R2 = 0.999), and 102.86% (R2 = 0.993), respectively. Likewise, for transcript levels, four dilutions of cDNA were made, and the primer efficiencies for the γ-actin reference gene and the genes of interest, pcbAB, pcbC, penDE, and phl, were 98.19% (R2 = 1.000), 96.78% (R2 = 0.997), 102.86% (R2 = 0.993), 96.38% (R2 = 0.999), and 88.57% (R2 = 0.981), respectively.
Biochemical techniques.
Cell extracts of P. chrysogenum were isolated as follows. After 5 days of growth in penicillin-producing medium, 1 ml of the culture was mixed with 1 ml 25% trichloroacetic acid (TCA) and frozen at −20°C. After being thawed on ice, cells were pelleted by centrifugation (10,000 × g for 10 min) and washed twice with cold (−20°C) 80% acetone. The pellet was air dried and solubilized in 200 μl solubilization buffer containing 1% sodium dodecyl sulfate (SDS) and 0.1 M NaOH, and subsequently, 50 μl 5× SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer was added. Samples were boiled for 5 min and centrifuged for 10 min at 10,000 × g, whereupon the supernatant was used for SDS-PAGE analysis. Western blots were stained with polyclonal antibodies raised against the indicated penicillin biosynthetic pathway proteins and transcription elongation factor EF1-α as a reference protein (6).
Transformation of P. chrysogenum with the DS-RED gene.
Protoplasts of P. chrysogenum DS17690 and the derivatives DS47274 and DS47276 (5) were isolated, and cells were cotransformed with plasmids pBBK-007 (10) and pBlue-AMDS, bearing the acetamidase gene (amdS) as a selection marker. Plasmid pBBK-007 contains the DS-RED gene fused with the peroxisomal targeting signal SKL under the control of the gpdA promoter of Aspergillus nidulans and the terminator of the penDE gene of P. chrysogenum. Transformants were selected on plates with acetamide as the sole nitrogen source (17). Positive colonies were selected based on the reddish DS-RED color.
Confocal fluorescence microscopy.
Confocal images were made using a Zeiss LSM510 confocal laser scanning microscope (CLSM; Zeiss, Netherlands). DS-RED fluorescence was analyzed by excitation of the mycelium with a 543-nm helium-neon ion laser and detection of fluorescence emission using a band-pass LP 560-nm filter. Positive DS-RED transformants were inoculated in penicillin-producing medium and grown for 96 h before the number of peroxisomes was counted.
Determination of metabolite concentrations.
The extracellular titers of phenoxyacetic acid and penicillin V in the culture medium were determined by high-pressure liquid chromatography (HPLC) using an isocratic flow of acetonitrile at 350 g/liter, KH2PO4 at 640 mg/liter, and H3PO4 at 340 mg/liter. Peaks were separated on a Platinum EPS 5-μm C18 column (Grace) at a flow of 1 ml/min and detected at a wavelength of 254 nm (4). Intracellular metabolite levels were analyzed after 5 days of growth in penicillin-producing medium, and 5 ml of the broth was washed 3 times with 15 ml of 60% MeOH (−40°C) and frozen in liquid N2 for storage (15). Freeze-dried cell-free extract (50 mg) was prepared in 1 ml 90°C MilliQ water to lyse the cells. Samples were firmly shaken for 20 s, rapidly cooled in ice water, and diluted 25 times prior to analysis. Intracellular metabolites were separated by liquid chromatography on a Waters SunFire column (2.1 by 150 mm) at 30°C at a flow of 0.25 ml/min using a sample injection of 25 μl. Eluates were analyzed by tandem mass spectrometry (MS/MS) on LTQ Orbitrap in positive-ion mode (m/z range of 200 to 1,000) with and without spiking (100 ng/ml) of the compound of interest.
RESULTS
Variation of the penicillin amplicon number.
