Abstract
We introduce a near-real-time optical imaging method that works via the detection of the intrinsic fluorescence of life forms upon excitation by deep-UV (DUV) illumination. A DUV (<250-nm) source enables the detection of microbes in their native state on natural materials, avoiding background autofluorescence and without the need for fluorescent dyes or tags. We demonstrate that DUV-laser-induced native fluorescence can detect bacteria on opaque surfaces at spatial scales ranging from tens of centimeters to micrometers and from communities to single cells. Given exposure times of 100 μs and low excitation intensities, this technique enables rapid imaging of bacterial communities and cells without irreversible sample alteration or destruction. We also demonstrate the first noninvasive detection of bacteria on in situ-incubated environmental experimental samples from the deep ocean (Lo'ihi Seamount), showing the use of DUV native fluorescence for in situ detection in the deep biosphere and other nutrient-limited environments.
Bacteria are widely recognized for living in extreme environments and as integral players in processes as varied as weathering, corrosion, environmental remediation, pathogenesis, and symbiosis (3, 4, 26). In most of these cases, surface-bound bacteria play key roles (1, 7, 19) and pose a particular challenge for researchers: the detection and imaging of life on reflective and/or fluorescent surfaces at the microbial (μm) scale (5, 12, 18). In environments ranging from the deep subsurface biosphere, dry deserts, and deep ice cores to hospitals and clean rooms, concentrations of bacteria, either as spores or active cells, can range from 109 to less than 1,000 cells/gram (14, 22, 24, 25, 29, 34). Finding and quantifying these microbes when they are on surfaces usually involves epifluorescence techniques, using dyes that bind to DNA or proteins, and examining the fluorescence of those dyes under UV or visible illumination (6, 8, 9, 16, 23, 31).
Current tagging methods offer a number of significant disadvantages. First, the mineral surfaces on which the microbes are found are often themselves highly fluorescent, making the microbes difficult or impossible to differentiate; second, the act of adding the fluorescent probe can alter the physical and chemical nature of the system; additionally, nonspecific binding can lead to overestimation of cell abundance (2, 18). Because of the problems associated with the fluorescence of minerals and staining to detect microbial cells, researchers typically resort to physically removing cells from surfaces and staining/counting them separately from their matrix (12). This is an inefficient process that involves both cell loss and the loss of information about the mineralogical context that may have an influence on the microbial ecology. More recently, cell staining of active cells with SYBR green 1 and a computer-assisted analysis method has demonstrated an ability to separate fluorescent cells from nonspecific binding (17). However, a label-free method to search for and quantify the distribution and abundance of bacteria on natural samples over multiple spatial scales has not been available.
Label-free optical approaches using Raman scattering methods have been offered as a nondestructive imaging solution (13, 27). However, these systems utilize laser energies greater than 1 × 109 joules/cm2, exceeding the energies necessary for chemical damage to the cell (33), require relatively flat surfaces for optimal collection efficiency, and can suffer from background fluorescence of the target and the substrate it may reside on.
In response to these challenges, we have developed an optical method that enables detection and imaging of single bacterial cells on natural and opaque surfaces and assessment of bacterial density and distribution of single cells to biofilms over spatial scales ranging from microns to centimeters. The method utilizes deep-UV (DUV) (<250-nm)-laser-induced native fluorescence of organic components intrinsic to the cell or spore while avoiding autofluorescence interference from the substrate. Here we show DUV native fluorescence as a near-real-time optical imaging method and demonstrate the first noninvasive detection of bacteria on in situ-incubated environmental experimental samples from the deep ocean (Lo'ihi Seamount) for which we correlate the bacterial biomass to distributions of the iron-oxide precipitates.
MATERIALS AND METHODS
Bacterial culture preparation.
Bacterial samples of Shewanella oneidensis MR1 were obtained from stock cultures from Ken Nealson's lab at USC. Bacterial spores of Bacillus pumilis were obtained from spore enrichments from Kasthuri Venkateswaran in the Planetary Protection group at JPL. The spores were used as the inoculum for the vegetative cells. Both genera were grown in LB for 24 h to a cell concentration of 107 cells/ml. For MR1 and B. pumilis vegetative cells, 1 ml of each was washed three times with decreasing concentrations of phosphate-buffered saline (PBS) to reduce crystallization of salt when the bacteria were deposited onto the samples. Washing the cells consisted of centrifuging 1 ml of the culture at 3,000 × g for 5 min, removal of the supernatant, and resuspension of the cells. Resuspension of the cells was initially in 0.9% PBS and then in 0.09% and ended with cells in 0.005% PBS.
