Abstract
During the aggregation of Dictyostelium cells, signaling through RasG is more important in regulating cyclic AMP (cAMP) chemotaxis, whereas signaling through RasC is more important in regulating the cAMP relay. However, RasC is capable of substituting for RasG for chemotaxis, since rasG− cells are only partially deficient in chemotaxis, whereas rasC−/rasG− cells are totally incapable of chemotaxis. In this study we have examined the possible functional overlap between RasG and RasC in vegetative cells by comparing the vegetative cell properties of rasG−, rasC−, and rasC−/rasG− cells. In addition, since RasD, a protein not normally found in vegetative cells, is expressed in vegetative rasG− and rasC−/rasG− cells and appears to partially compensate for the absence of RasG, we have also examined the possible functional overlap between RasG and RasD by comparing the properties of rasG− and rasC−/rasG− cells with those of the mutant cells expressing higher levels of RasD. The results of these two lines of investigation show that RasD is capable of totally substituting for RasG for cytokinesis and growth in suspension, whereas RasC is without effect. In contrast, for chemotaxis to folate, RasC is capable of partially substituting for RasG, but RasD is totally without effect. Finally, neither RasC nor RasD is able to substitute for the role that RasG plays in regulating actin distribution and random motility. These specificity studies therefore delineate three distinct and none-overlapping functions for RasG in vegetative cells.
The Ras subfamily proteins are monomeric GTPases that act as molecular switches, cycling between an active GTP-bound and an inactive GDP-bound state (17). Activation is controlled by guanine nucleotide exchange factors (GEFs), which catalyze the exchange of GDP for GTP, and inactivation regulated by GTPase-activating proteins (GAPs) that stimulate the hydrolysis of bound GTP to GDP (17). Activated Ras proteins stimulate numerous downstream signaling pathways that regulate a wide range of cellular processes, including proliferation, cytoskeletal function, chemotaxis, and differentiation (4). The complexity of this regulation has been emphasized by the discovery of the presence of a large number of Ras subfamily homologues in metazoan organisms (19) and elucidation of the roles played by each protein remains a formidable challenge. An important approach to this problem is an analysis of Ras protein function in organisms amenable to genetic analysis.
The Dictyostelium genome encodes 14 Ras subfamily members, an unusually large number for such a relatively simple organism (6, 25). Six of these have been partially characterized and have been shown to be involved in a wide variety of processes, including cell movement, polarity, growth, cytokinesis, chemotaxis, macropinocytosis, and multicellular development (5, 15, 23, 25). They exhibit considerable functional specificity, and even the two highly related proteins, RasD and RasG, perform different functions (23, 26). RasC and RasG are the best characterized of these proteins, and both are activated in response to cyclic AMP (cAMP) during aggregation (11). Although both proteins are involved in aggregation, signaling through RasC is more important for the regulation of the cAMP relay, whereas signaling through RasG is more important for cAMP-dependent chemotaxis, but there is some overlap of function (2, 3). Disruption of both the rasC and rasG genes results in a total loss of cAMP-mediated signaling, suggesting that all cAMP signal transduction in early development is partitioned between pathways that use either RasC or RasG (2, 3).
In addition to their roles in early development, both RasG and RasC have vegetative cell functions. Cells with a disrupted rasG gene were found to exhibit a reduced growth rate, which was most apparent when cells were grown in suspension, and were multinucleate, indicating a defect in cytokinesis (13, 23). In addition, rasG− cells exhibited reduced motility and polarity and an altered actin distribution. Vegetative rasC− cells had a less pronounced phenotype: changes in actin distribution and motility but normal growth and cytokinesis (16). Given that there was evidence for some overlap of function between RasG and RasC during early development, it was important to determine the extent of their functional overlap in vegetative cells.
In the present study, we have compared the potential overlap of RasG and RasC requirements for vegetative cell function in the recently generated isogenic rasC−, rasG−, and rasC−/rasG− strains (2, 3). In addition, the availability of stable rasG− and rasC−/rasG− strains has enabled us to determine to what extent RasD, a protein that is highly related to RasG but not present in wild-type vegetative cells, can substitute for loss of function of RasG.
MATERIALS AND METHODS
Growth of Dictyostelium discoideum.
Dictyostelium discoideum strain JH10 was grown at 22°C in HL5 medium (14.3 g of peptone, 7.15 g of yeast extract, 15.4 g of glucose, 0.96 g of Na2HPO4·7H2O, and 0.486 g of KH2PO4 per liter of water), supplemented with 50 mg of streptomycin sulfate (Sigma, St. Louis, MO)/ml and 100 μg of thymidine (Sigma)/ml, in tissue culture dishes (Nunc, Rochester, NY). The rasG− and rasC− cells were grown in HL5 medium supplemented with 50 mg of streptomycin sulfate/ml but without the thymidine supplement. rasC−/rasG− mutant cells were grown in HL5 medium, supplemented with 50 mg of streptomycin sulfate/ml and 10 μg of blasticidin S (Calbiochem, San Diego, CA)/ml. The rasG−/[rasG]:rasD and the rasC−/rasG−/[rasG]:rasD cells were grown in HL5 medium supplemented with 10 μg of G418 (Invitrogen, Carlsbad, CA)/ml. The cell numbers were determined microscopically by using a hemacytometer chamber. The cells were routinely grown to a density of ∼4 × 105 cells/cm2 and then harvested for further experimentation. At this density, they were still actively growing. For some experiments, cells were grown in HL5 medium in shake suspension, as previously described (24).