In a previous study, the high-yielding P. chrysogenum strain DS17690 was cured from its penicillin biosynthetic gene cluster-containing amplicons by protoplasting and plating without any selection (5). This yielded a series of strains (DS47272, DS47273, DS47274, DS47276, DS48082, and DS50652) that varied in their penicillin titers. Finally, a penicillin gene cluster-free strain (DS50652) was obtained by a directed deletion of the remaining single cluster in strain DS47274. To determine the penicillin biosynthetic gene cluster copy numbers for these DS17690-derived strains, gDNA was isolated, and quantitative PCR was performed on the pcbAB and pcbC genes. Primers amplifying the γ-actin gene and an intergenic region on gDNA were used as references. The sequenced ancestry strain Wisconsin54-1255 (19) was used as a reference, as it is known to contain a single copy of the penicillin gene cluster, while the cluster-free strain DS50652 indeed lacked the penicillin gene cluster. The number of penicillin biosynthetic gene clusters in strains DS47272, DS47273, DS47274, and DS47276 was 7, 3, 1, and 4 copies, respectively. The original strain DS17690 contained 8 copies (Fig. 1B). Furthermore, the copy number for the phl gene that is not part of the penicillin biosynthetic gene cluster amplicon was determined to exclude amplification or reduction events of critical genes in the various strains. All DS17690 derivatives, including Wisconsin54-1255, contained only 1 copy of the phl gene in their genomes (data not shown). These data demonstrate that in the selected strains, the number of penicillin biosynthetic gene cluster amplicons varies from 0 to 8.
Expression analysis.
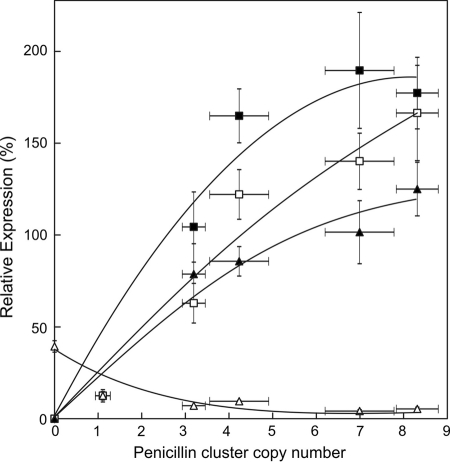
The transcript levels of the various penicillin biosynthetic genes (pcbAB, pcbC, and penDE) and phl were determined. Herein, the set of strains that vary in penicillin gene cluster copy number were grown for 5 days in penicillin-producing medium, and total RNA was isolated. In the qPCR experiments, γ-actin was used as a reference gene. For comparison, the expression of the analyzed genes in the strain with a single copy of the penicillin cluster (DS47274) was set at 12.5%, as it reflects one-eighth of the number of clusters present in the DS17690 strain. As expected, the expression levels of the three structural penicillin biosynthetic genes increased nearly linearly with the cluster number but started to saturate at very high amplicon cluster numbers (Fig. 2). In contrast, the expression of phl decreased with the copy number of the penicillin cluster.
FIG. 2.
Relative expression levels of the key genes of penicillin biosynthesis as a function of the genomic copy number of the penicillin cluster. Expression of pcbAB (closed squares), pcbC (closed triangles), penDE (open squares), and phl (open triangles). Transcript levels were normalized with the γ-actin gene, and the expression level in the single-copy strain DS47274 was set at 12.5% to relate to the 8-fold-increased copy number of the P. chrysogenum DS17690 strain. Because of the experimental error in the determination of the expression level in the single-copy strain, the maximum expression levels may scale beyond 100%.
Protein levels.
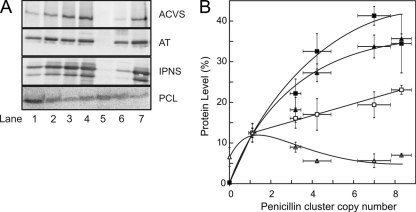
To correlate the transcriptome with the proteome, the relative protein levels of the three penicillin biosynthetic genes and PCL were analyzed by immunoblotting, using specific polyclonal antibodies directed against these proteins. Protein levels were determined in mycelia obtained after growth for 5 days in penicillin-producing medium, but essentially similar results were obtained after 7 days of fermentation. Cells were harvested and lysed, whereupon proteins were isolated by rapid trichloracetic precipitation. Immunoblots of the cell lysates were developed against the elongation factor EF1-α for normalization purposes. For comparison, the protein level in the single-copy strain DS47274 was again set at 12.5% (see above). Protein levels of ACVS (encoded by the pcbAB gene) and IPNS (encoded by the pcbC gene) increased with gene copy numbers and saturated at the higher gene cluster copy numbers (Fig. 3). Thus, the transcript and protein levels of these enzymes show a good correlation. Remarkably, the level of AT already seemed to saturate at a copy number above 1, and only a small increase was observed at higher gene cluster copy numbers (Fig. 3). This shows that the protein and transcript levels of AT do not correlate. Finally, the protein levels of PCL decrease in the strains with increasing penicillin biosynthetic cluster copy numbers, although less dramatically than the transcript levels (Fig. 2). These data indicate that the AT levels do not increase proportionally with the transcript levels and genomic copy numbers.