Substrate inoculation.
All samples were inoculated in the same manner. A total of 100 μl each of the bacteria (107 cells/ml) and spores was spread over each sample and allowed to dry prior to imaging. Prior to inoculation, the samples were imaged using the DUV microscope and/or the DUV large-scale raster scanner to get a background fluorescence reading.
DUV instrumentation.
The deep-UV microscope used in these experiments is a custom instrument. The base system is an Olympus microscope that has been modified with quartz optics, for excitation and transmission below 350 nm, and has been coupled to a 224.3-nm HeAg, hollow-cathode laser for DUV epi-illumination. The DUV laser power at the sample, after UV-enhanced turning and focusing optics, was maximized to <1 μJ/100-μs pulse. In addition to the deep-UV source, a white-light source from Olympus was attached to a secondary port on the microscope. A DUV-sensitive electron-multiplied charge-coupled device (EMCCD) was used for detection.
Illumination of the sample uses a custom 45-degree dichroic filter that injects both the DUV and the white-light sources through a 52× reflective objective (Ealing Optics). To acquire native fluorescence images, an electron-multiplied (EM) gain of 100× was used. In addition to increasing the sensitivity, the EMCCD was binned 2×. The image integration time was triggered by the DUV laser and matched to the maximum pulse duration of the DUV laser (100 μs). To further increase the signal-to-noise ratio (SNR), 10 pulses of the laser over 1 s (10 Hz) were used with on-chip accumulation. Bandpass filters, centered at 320, 340, and 360 nm, were used to observe the fluorescence. For the overlays, corresponding visible images using the white-light illumination were taken without a filter. The 52× objective observes a 150- by 150-μm area with white-light illumination. However, with the DUV laser, a 30-μm diameter spot is illuminated.
For each DUV image, the clock-induced noise from the EMCCD was removed and the grayscale values were colorized. The same processing was performed on the entire image on all native fluorescence microscope images by using Photoshop CS3. The overlays used only the nonzero regions of the native fluorescence images shown.
The DUV large-scale raster-scanning instrument combines a targeted UV chemical sensor (TUCS; Photon Systems, Inc.) and a 248.6-nm NeCu hollow cathode laser that illuminates the sample with a 4-μJ/60-μs pulse. For fluorescence detection, six discrete, gated, photomultiplier (PMT)-based detection bands at 280, 300, 320, 340, 360, and 380 nm were detected simultaneously and integrated for the 60-μs laser pulse duration.
The excitation laser is focused to a 200-μm spot that is translated over the sample at a rate that satisfies Nyquist sampling. Using the motor encoders, the maps are displayed in millimeters and provide spatial coordinates from a registered set of points to locate the same areas on the DUV microscope stage.
DAPI staining and imaging of mineral surfaces.
A total of 2.4 × 104 cells of MR1 were deposited on freshly split gypsum (calcium sulfate), siderite (iron carbonate), and basalt samples to compare single-cell detection capabilities of native fluorescence to those of DAPI (4′,6-diamidino-2-phenylindole) staining. The comparison between DUV-laser-induced native fluorescence of bacteria and DAPI staining was performed on the same mineral samples.
Prior to inoculation, the basalt and gypsum samples were analyzed for background fluorescence, with excitation at 224 nm (DUV microscope) and an Hg lamp (traditional epifluorescence microscope excitation filter center, 380 nm). For comparison between the DUV microscope and the Hg lamp, a 438-nm band on the DUV microscope was used. This filter overlaps the majority of the DAPI spectral band and shows the effect of excitation.
After the samples were inoculated with bacterial cells, they were first observed with the DUV microscope using both the white-light illumination and the DUV laser source. The white-light illumination enabled detection of bacterial features using morphology. These were illuminated using the 224.3-nm laser source in order to detect whether they fluoresced at 320, 340, and 360 nm, a spectral feature consistent with bacterial cells. After the DUV analysis, the same samples were stained using DAPI. Prior to staining, visible white-light illumination and DUV-induced native fluorescence images were used to locate single bacteria on the surface. A total of 10 ml of a working concentration of DAPI stain (0.3 mM) was added directly to the samples. Samples were imaged under a traditional epifluorescence microscope (Nikon E600) with DAPI filters and an Hg lamp for illumination.
Detection of microbes at multiple spatial scales.