Western blot analysis.
Cells were harvested by centrifugation and lysed in 1× Laemmli SDS-PAGE loading buffer (6× buffer; 350 mM Tris-Cl [pH 6.8], 10% SDS, 600 mM dithiothreitol, 0.012% [wt/vol] bromophenol blue, 30% glycerol). Samples were then boiled at 100°C for 5 min, and protein concentrations were determined by using a protein assay from Bio-Rad (Hercules, CA). Equal amounts of protein were fractionated by SDS-PAGE (21). After electrophoresis, the proteins were transferred onto nitrocellulose (Amersham) membranes, blocked with a 5% nonfat milk solution, and probed with the appropriate antibody; bound antibody was detected by using enhanced chemiluminescence (Amersham). The levels of Ras protein were quantified approximately by densitometry (VersaDoc, imaging system, model 5000; Bio-Rad). To confirm that proteins had been equally loaded, the gels were also stained with Coomassie blue solution (0.025% [wt/vol] Coomassie G-250, 10% [vol/vol] acetic acid).
Chemotaxis assays.
Folate chemotaxis was determined by using a previously described protocol (18). Actively growing cells were rinsed once and submerged in 20% HL5. At t = 0, an Eppendorf (Hamburg, Germany) Femtotip micropipette filled with 25 mM folate was positioned in the field of view, and cell movements were monitored by time-lapse microscopy.
Cytokinesis.
To determine the nuclear number, cells were grown in shake suspension in HL5 for 5 days. A total of 3 × 103 cells/cm2 were allowed to adhere to glass coverslips for 30 min and then washed three times with KK2. The cells were fixed with 3.7% formaldehyde for 10 min, and the fixed cells were washed an additional three times with KK2. The cells were then permeabilized with −20°C acetone, dried, and rehydrated with KK2, and the nuclei were stained with 1 μM DAPI (4′,6′-diamidino-2-phenylindole; Sigma). The stained cells were washed three times with phosphate-buffered saline (PBS), air dried, and mounted on glass slides with 50% glycerol. Epifluorescence images of random fields of view were captured by using an Olympus IX-70 inverted microscope, a DAGE-MTI CCD-100 camera (Michigan City, IN), and Scion (Frederick, MD) Image 4.0 software. The average number of nuclei per cell was obtained by counting 300 to 390 cells.
RasC and RasD activation assay.
After cAMP pulsing, the cells were harvested by centrifugation and resuspended at a density of 2 × 107 cells/ml in KK2 containing 1 mM caffeine. After 30 min, aliquots (2 ml) of the cell suspension were stimulated by the addition of cAMP to 15 μM. Cell suspensions (350 μl) were lysed at the indicated times by mixing them with an equal volume of 2× lysis buffer (20 mM sodium phosphate (pH 7.2; 2% Triton X-100, 20% glycerol, 300 mM NaCl, 20 mM MgCl2, 2 mM EDTA, 2 mM Na3VO4, and 10 mM NaF containing two tablets of Roche Complete protease inhibitor per 50 ml of buffer). The lysates were centrifuged for 10 min, and the protein concentrations of the supernatants were determined as described above. The GST-Byr2-Ras binding domain (RBD) was expressed in Escherichia coli and purified as described previously (11). Next, 400 μg of protein was incubated with 100 μg of GST-Byr2-RBD on glutathione-Sepharose beads (Amersham Biosciences) at 4°C for 1 h. The glutathione-Sepharose beads were harvested by centrifugation and washed three times in 1× lysis buffer. A portion (50 μl) of 1× SDS gel loading buffer was then added to the pelleted beads, and the suspension was boiled for 5 min. The samples were subjected to SDS-PAGE, and Western blots were probed with anti-RasC or anti-RasD specific antibodies. The total level of a specific Ras protein was determined by adding 10 μl of 6× SDS loading buffer to 50 μl of Dictyostelium lysate (1 mg/ml), ∼8.3 μg of protein was subjected to SDS-PAGE, and Western blots were probed with anti-RasC or anti-RasD specific antibodies.
RESULTS
Cell growth.