FIG. 3.
Protein levels for the key enzymes of penicillin biosynthesis as a function of the genomic copy number of the penicillin gene cluster. (A) Relative levels of the enzymes ACVS, AT, IPNS, and PCL, as determined by Western blotting and immunostaining using specific polyclonal antibodies. Samples used for Western blotting were normalized with antibodies directed at elongation factor EF1-α. Strains analyzed are as follows: lane 1, DS47274 (1 copy); lane 2, DS47273 (3 copies); lane 3, DS47276 (4 copies); lane 4, DS47272 (7 copies); lane 5, DS50652 (cluster free); lane 6, Wisconsin54-1255; and lane 7, DS17690 (8 copies). (B) Quantitation of the protein levels as a function of the biosynthetic gene cluster number. Protein levels of ACVS (closed squares), IPNS (closed triangles), AT (open squares), and PCL (open triangles). The protein level in the single-copy strain DS47274 was set at 12.5% to relate to the 8-fold-increased copy number of the P. chrysogenum DS17690 strain.
Quantitation of peroxisomes.
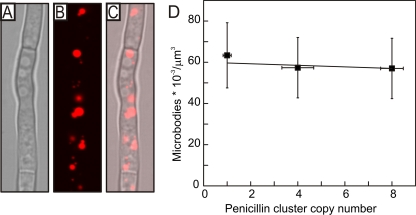
Since AT localizes to the peroxisomes, and because penicillin production levels increase with the number of cellular peroxisomes (10), we also determined if the number of these organelles varied in the set of strains. Herein, we selected three different strains, i.e., P. chrysogenum DS17690, DS47276, and DS47274 containing 8, 4, and 1 penicillin gene clusters, respectively. The strains were cotransformed with plasmids pBlue-AMDS and pBBK-007, containing the AmdS- and DS-RED-encoding genes as markers, respectively. The DS-RED protein was fused at its C terminus to the SKL amino acid sequence, which targets the protein to the peroxisomes, thereby allowing the detection of the organelles by fluorescence microscopy. For penicillin gene cluster copy number determination, gDNA was isolated from two transformants each that were selected on the basis of the DS-RED staining of the colony. The DS-RED strains that showed the same copy numbers as those obtained before the transformation were selected. The strains were grown for 4 days in penicillin-producing medium and analyzed by confocal laser scanning microscopy (Fig. 4 B). The CLSM analysis indeed showed the DS-RED staining of the peroxisomes, while counting revealed that the strains do not differ significantly in the number of peroxisomes (Fig. 4D). These data demonstrate that the number of peroxisomes does not change in the set of P. chrysogenum strains that vary in penicillin biosynthetic gene cluster numbers.
FIG. 4.
Quantitation of the peroxisome numbers in P. chrysogenum strains that vary in the number of penicillin biosynthetic gene clusters. (A to C) Visualization of hyphae and peroxisomes through DS-RED fluorescence, as recorded by CLSM of strain DS47274 containing a single penicillin gene cluster, as shown in the following images: bright-field image (A), DS-RED fluorescence image (B), and merged image (C). (D) The numbers of peroxisomes per 1,000 μm3 in the DS47274, DS47276, and DS17690 strains were counted and plotted against the penicillin cluster copy numbers. The hyphae of the different cells had identical diameters and dimensions.
Extra- and intracellular penicillin pathway metabolites and penicillin V production.