In order to test the sensitivity of this technique at different spatial scales, approximately 1 × 106 MR-1 and B. pumilis cells and approximately 1 × 104 B. pumilis spores were distributed over a steel plate such that a concentration gradient was formed along one axis. The gradient was formed by depositing 100 ml of each sample with a pipettor and increasing the amount of material released as the pipettor was moved across the plate. A 1,000-cm2 area of the plate was mapped using a large-scale DUV raster scanner. Fluorescing regions of the plate were then imaged using the deep-UV microscope to determine the relative abundance of cells and spores.
RESULTS AND DISCUSSION
Native fluorescence of microbes and substrates.
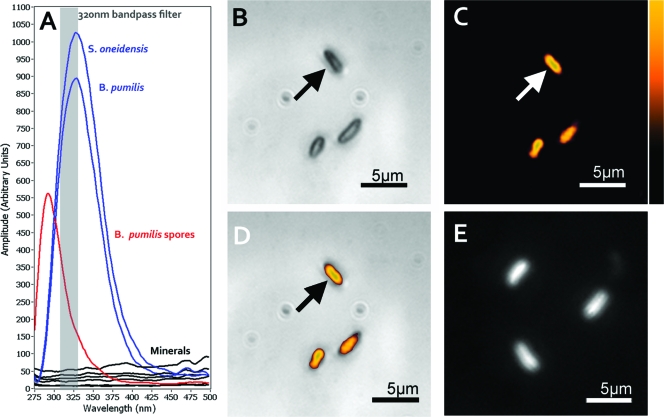
Excitation at wavelengths between 200 and 250 nm results in fluorescence responses that can be used to distinguish biological samples from many other naturally occurring or anthropomorphically generated environmental fluorophores (5). This response derives from the combinatorial absorption and fluorescence signatures of intrinsic proteins, free amino acids, nucleic acids, flavins, and other aromatic compounds that have been concentrated in the cells; e.g., Bacillus spores exhibit a unique signature, hypothesized to be due to the high concentrations of dityrosine in the spore coat (10, 20). Although chromophores associated to specialized bacteria can extend the range of fluorescence to beyond 600 nm, the majority of the intrinsic fluorescence signature associated to all bacterial cells or spores peaks in a UV range between 270 and 400 nm (Fig. 1 A).
FIG. 1.
Label-free and DAPI imaging of bacterial cells on a freshly cleaved gypsum surface. (A) Emission spectra of a variety of minerals and rocks (siderite, gypsum, orthoclase, and basalt) (this study) (black) and bacterial cells (blue) and spores (red) with 224-nm excitation (5). The gray bar shows the spectral band used to obtain the native fluorescence images. (B) White-light-illuminated visible image of the gypsum surface with three putative bacteria. (C) Deep-UV native fluorescence image of image shown in panel B, showing that the objects correlate to bacterial cells. (D) Overlay of the native fluorescence (shown in panel C) and visible image. (E) UV image of the gypsum sample after DAPI.
The visible image showed morphological features consistent with the morphology of MR-1 (Fig. 1B). Native fluorescence images showed 320-nm emissions from these same features and no detectable fluorescence from gypsum. Although a single 100-μs pulse can be used to image the cells (see movie in the supplemental material), 10 pulses were accumulated on the chip to increase the signal-to-noise ratio to 6.8, with a signal-to-background contrast ratio of >2,000:1. The native fluorescence images compare favorably to those of DAPI staining, with respect to the localization of microbes on the gypsum surface (Fig. 1B to E), and do not result in the background fluorescence seen in the DAPI-stained mineral sample (Fig. 1E).
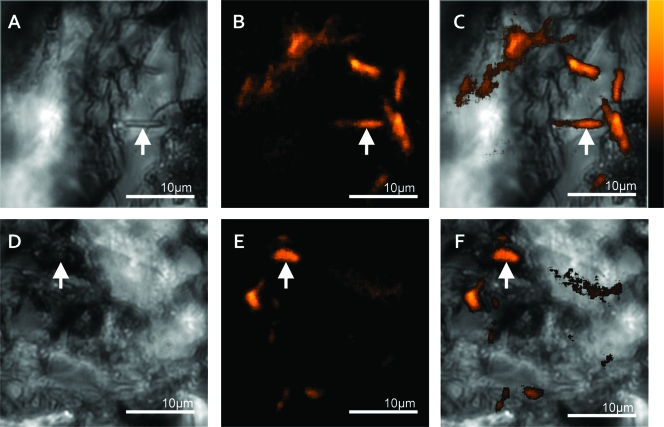
To demonstrate the ability to extend this to localizing bacteria on opaque and rough surfaces, a similar concentration of MR-1 cells was introduced to cut basalt and freshly split siderite surfaces. These cases represent more real-world samples in which visible white-light illumination was not definitive in identifying the location of microbes on these surfaces (Fig. 2 A and D). Visible features with morphologies that suggest a bacterium within a crevice were definitively shown as bacterial fluorescence in the native fluorescence images. In addition, other bacterial cells not apparent in the visible images were detected on both of these opaque surfaces (Fig. 2B, C, E, and F). These images represent the first use of DUV-induced native fluorescence to noninvasively image single bacterial cells on rough, natural, opaque surfaces. Although the use of DAPI staining is an established method for localizing bacteria in similar mineral samples, in the case of deep-UV native fluorescence, there was no need to alter the samples chemically, making them available for further investigations.