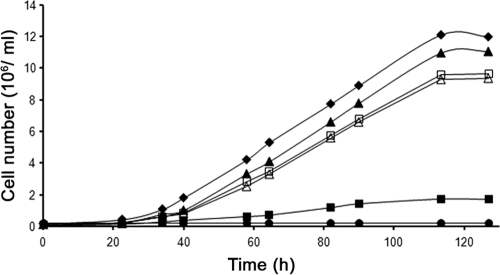
Since preliminary results had indicated that rasC−/rasG− cells grew less efficiently than either rasG− or rasC− cells (2), we examined the growth properties of these cell lines in more detail. The JH10 derived rasG− cells grew slightly slower than the parental JH10 cells (13.5 h versus 12.5 h doubling times) on a plastic surface (data not shown) but grew considerably slower in shaken suspension and ceased growth at much lower cell density (Fig. 1), a defect similar to that reported previously for the AX2 and AX3 derived rasG− cells (13, 23). rasC−/rasG− cells grew at the same rate as rasG− cells on a plastic surface (data not shown) but exhibited no growth in shaken suspension (Fig. 1). However, although these cells did not grow in suspension, they retained full viability after 5 days, as determined by plating cells onto bacterial lawns (data not shown). Although the loss of the rasC gene in Ax2 cells resulted in slight effects in growth (16), the growth of the JH10 derived rasC− strain was indistinguishable from that of JH10 cells, either in suspension (Fig. 1) or on plastic (data not shown).
Fig. 1.
Cell growth in suspension culture. JH10 (⧫), rasC− (▴), rasG- (▪), rasC-/rasG- (•), rasG-/[rasG]:rasD (□), and rasC-/rasG-/[rasG]:rasD (▵) strains were transferred from petri plates into axenic medium at time zero and counted at intervals thereafter. The plotted values are the means of duplicate cell counts. The data plotted are for a single experiment, but similar data were obtained for three independent experiments.
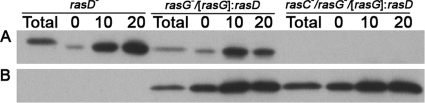
These results emphasize the importance of RasG for the growth of cells in suspension (13, 23). Since cells that lack both RasG and RasC (rasC−/rasG− cells) are incapable of any growth in suspension, it is possible that RasC plays a role during growth, in the absence of RasG. However, it was shown earlier that the AX2- and AX3-derived rasG− cells, strains IR15 and IR17, exhibited an appreciable increase in the vegetative cell level of RasD compared to parental cells, and it was conjectured that this increase in RasD was important to maintain viability (13, 23). It is therefore possible that the reduced growth of the rasC−/rasG− cells in suspension could be due to a reduced level of RasD in these cells rather than to the absence of RasC. In fact, as shown in Fig. 2, there is a twofold reduction in the level of RasD in the rasC−/rasG− cells compared to rasG− cells. The specificity of the RasD antibody used in these studies is evidenced by the fact that rasD− cells exhibit a total absence of RasD protein (Fig. 2). Thus, the complete absence of growth in suspension of the rasC−/rasG− cells could be the result of the lowered levels of RasD in these cells. Interestingly, vegetative cells rasC− cells did not exhibit the enhanced level of RasD, so the minimal decrease of growth in these cells cannot be explained by increased levels of RasD (Fig. 2).
Fig. 2.
Western blot analysis of RasD protein level. Cell lysates from the JH10, rasG-, rasG-/[rasG]:rasD, rasC-/rasG-, rasC-/rasG-/[rasG]:rasD, rasC-, and rasD- strains were probed with RasD specific antibody. For the rasG-/[rasG]:rasD and the rasC-/rasG-/[rasG]:rasD cells, lanes “a,” “b,” and “c” refer to independently clonal isolates.
In order to further determine the effects of RasD on vegetative cell functions, we derived rasG− and rasC−/rasG− cell lines that ectopically expressed the rasD gene under the control of the rasG promoter, to generate similar, elevated levels of RasD in vegetative cells (Fig. 2), a 2.5-fold increase for the rasG− cells and a 5-fold increse for the rasC−/rasG− cells. In both cases, ectopic expression of rasD resulted in growth in suspension that was similar to that of JH10 cells (Fig. 1), arguing convincingly that RasD was capable of replacing the RasG requirement. This result also emphasizes the fact that RasC is not essential for growth in cells containing RasG or RasD and that any effects of RasC on growth are subtle (16).
We showed previously that rasC−/rasG− cells were defective in the induction of early gene expression that occurs following starvation, but that the expression of the carA gene from the actin 15 promoter restored early gene expression (3). These rasC−/rasG−/[act15]:carA cells exhibited identical vegetative cell phenotypes to the rasC−/rasG− cells in these and in all subsequent experiments. No data for the rasC−/rasG−/[act15]:carA cells have therefore been included.
Cytokinesis.
It was shown previously that the IR15 and IR17 cells that grew poorly in shaken suspension, were multinucleate, indicating a defect in cytokinesis (13, 23). When the JH10-derived strains were grown in shaken suspension for 5 days and then subjected to nuclear staining, the majority of the JH10 and rasC− cells contained no more than two nuclei (Fig. 3 and Table 1). In contrast, both rasG− and rasC−/rasG− cells were multinucleate (Fig. 3), with an average number of nuclei of greater than 4 (Table 1). Since there is little or no difference in cytokinesis between the rasG− and the rasC−/rasG− cells, it is apparent that RasC does not make a significant contribution to the regulation of cytokinesis. Expression of RasD in the rasG− and rasC−/rasG− cells resulted in a complete correction of the multinucleate phenotype, indicating that RasD can totally replace the role of RasG in regulating cytokinesis (Fig. 3 and Table 1).