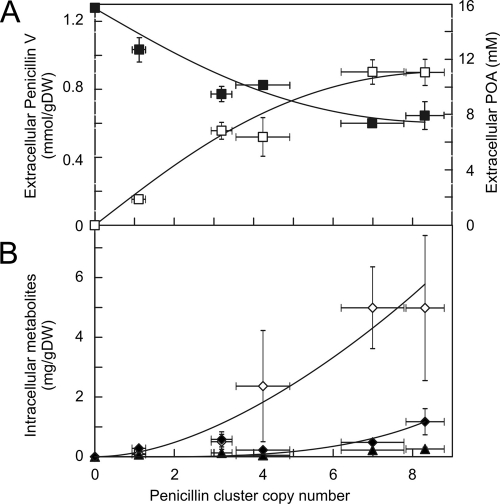
To relate the biosynthetic gene cluster copy number to β-lactam production, the extracellular level of penicillin V was determined after 5 and 7 days of growth. The production levels were corrected for growth differences by dry weight determination. Penicillin V production increased with the penicillin gene cluster copy number but saturated at higher copy numbers (Fig. 5 A), much akin to the trend shown by the protein and transcript levels. In addition, intracellular levels of the key intermediates of biosynthesis were determined by liquid chromatography (LC)-MS/MS. Remarkably, isopenicillin N accumulated at a significantly higher gene cluster copy number (Fig. 5B), i.e., from 0 to up to 5 mg IPN/g (dry weight) of cells. There was barely any accumulation of the tripeptide ACV (up to 0.26 mg/g [dry weight] of cells), while only smaller amounts of penicillin V stayed within the cells of the higher-copy-number strains (up to 1.17 mg/g [dry weight] of cells). Importantly, the analysis revealed no significant levels in the β-lactam nucleus of 6-aminopenicillanic acid (6-APA) (data not shown) or any other β-lactam-derived by-products, neither intra- nor extracellularly. With the DS17690 strain, the intracellular and extracellular penicillin V/IPN ratios were 0.23 and 44, respectively. Thus, the accumulation of IPN within cells indicates that the conversion of IPN into penicillin V is a rate-determining step at high gene cluster copy numbers. This enzymatic conversion involves AT and PCL.
FIG. 5.
Production of penicillin V and the intracellular accumulation of β-lactam intermediates in P. chrysogenum strains that vary in the number of biosynthetic gene clusters after 5 days of fermentation. (A) Extracellular penicillin V production (open squares) and phenoxyacetic acid (POA) levels (closed squares). (B) Intracellular levels of ACV (closed triangles), IPN (open diamonds), and penicillin V (closed diamonds). gDW, gram (dry weight) of cells.
Comparison of the single-biosynthetic-gene-copy strains Wisconsin54-1255 and DS47274.
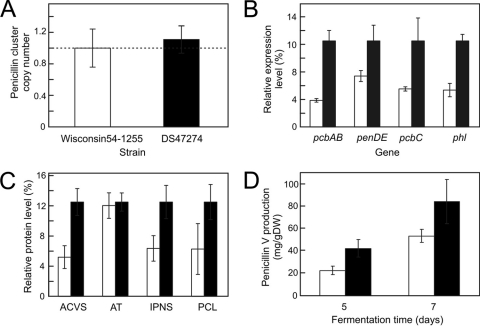
The industrial strain DS17690 was derived from the ancestor strain Wisconsin54-1255 via an extensive classical strain improvement program that lasted at least 30 years. Wisconsin54-1255 contains only a single penicillin biosynthetic gene cluster, which is the same number as that of the DS47274 strain, derived directly from strain DS17690 through amplicon curing (Fig. 6 A). However, strain DS47274 produces two times as much penicillin V as Wisconsin54-1255 in the shaken flask fermentations after 5 days (Fig. 6D). Remarkably, the expression of all analyzed penicillin biosynthetic pathway genes, pcbAB, pcbC, penDE, and phl, is also increased by more than 2-fold in the DS47274 strain compared to that in Wisconsin54-1255 (Fig. 6B). Likewise, the protein levels of ACVS, IPNS, and PCL increased in the DS47274 strain (Fig. 6C). However, both strains showed the same levels for the AT enzyme, despite the 2-fold difference in transcript levels. Overall, these data suggest that DS47274 is more efficient than Wisconsin54-1255, due to regulatory mutations that cause the upregulation of the key enzymes in penicillin biosynthesis and likely other mutations that affect the overall performance.
FIG. 6.