FIG. 2.
Bacterial cells on unprepared substrates (siderite and basalt). (A) White-light-illuminated visible image of the siderite surface. The arrow points to a morphology consistent with a bacterium. (B) Deep-UV native fluorescence image of image shown in panel A, showing that the morphology indicated by the arrow in the visible image is a bacterium with other previously undetected bacterial cells surrounding it. (C) Overlay of the native fluorescence (shown in panel B) and visible image (shown in panel A). (D) White-light-illuminated visible image of a basalt surface. The arrow indicates the location of one of the bacterial cells observed in panel E. (E) Deep-UV native fluorescence image of image shown in panel D, showing the location of bacterial cells. The arrow points to a bacterium whose morphology cannot be seen in panel D. (F) Overlay of the native fluorescence (shown in panel E) and visible image (shown in panel D). In some of the fluorescence images, speckled features are bacterial cells beyond the depth of focus of the microscope.
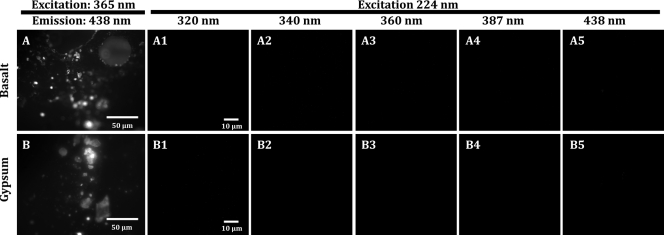
Results from the uninoculated minerals showed that with excitation at 224 nm, mineral fluorescence in the UV was minimal or nonexistent and thus did not obscure fluorescence from bacterial cells and spores. DUV-excited images of gypsum and basalt samples observed at multiple locations revealed no detectable mineral fluorescence at emission wavelengths from 290 nm to 450 nm (Fig. 3). In contrast, with excitation with a traditional epifluorescence microscope at 365 nm and emission at 450 nm (DAPI filter cube), these same unstained samples produced a diffuse fluorescence background, with some areas containing highly fluorescent mineral grains (Fig. 3). This was likely the result of absorption efficiencies associated to trace elements present in the minerals.
FIG. 3.
Mineral fluorescence comparison between DUV (224-nm) and Hg (365-nm) lamp for uninoculated basalt (A) and gypsum (B). Panels A1 and B1 are fluorescence images of basalt and gypsum with an excitation at 365 nm and emission at 435 nm (e.g., DAPI filter). The same samples were illuminated at 224 nm with images collected at 320, 340, 360, 387, and 438 nm (A2 to 5 and B2 to 5). Panels A2 to 5 show no fluorescence from the basalt, and panels B2 to 5 show no fluorescence from the gypsum, even at the 438-nm band, which is similar to the DAPI filter.
Spatial scalability and sensitivity of native fluorescence-based microbial detection.
The DUV microscope configuration enables a resolution of 300 nm over a 700-μm2 area; a large-area raster-scanning configuration enables a resolution of 200 μm with which the total observable area is limited by the 1,000-mm by 500-mm raster tracks. With the multiple scales that this method enables, bacterial distributions over large areas can be easily observed and cell-substrate interactions can be visualized.
Using only ∼4 μJ/pulse, the DUV-induced native fluorescence results in a strong signal response from single microbial cells. It is therefore possible to spatially scale this approach over three orders of magnitude while maintaining single-cell sensitivity. In contrast to DUV-induced native fluorescence, Raman spectra have shown an ability to detect and characterize bacterial cells using vibrational bonds, but the comparatively weak scattering cross sections and current methods for Raman imaging make the technique impractical for bacteria widely distributed over natural, rough surfaces. Compared to current Raman imagers, the DUV system demonstrated here provided scan rates that were more than 7 to 9 orders of magnitude faster.