Fig. 3.
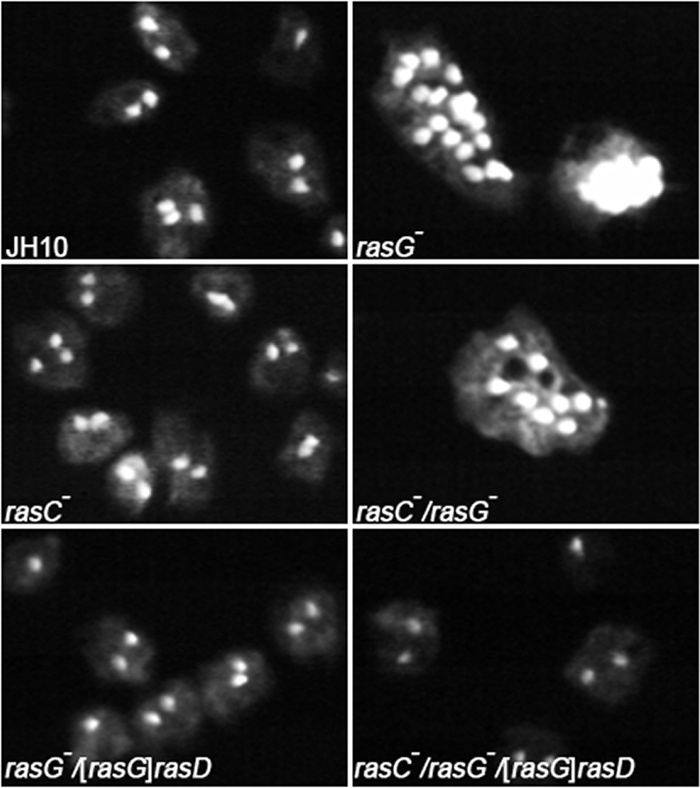
Nuclear staining. Cells were grown in shaken suspension for 5 days and allowed to adhere to glass coverslips for 30 min, washed, and then fixed with formaldehyde. Fixed cells were stained with DAPI as described in Materials and Methods. Epifluorescence images of random fields of view were captured by using an Olympus IX-70 inverted microscope. The cells shown are representative of all of the cells in the population.
Table 1.
Number of nuclei for Dictyostelium cells grown in suspension
| Strain genotype | No. of cells | Mean no. of nuclei per cell ± SDa |
|---|---|---|
| JH10 (wild type) | 303 | 1.82 ± 0.05 |
| rasG− | 315 | 4.81 ± 0.55 |
| rasC− | 360 | 1.85 ± 0.06 |
| rasC−/rasG− | 389 | 4.22 ± 0.16 |
| rasG−/[rasG]rasD | 308 | 1.81 ± 0.06 |
| rasC−/rasG−/[rasG]rasD | 305 | 1.83 ± 0.05 |
Values were calculated by counting the number of nuclei in 300 to 390 cells.
Cells growing on plastic did not exhibit a defect in cytokinesis (data not shown). Thus, the slight decrease in growth rate that occurs when rasG- and rasC-/rasG- cells are grown on plastic is independent of the effects on cytokinesis. Expression of RasD in the rasG- and rasC-/rasG- cells restored growth on plastic to those exhibited by the parental JH10 cells.
Chemotaxis to folate.
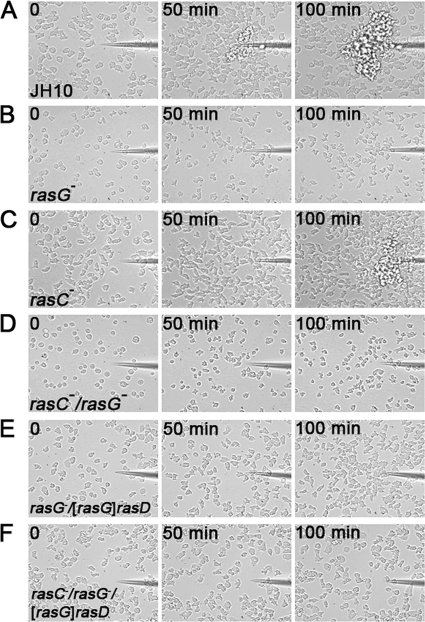
Bacteria secrete folic acid, and this molecule chemotactically attracts vegetative amoebae, resulting in successful predation and feeding (1). The role of RasG and RasC in folate chemotaxis was determined by measuring the movement of cells toward a folate filled micropipette. As shown in Fig. 4A and C, a significant number of JH10 and rasC- cells had moved toward the micropipette after 50 min, with the rasC- cells only slightly slower than the JH10 cells. In contrast, movement of the rasG- cells toward the micropipette was only detectable after 100 min (Fig. 4B), and the rasC-/rasG- cells exhibited no obvious migration (Fig. 4D).