Comparison of two single penicillin biosynthetic gene cluster strains, P. chrysogenum Wisconsin54-1255 (white bars) and DS47274 (black bars). (A) Quantification of the penicillin cluster copy number; (B) relative expression levels of pcbAB, penDE, pcbC, and phl; (C) relative protein levels of ACVS, AT, IPNS, and PCL; (D) extracellular levels of penicillin after 5 and 7 days. Samples were taken after 5 days of fermentation, unless indicated otherwise. The transcript and protein levels of DS47274 were set at 12.5%, as described in the legends to Fig. 2 and 3.
DISCUSSION
Industrial β-lactam production processes depend on strains of Penicillium chrysogenum that have been subjected to an intense classical strain improvement (CSI) program involving random mutagenesis, screening, and selection. Strain DS17690, used in this study, was developed from the Wisconsin54-1255 lineage over a period of many years. One of the most remarkable results of CSI is the amplification of the penicillin biosynthetic gene cluster between tandem repeats. The conserved amplicon-flanking hexanucleotides might be hot spots for site-specific recombination that results in the amplification of the penicillin cluster (2). However, the region might also be responsible for the genetic instability of the strains with amplified biosynthetic gene clusters. We noted that after protoplasting, gene clusters are readily lost, while only the last remaining cluster required a directed deletion approach for removal (5). At the same time, this offered us the possibility to “reverse” study the effect of penicillin gene cluster amplification by obtaining a series of isogenic strains that differ only in the number of biosynthetic gene clusters.
Previously, penicillin production levels among different strains derived from CSI were compared (12, 14). However, those reports differ fundamentally from this study, as they did not take into account the many other alterations that occurred during CSI, except for amplicon amplification. In a recent analysis of the genome sequence of P. chrysogenum Wisconsin54-1255 and a transcriptomic comparison of that strain with strain DS17690 (19), the presence of multiple mutations and alterations introduced by the CSI program was readily demonstrated. For instance, CSI resulted in the upregulation of genes involved in the biosynthesis of α-aminoadipate, cysteine, valine, and various transporters, in addition to the downregulation of many secondary metabolite biosynthesis gene clusters (19). Also, the number of peroxisomes among strains in a lineage varies, and this may have a direct impact on the productivity (8). Therefore, only through the use of isogenic strains can exclusive insight into the gene copy number dose effect of the penicillin gene cluster on β-lactam production be obtained.
Quantitation of the genomic copy number of the biosynthesis gene clusters showed that the high-yielding strain DS17690 contains 8 copies, a number that was almost stepwise reduced to 0 in a series of derived strains. Transcription of the biosynthetic genes in these strains appeared to be very efficient and increased almost linearly with the cluster copy number, only saturating at (very) high copy numbers. Based on our qPCR analysis referenced against γ-actin, it appeared that pcbAB is expressed at lower levels than the other two cluster genes, pcbC and penDE. This is in contrast to recent microarray experiments that showed that penDE is expressed at a 2.5-times-lower level than pcbAB and pcbC (19). The apparent discrepancy is most likely caused by differences in growth conditions and/or penicillin production medium. Although the protein levels of ACVS and IPNS correlated well with the transcript levels, this was not the case for AT. While a substantial level of AT protein is produced by the single-cluster-copy-number strain, only a marginal 2-fold increase occurred in the strain having an 8-fold-higher gene copy number. Importantly, this phenomenon was accompanied by a substantial increase in the intracellular level of isopenicillin N, suggesting that at high penicillin gene cluster copy numbers, the AT activity becomes limiting for production. Why do the AT protein levels not correlate with the penDE transcript levels? Possibly, there are limitations at the level of penDE translation, import into the peroxisome, maturation of the AT, and/or the stability of the protein that may cause this large discrepancy in the protein level. AT contains the PTS1 peroxisomal targeting sequence ARL, which is less optimal than the consensus sequence SKL (7). A replacement of the ARL targeting sequence by SKL might boost the production of penicillin.