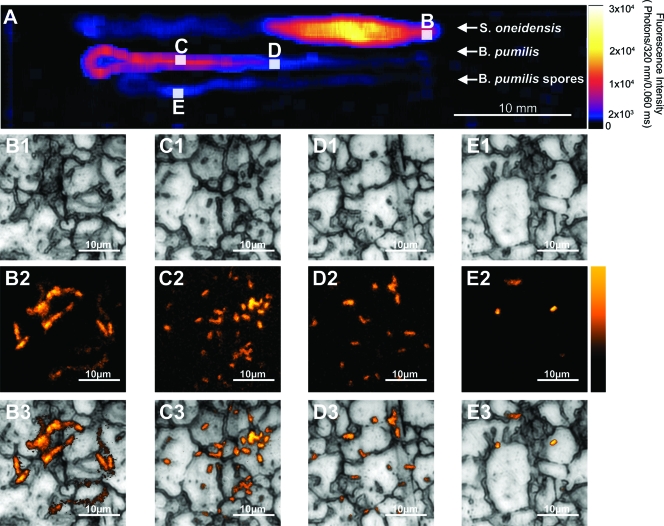
Results of imaging done with the steel plate indicate that the fluorescence from the scanner comes from the bacterial cells and spores and not from the background (Fig. 4 A). Additionally, there is a correlation between the fluorescence intensity of the raster scanner and the concentration of individual cells/spores, giving the large-scale methodology a high level of detection capability (Fig. 4B to E).
FIG. 4.
Label-free imaging of bacteria from the macroscopic to the microscopic using deep-UV native fluorescence. (A) Raster-scanned image of the plate after inoculation with bacteria. The color bar represents the fluorescence intensity by the number of 320-nm photons emitted in a 60-μs pulse. These data were collected using the DUV raster scanner. The images are of bacteria dried on a stainless steel plate over an area of 600 mm2 (50 by 12 mm). The intensity of the signal is proportional to the density of the cells and spores in the area. (B to E) High-resolution visible and native fluorescence images of the plate were taken with a DUV fluorescence microscope (excitation, 224 nm; emission, 320 nm) at areas indicated by white squares, indicating that the macroscopic signal comes from few to many single cells or spores. (B) S. oneidensis from the edge of a high-concentration region; (C) B. pumilis cells from a region of high concentration; (D) B. pumilis cells from a region of low concentration; (E) B. pumilis spores from a region of very low concentration. (B1 to E1) Visible images of the stainless steel surface. Panels B2 to E2 are native fluorescence images of the areas shown in panels B1 to E1, and panels B3 to E3 are overlays of the native fluorescence and visible images, showing that the bacteria are preferentially attaching at grain boundaries.
Application to the detection of microbes on environmental samples.
These laboratory-prepared samples demonstrate that DUV excitation provides a noncontact, noninvasive optical method for detection of bacterial cells. However, in environmental samples, cells may not exist as simple cell accumulations at the surface but rather may be present in complex three-dimensional cell-organic-mineral biofilm matrixes. As such, we used DUV native fluorescence to describe the macroscopic community structure and single-cell features on basalt thick sections that were incubated as an in situ colonization experiment at 980 m below sea level at the Lo'ihi Seamount. Similar experiments have been conducted in the deep sea (11, 15, 30, 32), corroborating the general observations (adhesion patterns, cell densities, and secondary mineral associations) made here. We present here the first example of using DUV-induced native fluoresce as a noninvasive and label-free method to map the distribution of bacterial populations and image discrete bacteria and bacterial communities from a natural environment.