Fig. 4.
Chemotaxis in a folate spatial gradient. The indicated cells were grown to a density of ∼4 × 105 cells/cm2 in Nunc dishes. At t = 0, a micropipette filled with 25 mM folate was positioned in the field of view, and cell movements were monitored by time-lapse microscopy. The results of single experiments are shown, but the results for each strain were highly reproducible.
These results were quantified by calculating chemotactic indices and instantaneous velocities, using OpenLab software. These calculations revealed a considerable reduction in the chemotactic indices for the rasG- cells relative to JH10 cells, a slight reduction in the value for the rasC- cells, and negligible values for the rasC-/rasG- cells (Table 2). Calculation of the rates of instantaneous velocity revealed similar values for JH10 and rasC- cells, but lower values for rasG- and rasC-/rasG- strains (Table 2). These results indicate that RasG is significantly more important than RasC for both vegetative cell motility and chemotaxis to folate. In fact, since motility was not further reduced in the double-null strain relative to the rasG- strain, these results indicate that RasC is relatively unimportant for motility. In contrast, chemotaxis to folate was considerably reduced in rasC-/rasG- cells relative to the rasG- cells, suggesting that RasC does make a contribution to folate chemotaxis, a situation similar to that observed for chemotaxis to cAMP by aggregating cells.
Table 2.
Chemotaxis analysis of Dictyostelium cells in a spatial folate gradient
| Strain genotype | No. of cells | Mean ± SD |
|
|---|---|---|---|
| Instantaneous velocity (μm/min) | Chemotaxis indexa | ||
| JH10 (wild type) | 30 | 13.70 ± 0.93 | 0.90 ± 0.05 |
| rasG− | 31 | 6.85 ± 0.69 | 0.31 ± 0.05 |
| rasC− | 32 | 11.78 ± 0.82 | 0.70 ± 0.06 |
| rasC−/rasG− | 28 | 7.00 ± 0.75 | 0.06 ± 0.02 |
| rasG−/[rasG]rasD | 27 | 7.02 ± 0.64 | 0.32 ± 0.05 |
| rasC−/rasG−/[rasG]rasD | 29 | 7.34 ± 0.76 | 0.07 ± 0.03 |
The chemotaxis index was calculated as the net distance traveled toward the source of chemoattractant divided by the total distance traveled in that time period.
An increase in the level of RasD in the rasG- and rasC-/rasG- cells did not improve either cell motility or folate chemotaxis (Fig. 4 and Table 2), indicating that RasD is unable to replace the RasG requirement for either function.
Ras activation in response to cAMP.
The rasG-/[rasG]:rasD cells exhibited the same delayed aggregation as the rasG- strain, and the rasC-/rasG-/[rasG]:rasD cells failed to aggregate, just like the rasC-/rasG- strain (data not shown). Similarly, chemotaxis to cAMP was identical for the rasG-/[rasG]:rasD and rasG- cells and for the rasC-/rasG-/[rasG]:rasD and rasC-/rasG- strains (data not shown). These results indicate that RasD is also unable to perform the aggregation specific functions of RasG and suggest the possibility that RasD, unlike RasG and RasC, might not be activated in response to cAMP. To test this possibility, the rasG-/[rasG]:rasD and rasC-/rasG-/[rasG]:rasD cells were pulsed with cAMP for 6 h and, after subsequent stimulation with cAMP, the levels of activated RasC and RasD were determined. As shown in Fig. 5A, a robust activation of RasC occurred in these cells in response to cAMP, as has been observed previously with other cells (11). RasD was also activated, although the basal level of activated RasD was quite high, and the magnitude of the activation in response to cAMP was less (Fig. 5B).
Fig. 5.
RasC and RasD activation in response to cAMP. Extracts from rasD-, rasG-/[rasG]:rasD, and rasC-/rasG-/[rasG]:rasD cells that had been pulsed with cAMP and then subjected to a single cAMP stimulus for the indicated times (in seconds) were bound to GST-Byr2-RBD as described in Materials and Methods. The bound material was analyzed by Western blotting, using antibodies specific for RasC (A) or RasD (B). The “Total” lanes represent the level of Ras protein in 8.3 μg of lysate protein used for the pull-down assay.
DISCUSSION
Signal transduction through RasG and RasC plays an important role in the aggregation of D. discoideum during early development. Signaling through RasG is more important for chemotaxis to cAMP, while signaling through RasC is more important for the cAMP relay, but the two proteins do have overlapping functions (2, 3). An important question that remained to be answered is whether signaling through RasG and RasC overlaps in vegetative cells and the experiments described in the present study address this question. An analysis of the various vegetative cell properties of rasC-, rasG-, and rasC-/rasG- cells revealed that, in general, RasG is more important for vegetative cell functions than RasC, in that phenotypic defects for vegetative rasG-cells are more pronounced than those for rasC- cells. However, some of the defects observed for rasG- cells are enhanced in rasC-/rasG- cells, suggesting the possibility that RasC can at least partially substitute for RasG in the transmission of some signals.