The various isogenic DS17690 strains did not differ in the numbers of peroxisomes, but the numbers were almost 3-fold higher than that for the NRRL1951 strain (10). This has been attributed to the upregulation of pex11, a peroxisome proliferation gene (8). Possibly, the AT protein level produced by the single-copy strain already challenges the import capacity of the peroxisomes. Another factor that might potentially be limiting is the activity of the phenoxyacetic acid CoA ligase (PCL), one of the side chain-activating enzymes that also localizes to the peroxisome (9). This enzyme is encoded by the single-copy phl gene, which localizes outside the penicillin biosynthetic gene cluster. In shaken culture, the phl transcript levels decreased with increasing cluster copy numbers, and a similar but weaker effect for the PCL protein levels was observed. Although phl expression is induced by phenoxyacetic acid and the alternative side chain, phenylacetic acid, these organic acids seem to be only weak inducers, as substantial amounts of phenoxyacetic acid remained in the medium, even at high gene copy numbers (Fig. 5A) (19). However, it seems less likely that the activity of PCL and related enzymes is limiting at high penicillin gene copy numbers, as we did not notice any intra- or extracellular accumulation of 6-APA or 8-hydroxy penicillanic acid, which is produced from 6-APA upon reaction with CO2.
In summary, we conclude that in a strain optimized by classical strain improvement, the production of penicillin saturates at high copy numbers of the biosynthetic genes and that the major limitation resides in the production of sufficient AT protein. Therefore, further “overexpression” by increasing the penicillin cluster copy number will likely not result in improved levels of penicillin, and this necessitates more unconventional approaches in future metabolic engineering programs, for instance, the improvement of AT targeting to the peroxisomes, the stabilization and maturation of the enzyme, or possibly, an increased number of peroxisomes. Reconstruction of the β-lactam biosynthetic pathway through synthetic biology, employing refactoring tools aimed at the optimization of the expression and targeting of the biosynthetic enzymes, may result in genetically more stable strains and a better balance among the various enzymes involved in β-lactam biosynthesis, with a potential for enhanced levels of production.
Acknowledgments
This project was financially supported by the Netherlands Ministry of Economic Affairs and the B-Basic partner organizations (www.b-basic.nl) through B-Basic, a public-private NWO-ACTS (Advanced Chemical Technologies for Sustainability) program.
We acknowledge DSM Anti-Infectives for providing the strains and the LC-MS/MS analysis, and M. Veenhuis and I. van der Klei for performing the confocal fluorescence microscopy.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Elander, R. P. 2003. Industrial production of beta-lactam antibiotics. Appl. Microbiol. Biotechnol. 61:385-392. [DOI] [PubMed] [Google Scholar]
- 2.Fierro, F., J. L. Barredo, B. Diez, S. Gutierrez, F. J. Fernandez, and J. F. Martin. 1995. The penicillin gene cluster is amplified in tandem repeats linked by conserved hexanucleotide sequences. Proc. Natl. Acad. Sci. U. S. A. 92:6200-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harju, S., H. Fedosyuk, and K. R. Peterson. 2004. Rapid isolation of yeast genomic DNA: bust n' grab. BMC Biotechnol. 4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris, D. M., J. A. Diderich, Z. A. van der Krogt, M. A. Luttik, L. M. Raamsdonk, R. A. Bovenberg, W. M. van Gulik, J. P. van Dijken, and J. T. Pronk. 2006. Enzymic analysis of NADPH metabolism in beta-lactam-producing Penicillium chrysogenum: presence of a mitochondrial NADPH dehydrogenase. Metab. Eng. 8:91-101. [DOI] [PubMed] [Google Scholar]