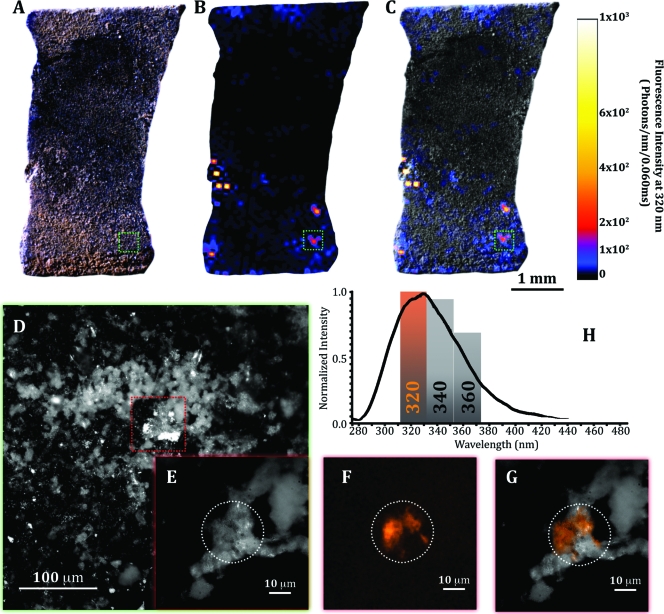
A raster-scanned DUV map over the 6.5- by 4.5-mm chip showed the spatial heterogeneity of bacterial growth in which signatures were spatially associated to the edge of iron-oxide mineral features (Fig. 5 A to C). The densest regions of these oxides had very low fluorescence intensity, indicating very low biomass. The intense signatures were on the periphery of the oxides, suggesting that either other bacteria were buried within the oxides or had migrated as these secondary minerals formed.
FIG. 5.
Label-free imaging of bacterial communities from the Lo'ihi Seamount. (A) Visible image of a basalt chip incubated at the Lo'ihi Seamount (980 m below sea level). Lighter regions are iron oxides that formed during incubation. (B) Raster-scanned DUV native fluorescence image (6 by 4 mm) of the biomass distribution over the basalt chip (emission at 320 nm). The color ramp is the intensity in photons/nm. (C) An overlay of the visible image and the native fluorescence image, indicating that the biomass is located at the periphery of the dense iron oxide regions. (D) Visible image of the Y-shaped iron oxide feature, at a higher magnification, from the region identified by the green box in panels A to C. Visible image (E), DUV native fluorescence image at 320 nm (F), and overlay (G) of the Y-shaped feature from the red boxed area shown in panel D, showing bacterial communities attached to the oxide formations. The white circle indicates the area illuminated by the DUV laser. (H) Spectroscopic comparison of a DUV native fluorescence spectrum of bacterial cells (S. oneidensis) compared to relative fluorescence intensities of the bacterial mass at 320, 340, and 360 nm.
Using the raster scan images as a guide (Fig. 5A to C), visible and DUV microscopic images were acquired from regions of both low and high fluorescence intensity. Images collected from the low-fluorescence regions consisted of cell abundances of approximately 1,000 cells/mm2. However, for the dense high-fluorescence regions, the cell abundance was closer to 5,000 to 10,000 cells/mm2. One particularly dense region had a distinct Y-shaped morphological feature in the fluorescence data (Fig. 5D and E). A DUV microscopic image of this region shows that the signature emanated from a biofilm attached to an iron oxide formation (Fig. 5F and G). Although single cells cannot easily be deciphered from the image, the 300-μm2 area covered by the bacteria suggests that there are 60 to 150 cells. The local cell density limits distinct cellular morphology and requires spectral data to confirm that these are bacterial cells. In addition to the 320-nm band, DUV images were also taken using 340- and 360-nm bands to show that the fluorescence of the biofilm correlates to a high-resolution fluorescence spectrum of bacterial cells (Fig. 5H).
A DUV source enables the detection of microbes in their native state on natural materials, avoiding background autofluorescence and without the need for fluorescent dyes or tags. Excitation in the DUV offers real-time detection of bacteria on mineral and metal surfaces at spatial scales ranging from tens of centimeters to micrometers and from communities to single cells, without irreversible sample alteration or destruction. DUV native fluorescence is an ideal tool for in situ detection of bacteria, extending from use in the deep biosphere to other nutrient-limited environments.
The capability of the DUV methodology to image over many spatial scales provides both contextual and high-resolution images of bacterial cells. The combination enables not only correlation of bacterial populations to surface variables, including texture, mineralogy, and stress regions on a microscopic scale, but large-scale correlations, such as chemical gradients found in ice cores and rock strata. In addition, the combination of the high sensitivity and wide-area imaging capability provides a search tool for bacterial cells that may be surviving at the limits of life, such as the oceanic lithosphere and the deep subsurface, or for life detection on other planetary bodies (21, 28, 29). Deep-UV-induced fluorescence detection of cells in the environments would allow targeted analysis by secondary methods, enabling the location of potential life forms and allowing the use of subsequent, more-specific spectroscopic, chemical, and biological methods. For example, combining spectroscopic methods such as Raman and native fluorescence enables rapid triaging of a sample and identifies specific regions over which Raman and resonance Raman imaging would provide further characterization of the cell(s) as well as the substrate.