The other important question examined in the present study is the extent to which RasD can substitute for RasG in vegetative cells, since RasG and RasD exhibit a high level of identity, with only three differences in the initial 110 amino acids of their sequences (20). RasG is expressed in vegetative cells with levels decreasing during the differentiation process, whereas RasD is barely detectable in vegetative cells with levels increasing during development (26). We therefore examined this question by expressing RasD under the control of the rasG promoter in vegetative rasC-/rasG- and rasG- cells.
One vegetative cell function where rasC-/rasG- cells exhibited a greater defect than rasG- cells is growth in cell suspension, suggesting the possibility that RasC is able to substitute for RasG for growth in suspension. However, rasC-/rasG- cells contain less RasD than rasG- cells, so the difference in growth between the two mutants could be due to this reduction in the level of RasD. Consistent with this latter interpretation, the growth of the rasG-/[rasG]:rasD and the rasC-/rasG-/[rasG]:rasD cells in suspension was identical to that of the JH10 parental strain, suggesting that RasD is more important than RasC for growth. The defect in suspension growth of the rasG- and rasC-/rasG- cells is probably related to a defect in cytokinesis, since both mutants are multinucleate under these conditions. rasG- and rasC-/rasG- cells expressing rasD were not multinucleate when grown in suspension, indicating that RasD can also fulfill the role played by RasG in regulating cytokinesis.
Another vegetative cell property clearly more defective in rasC-/rasG- cells compared to rasG- cells was chemotaxis to folate. Folate chemotaxis was only slightly reduced in the rasC- strain but considerably reduced in rasG-cells, indicating that signaling through RasG is more important for this process. However, since chemotaxis was completely absent in rasC-/rasG- cells, it is clear that RasC can partially replace the role of RasG in cells containing no RasG. Increased expression of rasD in the rasC-/rasG- and rasG- cells resulted in no improvement in the ability of these cells to perform folate chemotaxis, indicating that RasD is incapable of substituting for RasG in the signal transduction pathways required for chemotaxis to folate.
The requirements of RasG and RasC for folate chemotaxis in vegetative cells therefore appear to be completely analogous to their requirements for cAMP chemotaxis in aggregating cells. Although cAMP and folate stimulate cells through different cell surface receptors, coupled to heterotrimeric G proteins that contain different Gα subunits, the downstream Ras-dependent signaling pathways appear to be similar if not identical. Similarly, in addition to its inability to substitute for RasG/RasC in folate chemotaxis, RasD is not able to rescue the aggregation and cAMP chemotaxis defects exhibited by the rasG- and rasC-/rasG- cells (data not shown). This failure of RasD to substitute for RasG during aggregation is presumably due to an inability of RasD to interact with the relevant downstream effector for RasG, since RasD is clearly activated in response to cAMP. However, the steady-state levels of activated RasD that exist prior to cAMP stimulation are considerably higher than the levels observed for activated RasG and RasC (3), and the failure of RasD to substitute for RasG or RasC during aggregation might be due to the presence of the high level of activated RasD in unstimulated cells.
In contrast to the evidence in favor of an overlap of signaling function for RasG and RasC in folate chemotaxis, there is no evidence to suggest an overlap of signaling function for the two proteins in random vegetative cell motility. Thus, instantaneous velocity measurements for the rasC-/rasG- and rasG- cells were lowered to the same extent, relative to the parental JH10 cells. In addition, there is no evidence to suggest that RasD can replace the requirement for RasG for random motility. Thus, instantaneous velocity measurements for the rasG-/[rasG]:rasD and rasC-/rasG-[rasG]:rasD cells were identical to those of the rasG- and rasC-/rasG- cells. A recent report implicated a Ras protein in the regulation of Dictyostelium vegetative cell motility, without establishing its identity (22). The present study indicates that RasG might be the Ras protein involved.
The effect of rasG deletion on random motility corresponds closely to the effect of rasG deletion on cell shape and actin distribution. rasG- cells were shown previously to exhibit a rounded morphology and an abnormal actin distribution (23), and rasC-/rasG- cells exhibit similar properties (data not shown). Normal cell shape and actin distribution were similarly not restored by the expression of RasD in these cells (data not shown). The actin cytoskeleton plays important roles in cytokinesis, cell morphology, and cell migration in mammalian cells, but each of these functions require distinct protein components (8). In Dictyostelium, signaling through RasG clearly impacts actin distribution and cell shape, but the specificity studies presented here indicate that this is distinct from its role in regulating chemotaxis and cell growth.
These Ras specificity studies clearly delineate at least three distinct roles that RasG plays in signal transduction in vegetative cells of Dictyostelium. First, signaling through RasG is important for growth and cytokinesis, and this requirement can be replaced by RasD but not by RasC. Second, signaling through RasG is important in regulating chemotaxis to folate, and this function can be partially replaced by RasC but not by RasD. Finally, signaling through RasG functions in regulating random cell motility, and neither RasD nor RasC is capable of substituting for RasG for this function. The components of the pathways involved in the transmission of the signals for these three vegetative cell RasG functions are still unknown.