- 5.Harris, D. M., Z. A. van der Krogt, P. Klaassen, L. M. Raamsdonk, S. Hage, M. A. van den Berg, R. A. Bovenberg, J. T. Pronk, and J. M. Daran. 2009. Exploring and dissecting genome-wide gene expression responses of Penicillium chrysogenum to phenylacetic acid consumption and penicillin G production. BMC Genomics 10:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiel, J. A., V. I. Titorenko, I. J. van der Klei, and M. Veenhuis. 2007. Overproduction of translation elongation factor 1-alpha (eEF1A) suppresses the peroxisome biogenesis defect in a Hansenula polymorpha pex3 mutant via translational read-through. FEMS Yeast Res. 7:1114-1125. [DOI] [PubMed] [Google Scholar]
- 7.Kiel, J. A., M. A. van den Berg, F. Fusetti, B. Poolman, R. A. Bovenberg, M. Veenhuis, and I. J. van der Klei. 2009. Matching the proteome to the genome: the microbody of penicillin-producing Penicillium chrysogenum cells. Funct. Integr. Genomics 9:167-184. [DOI] [PubMed] [Google Scholar]
- 8.Kiel, J. A., I. J. van der Klei, M. A. van den Berg, R. A. Bovenberg, and M. Veenhuis. 2005. Overproduction of a single protein, Pc-Pex11p, results in 2-fold enhanced penicillin production by Penicillium chrysogenum. Fungal Genet. Biol. 42:154-164. [DOI] [PubMed] [Google Scholar]
- 9.Koetsier, M. J., P. A. Jekel, M. A. van den Berg, R. A. Bovenberg, and D. B. Janssen. 2009. Characterization of a phenylacetate-CoA ligase from Penicillium chrysogenum. Biochem. J. 417:467-476. [DOI] [PubMed] [Google Scholar]
- 10.Meijer, W. H., L. Gidijala, S. Fekken, J. A. Kiel, M. A. van den Berg, R. Lascaris, R. A. Bovenberg, and I. J. van der Klei. 2010. Peroxisomes are required for efficient penicillin biosynthesis in Penicillium chrysogenum. Appl. Environ. Microbiol. 76:5702-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller, W. H., R. A. Bovenberg, M. H. Groothuis, F. Kattevilder, E. B. Smaal, L. H. Van der Voort, and A. J. Verkleij. 1992. Involvement of microbodies in penicillin biosynthesis. Biochim. Biophys. Acta 1116:210-213. [DOI] [PubMed] [Google Scholar]
- 12.Muller, W. H., T. P. van der Krift, A. J. Krouwer, H. A. Wosten, L. H. van der Voort, E. B. Smaal, and A. J. Verkleij. 1991. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J. 10:489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newbert, R. W., B. Barton, P. Greaves, J. Harper, and G. Turner. 1997. Analysis of a commercially improved Penicillium chrysogenum strain series: involvement of recombinogenic regions in amplification and deletion of the penicillin biosynthesis gene cluster. J. Ind. Microbiol. Biotechnol. 19:18-27. [DOI] [PubMed] [Google Scholar]
- 14.Penalva, M. A., R. T. Rowlands, and G. Turner. 1998. The optimization of penicillin biosynthesis in fungi. Trends Biotechnol. 16:483-489. [DOI] [PubMed] [Google Scholar]
- 15.Seifar, R. M., Z. Zhao, J. van Dam, W. van Winden, W. van Gulik, and J. J. Heijnen. 2008. Quantitative analysis of metabolites in complex biological samples using ion-pair reversed-phase liquid chromatography-isotope dilution tandem mass spectrometry. J. Chromatogr. A 1187:103-110. [DOI] [PubMed] [Google Scholar]
- 16.Smith, D. J., J. H. Bull, J. Edwards, and G. Turner. 1989. Amplification of the isopenicillin N synthetase gene in a strain of Penicillium chrysogenum producing high levels of penicillin. Mol. Gen. Genet. 216:492-497. [DOI] [PubMed] [Google Scholar]
- 17.Snoek, I. S., Z. A. van der Krogt, H. Touw, R. Kerkman, J. T. Pronk, R. A. Bovenberg, M. A. van den Berg, and J. M. Daran. 2009. Construction of an hdfA Penicillium chrysogenum strain impaired in non-homologous end-joining and analysis of its potential for functional analysis studies. Fungal Genet. Biol. 46:418-426. [DOI] [PubMed] [Google Scholar]
- 18.Theilgaard, H., M. van den Berg, C. Mulder, R. Bovenberg, and J. Nielsen. 2001. Quantitative analysis of Penicillium chrysogenum Wis54-1255 transformants overexpressing the penicillin biosynthetic genes. Biotechnol. Bioeng. 72:379-388. [DOI] [PubMed] [Google Scholar]
- 19.van den Berg, M. A., R. Albang, K. Albermann, J. H. Badger, J. M. Daran, A. J. Driessen, C. Garcia-Estrada, N. D. Fedorova, D. M. Harris, W. H. Heijne, V. Joardar, J. A. Kiel, A. Kovalchuk, J. F. Martin, W. C. Nierman, J. G. Nijland, J. T. Pronk, J. A. Roubos, I. J. van der Klei, N. N. van Peij, M. Veenhuis, H. von Dohren, C. Wagner, J. Wortman, and R. A. Bovenberg. 2008. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 26:1161-1168. [DOI] [PubMed] [Google Scholar]