Supplementary Material
Acknowledgments
We thank L. Beegle, S. Feldman, and M. Pelletier for insightful technical and scientific discussions, K. Venkateshwaran and M. T. LaDuc for spore suspensions, and C. T. Mogensen for support in development of the mapping platform.
This work was supported by the Air Force under an AFOSR MURI grant (to K.H.N.), a Defense University Research Industry Program (DURIP; USC/Photon Systems Inc.), NASA Planetary Protection Research, and the JPL Icy Worlds node of the NASA Astrobiology Institute (NAI).
This research was carried out at the Jet Propulsion Laboratory/California Institute of Technology under a contract with the National Aeronautics and Space Administration, at the University of Southern California, and at Photon Systems, Inc.
Footnotes
Published ahead of print on 3 September 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Banfield, J. F., and K. H. Nealson (ed.). 1997. Geomicrobiology: interactions between microbes and minerals, p. 448. Mineralogical Society of America, Washington, DC.
- 2.Barra Caracciolo, A., P. Grenni, C. Cupo, and S. Rossetti. 2005. In situ analysis of native microbial communities in complex samples with high particulate loads. FEMS Microbiol. Lett. 253:55-58. [DOI] [PubMed] [Google Scholar]
- 3.Beech, I. B., J. A. Sunner, C. R. Arciola, and P. Cristiani. 2006. Microbially-influenced corrosion: damage to prostheses, delight for bacteria. Int. J. Artif. Organs 29:443-452. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, P., J. Rogers, and W. Choi. 2001. Silicates, silicate weathering, and microbial ecology. Geomicrobiol. J. 18:3-19. [Google Scholar]
- 5.Bhartia, R., W. F. Hug, E. C. Salas, R. D. Reid, K. K. Sijapati, A. Tsapin, W. Abbey, K. H. Nealson, A. L. Lane, and P. G. Conrad. 2008. Classification of organic and biological materials with deep ultraviolet excitation. Appl. Spectrosc. 62:1070-1077. [DOI] [PubMed] [Google Scholar]
- 6.Chan, C., G. De Stasio, S. Welch, M. Girasole, B. Frazer, M. Nesterova, S. Fakra, and J. Banfield. 2004. Microbial polysaccharides template assembly of nanocrystal fibers. Science 303:1656-1658. [DOI] [PubMed] [Google Scholar]
- 7.Croal, L., J. Gralnick, D. Malasarn, and D. Newman. 2004. The genetics of geochemistry. Annu. Rev. Genet. 38:175-202. [DOI] [PubMed] [Google Scholar]
- 8.Daley, R. J., and J. E. Hobbie. 1975. Direct counts of aquatic bacteria by a modified epifluorescence technique. Limnol. Oceanogr. 20:875-882. [Google Scholar]
- 9.Dang, H., and C. Lovell. 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards, K., T. McCollom, H. Konishi, and P. Buseck. 2003. Seafloor bioalteration of sulfide minerals: results from in situ incubation studies. Geochim. Cosmochim. Acta 67:2843-2856. [Google Scholar]
- 12.Epstein, S., D. Alexander, K. Cosman, A. Dompe, S. Gallagher, J. Jarsobski, E. Laning, R. Martinez, G. Panasik, C. Peluso, R. Runde, and E. Timmer. 1997. Enumeration of sandy sediment bacteria: are the counts quantitative or relative? Mar. Ecol. Prog. Ser. 151:11-16. [Google Scholar]
- 13.Kalasinsky, K., T. Hadfield, A. Shea, and V. Kalasinsky. 2007. Raman chemical imaging spectroscopy reagentless detection and identification of pathogens: signature development and evaluation. Anal. Chem. 79:2658-2673. [DOI] [PubMed] [Google Scholar]
- 14.La Duc, M., R. Kern, and K. Venkateswaran. 2004. Microbial monitoring of spacecraft and associated environments. Microb. Ecol. 47:150-158. [DOI] [PubMed] [Google Scholar]
- 15.Lovley, D. R. (ed.) 2000. Environmental microbe-metal interactions, p. 199-395. ASM Press, Washington, DC.