Two potential downstream effectors of RasG signal transduction have been described in the literature, Ras interacting protein 3 (Rip3) and phosphatidylinositol 3-kinase (PI3K). Rip3 was identified first as an effector for RasG by a yeast two hybrid analysis (14), and this designation has since been supported by pull-down assays (10). Rip3 is a component of the TORC2 complex that plays a role in cAMP relay and in cAMP chemotaxis during aggregation. Surprisingly, loss of RasC has a much more dramatic impact on the functioning of the TORC2 complex than the loss of RasG (12), but RasC does not interact with Rip3 in either yeast two-hybrid or pull-down assays (10, 14). Furthermore, the aggregation defect exhibited by ripA- cells closely resembles that of rasC- cells but is very different from that of rasG- cells (25). Thus, although there is clearly binding evidence suggesting that Rip3 is an effector for RasG, functional or genetic evidence for such a role is lacking.
The second presumptive RasG effector is PI3K, an enzyme that has been implicated in playing important regulatory roles in chemotaxis, random motility, and cytokinesis (7, 9, 22). Dictyostelium expresses six PI3K proteins, five of which contain a Ras-binding domain (RBD). The two PI3Ks that account for most of the PI3K activity in the cell, PI3K1 and PI3K2, bind RasG, RasC, and RasD (7, 11), properties not consistent with a selective role for PI3K in determining the specificity of signaling through the Ras proteins described here. However, a detailed analysis of the binding affinities of the various Ras proteins to PI3K1 and PI3K2 has not been conducted. Similarly, there is no information on the binding of the Ras proteins to PI3K3 to PI3K5. Therefore, the various PI3K isoforms might exhibit differential Ras binding and thus play a differential role in signal propagation through RasG in vegetative cells, and such a possibility needs to be studied further.
It is also conceivable that a differential subcellular localization of RasG and PI3K might explain the distinct Ras signal transduction specificities described here. Thus, a PI3K isoform activated at the leading edge of the cell might be able to propagate signals by interacting with RasG and RasC during chemotaxis but that the RasD expressed in vegetative cells might not be localized at the leading edge. Similarly, a PI3K isoform interacting with RasG at other plasma membrane localization, and involved in random motility, might not be accessible to either RasC or RasD. Finally, a PI3K isoform interacting with RasG at the poles or cleavage furrows of dividing cells and involved in regulating cytokinesis might be accessible to RasD but inaccessible to RasC. The C-terminal regions of RasG, RasC, and RasD are significantly different, and these differences could account for differential localization of the three Ras proteins in the cell. However, while the idea of differential subcellular localizations for PI3K isoforms and Ras proteins is conceivable, it must be emphasized that there is currently no evidence to support this scenario.
It is possible that the different RasG signaling functions described here are transmitted through other downstream effectors that have not yet been described, and efforts are under way to identify novel Ras interacting proteins that are specific for RasG.
ACKNOWLEDGMENT
This research was funded by a grant to G. Weeks from the Canadian Institute of Health Research.
Footnotes
Published ahead of print on 10 September 2010.
REFERENCES
- 1.Blusch J. H., Nellen W. 1994. Folate responsiveness during growth and development of Dictyostelium: separate but related pathways control chemotaxis and gene regulation. Mol. Microbiol. 11:331–335 [DOI] [PubMed] [Google Scholar]
- 2.Bolourani P., Spiegelman G. B., Weeks G. 2006. Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol. Biol. Cell 17:4543–4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolourani P., Spiegelman G. B., Weeks G. 2008. Rap1 activation in response to cAMP occurs downstream of ras activation during Dictyostelium aggregation. J. Biol. Chem. 283:10232–10240 [DOI] [PubMed] [Google Scholar]
- 4.Campbell S. L., Khosravi-Far R., Rossman K. L., Clark G. J., Der C. J. 1998. Increasing complexity of Ras signaling. Oncogene 17:1395–1413 [DOI] [PubMed] [Google Scholar]
- 5.Chubb J. R., Insall R. H. 2001. Dictyostelium: an ideal organism for genetic dissection of Ras signaling networks. Biochim. Biophys. Acta 1525:262–271 [DOI] [PubMed] [Google Scholar]
- 6.Eichinger L., Pachebat J. A., Glockner G., Rajandream M. A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Tunggal B., Kummerfeld S., Madera M., Konfortov B. A., Rivero F., Bankier A. T., Lehmann R., Hamlin N., Davies R., Gaudet P., Fey P., Pilcher K., Chen G., Saunders D., Sodergren E., Davis P., Kerhornou A., Nie X., Hall N., Anjard C., Hemphill L., Bason N., Farbrother P., Desany B., Just E., Morio T., Rost R., Churcher C., Cooper J., Haydock S., van Driessche N., Cronin A., Goodhead I., Muzny D., Mourier T., Pain A., Lu M., Harper D., Lindsay R., Hauser H., James K., Quiles M., Madan Babu M., Saito T., Buchrieser C., Wardroper A., Felder M., Thangavelu M., Johnson D., Knights A., Loulseged H., Mungall K., Oliver K., Price C., Quail M. A., Urushihara H., Hernandez J., Rabbinowitsch E., Steffen D., Sanders M., Ma J., Kohara Y., Sharp S., Simmonds M., Spiegler S., Tivey A., Sugano S., White B., Walker D., Woodward J., Winckler T., Tanaka Y., Shaulsky G., Schleicher M., Weinstock G., Rosenthal A., Cox E. C., Chisholm R. L., Gibbs R., Loomis W. F., Platzer M., Kay R. R., Williams J., Dear P. H., Noegel A. A., Barrell B., Kuspa A. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funamoto S., Meili R., Lee S., Parry L., Firtel R. A. 2002. Spatial and temporal regulation of 3-phosphoinositides by PI3-kinase and PTEN mediates chemotaxis. Cell 109:611–623 [DOI] [PubMed] [Google Scholar]
- 8.Hall A. 2009. The cytoskeleton and cancer. Cancer Metastasis Rev. 28:5–14 [DOI] [PubMed] [Google Scholar]
- 9.Janetopoulos C., Borleis J., Vazquez F., Iijima M., Devreotes P. 2005. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev. Cell 8:467–477 [DOI] [PubMed] [Google Scholar]
- 10.Kae H., Kortholt A., Rehmann H., Insall R. H., Van Haastert P. J., Spiegelman G. B., Weeks G. 2007. Cyclic AMP signaling in Dictyostelium: G-proteins activate separate Ras pathways using specific RasGEFs. EMBO Rep. 8:477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kae H., Lim C. J., Spiegelman G. B., Weeks G. 2004. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 5:602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamimura Y., Xiong Y., Iglesias P. A., Hoeller O., Bolourani P., Devreotes P. N. 2008. PIP3-independent activation of TorC2 and PKB at the cell's leading edge mediates chemotaxis. Curr. Biol. 18:1034–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khosla M., Spiegelman G. B., Insall R., Weeks G. 2000. Functional overlap of the Dictyostelium RasG, RasD, and RasB proteins. J. Cell Sci. 113 (Pt. 8):1427–1434 [DOI] [PubMed] [Google Scholar]
- 14.Lee S., Parent C. A., Insall R., Firtel R. A. 1999. A novel Ras-interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium. Mol. Biol. Cell 10:2829–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim C. J., Spiegelman G. B., Weeks G. 2001. RasC is required for optimal activation of adenylyl cyclase and Akt/PKB during aggregation. EMBO J. 20:4490–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim C. J., Zawadzki K. A., Khosla M., Secko D. M., Spiegelman G. B., Weeks G. 2005. Loss of the Dictyostelium RasC protein alters vegetative cell size, motility, and endocytosis. Exp. Cell Res. 306:47–55 [DOI] [PubMed] [Google Scholar]
- 17.Mitin N., Rossman K. L., Der C. J. 2005. Signaling interplay in Ras superfamily function. Curr. Biol. 15:R563–R574 [DOI] [PubMed] [Google Scholar]
- 18.Palmieri S. J., Nebl T., Pope R. K., Seastone D. J., Lee E., Hinchcliffe E. H., Sluder G., Knecht D., Cardelli J., Luna E. J. 2000. Mutant Rac1B expression in Dictyostelium: effects on morphology, growth, endocytosis, development, and the actin cytoskeleton. Cell Motil. Cytoskeleton 46:285–304 [DOI] [PubMed] [Google Scholar]
- 19.Reuther G. W., Der C. J. 2000. The Ras branch of small GTPases: Ras family members don't fall far from the tree. Curr. Opin. Cell Biol. 12:157–165 [DOI] [PubMed] [Google Scholar]
- 20.Robbins S. M., Williams J. G., Jermyn K. A., Spiegelman G. B., Weeks G. 1989. Growing and developing Dictyostelium cells express different ras genes. Proc. Natl. Acad. Sci. U. S. A. 86:938–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22.Sasaki A. T., Janetopoulos C., Lee S., Charest P. G., Takeda K., Sundheimer L. W., Meili R., Devreotes P. N., Firtel R. A. 2007. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J. Cell Biol. 178:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuxworth R. I., Cheetham J. L., Machesky L. M., Spiegelmann G. B., Weeks G., Insall R. H. 1997. Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J. Cell Biol. 138:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watts D. J., Ashworth J. M. 1970. Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem. J. 119:171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weeks G., Gaudet P., Insall R. H. 2005. The small GTPase superfamily, p. 211–234 InLoomis W. F., Kuspa A. (ed.), Dictyostelium genomics. Horizon Bioscience, Norfolk, United Kingdom [Google Scholar]
- 26.Wilkins A., Khosla M., Fraser D. J., Spiegelman G. B., Fisher P. R., Weeks G., Insall R. H. 2000. Dictyostelium RasD is required for normal phototaxis, but not differentiation. Genes Dev. 14:1407–1413 [PMC free article] [PubMed] [Google Scholar]