- 16.Moreau, J. W., P. K. Weber, M. C. Martin, B. Gilbert, I. D. Hutcheon, and J. F. Banfield. 2007. Extracellular proteins limit the dispersal of biogenic nanoparticles. Science 316:1600-1603. [DOI] [PubMed] [Google Scholar]
- 17.Morono, Y., T. Terada, N. Masui, and F. Inagaki. 2009. Discriminative detection and enumeration of microbial life in marine subsurface sediments. ISME J. 3:503-511. [DOI] [PubMed] [Google Scholar]
- 18.Nadeau, J. L., N. N. Perreault, T. D. Niederberger, L. G. Whyte, H. J. Sun, and R. Leon. 2008. Fluorescence microscopy as a tool for in situ life detection. Astrobiology 8:859-875. [DOI] [PubMed] [Google Scholar]
- 19.Nealson, K., and F. Popa. 2005. Introduction and overview: what do we know for sure? Am. J. Sci. 305:449-466. [Google Scholar]
- 20.Pandey, N., and A. Aronson. 1979. Properties of the Bacillus subtilis spore coat. J. Bacteriol. 137:1208-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parkes, R., G. Webster, B. Cragg, A. Weightman, C. Newberry, T. Ferdelman, J. Kallmeyer, B. Jorgensen, I. Aiello, and J. Fry. 2005. Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature 436:390-394. [DOI] [PubMed] [Google Scholar]
- 22.Parkes, R. J., B. A. Cragg, S. J. Bale, J. M. Getlifff, K. Goodman, P. A. Rochelle, J. C. Fry, A. J. Weightman, and S. M. Harvey. 1994. Deep bacterial biosphere in Pacific Ocean sediments. Nature 371:410-413. [Google Scholar]
- 23.Patel, A., R. T. Noble, J. A. Steele, M. S. Schwalbach, I. Hewson, and F. J. A. 2007. Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR green I. Nat. Protoc. 2:269-276. [DOI] [PubMed] [Google Scholar]
- 24.Priscu, J., E. Adams, W. Lyons, M. Voytek, D. Mogk, R. Brown, C. McKay, C. Takacs, K. Welch, C. Wolf, J. Kirshtein, and R. Avci. 1999. Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 286:2141-2144. [DOI] [PubMed] [Google Scholar]
- 25.Rogers, J., and P. Bennett. 2004. Mineral stimulation of subsurface microorganisms: release of limiting nutrients from silicates. Chem. Geol. 203:91-108. [Google Scholar]
- 26.Rosan, B., and R. J. Lamont. 2000. Dental plaque formation. Microbes Infect. 2:1599-1607. [DOI] [PubMed] [Google Scholar]
- 27.Rösch, P., M. Harz, M. Schmitt, K. Peschke, O. Ronneberger, H. Burkhardt, H. Motzkus, M. Lankers, S. Hofer, H. Thiele, and J. Popp. 2005. Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations. Appl. Environ. Microbiol. 71:1626-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roussell, E. G., M.-A. Cambon-Bonavita, J. Querellou, B. A. Cragg, G. Webster, D. Prieur, and R. J. Parkes. 2008. Extending the sub-sea-floor biosphere. Science 320:1046. [DOI] [PubMed] [Google Scholar]
- 29.Santelli, C. M., B. N. Orcutt, E. Banning, W. Bach, C. L. Moyer, M. L. Sogin, H. Staudigel, and K. J. Edwards. 2008. Abundance and diversity of microbial life in ocean crust. Nature 453:653-656. [DOI] [PubMed] [Google Scholar]
- 30.Sylvan, J., A. Turner, and K. J. Edwards. 2010. Microbe metal interactions on endolithic seafloor basalts. In Mineral-microbe interactions, in press. ASM Press, Washington, DC.
- 31.Teal, T. K., D. P. Lies, B. J. Wold, and D. K. Newman. 2006. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl. Environ. Microbiol. 72:7324-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Templeton, A. S., E. J. Knowles, D. L. Eldridge, B. W. Arey, A. C. Dohnalkova, S. M. Webb, B. E. Bailey, B. M. Tebo, and H. Staudigel. 2009. A seafloor microbial biome hosted within incipient ferromanganese crusts. Nat. Geosci. 2:872-876. [Google Scholar]
- 33.Vermeulen, N., and W. Keeler. 2007. The bactericidal effect of ultraviolet and visible light on Escherichia coli. Biotechnol. Bioeng. 99:550-556. [DOI] [PubMed] [Google Scholar]
- 34.Warren-Rhodes, K. A., K. L. Rhodes, S. B. Pointing, S. A. Ewing, D. C. Lacap, B. Gomez-Silva, R. Amundson, E. I. Friedmann, and C. P. McKay. 2006. Hypolithic cyanobacteria, dry limit of photosynthesis, and microbial ecology in the hyperarid Atacama Desert. Microb. Ecol. 52:389-398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.