Abstract
The organization and control of polarized growth through the cell cycle of Schizosaccharomyces pombe, a single-celled eukaryote, have been studied extensively. We have investigated the changes in these processes when S. pombe differentiates to form multicellular invasive mycelia and have found striking alterations to the behavior of some of the key regulatory proteins. Cells at the tips of invading filaments are considerably more elongated than cells growing singly and grow at one pole only. The filament tip follows a strict direction of growth through multiple cell cycles. A group of proteins involved in the growth process and actin regulation, comprising Spo20, Bgs4, activated Cdc42, and Crn1, are all concentrated at the growing tip, unlike their distribution at both ends of single cells. In contrast, several proteins implicated in microtubule-dependent organization of growth, including Tea1, Tea4, Mod5, and Pom1, all show the opposite effect and are relatively depleted at the growing end and enriched at the nongrowing end, although Tea1 appears to continue to be delivered to both ends. A third group acting at different stages of the cell cycle, including Bud6, Rga4, and Mid1, localize similarly in filaments and single cells, while Nif1 shows a reciprocal localization to Pom1.
The fission yeast Schizosaccharomyces pombe has been used extensively as a model to study the control and morphology of a eukaryotic cell cycle. As a simple rod-shaped cell, it grows by elongation and divides by medial fission. Growth at each end is strictly controlled during the cycle. After division, growth occurs only at the “old” end, but after S phase is completed, growth commences at the “new” end, a stage known as new-end take-off (NETO). The molecular basis of this ordered and polarized growth, which involves the actin cytoskeleton at the cell tips and microtubules to impose longer-range order, has been studied intensively (12). The Tea1 protein plays a key role in this process. It is delivered to cell tips on the plus ends of growing microtubules and accumulates at both tips even when only one end is growing. It and associated proteins in turn regulate, among other factors, the For3 formin and the GTP-binding protein Cdc42, both of which are involved in regulating actin-dependent tip growth. The site of cell division is marked by the anilin-related Mid1, while the Pom1 kinase interacts with all of these, apparently forming a concentration gradient that controls the timing of cell division (13, 20). These and other proteins form a group of reinforcing functional modules which combine to produce the characteristically uniform pattern of growth of the S. pombe cell.
Many fungi, however, can adopt a variety of cellular patterns for growth: for example, switching between a single-celled form and hyphal or pseudohyphal structures which can extend across a surface or invade the growth medium. This switching between forms may be a necessary component of some fungal infections (10, 21). S. pombe was long considered to adopt only single-celled growth. However, we found that it could be induced to invade the medium, forming elaborate elongated and branching structures very different from the well-known growth pattern (1). Invasion requires high cell density and can be encouraged by a combination of low nitrogen levels and abundant carbon (1, 5). We identified several genes necessary for different stages of the process (5), while the nutritional effect requires the cyclic AMP signaling pathway (1). Among the proteins required for invasion was For3, and inhibition of actin assembly with latrunculin A likewise prevented invasive growth. In contrast, loss of Tea1 or associated proteins did not prevent invasion but resulted in formation of deformed filaments (5).
Given the extensive knowledge of the factors controlling single-celled growth in S. pombe, this invasive response therefore presents an excellent opportunity to investigate how a eukaryotic cell differentiates at the molecular level to produce a radically different multicellular morphology. A requirement to exploit this system had been to visualize the invading cells and the molecules therein in real time as the invasion process proceeds. However, this presented several obstacles: the invading cells are buried in agar, only a small proportion of cells initiate the process at the cell surface, and they are obscured by the large mass of cells above.
Here we describe a system that overcomes these difficulties. We delineate the changes in cell dimensions and growth pattern resulting in mycelial formation and report striking alterations in the intracellular distributions of several groups of key proteins known to control the different stages of ordered polarized growth in this organism.
MATERIALS AND METHODS
Yeast strains and media.
The S. pombe strains used in this study are listed in Table 1. The media, rich (YES), G418 selection (YES medium plus G418), minimal (EMM), mating (SPAS), and nitrogen-limited (LNB) media, and methods for strain construction were as described previously (1, 5, 19).
Table 1.
S. pombe strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source | Reference |
|---|---|---|---|
| Strains | |||
| 972 | h− | Our collection | |
| 556 | h+ade6-216 ura4-D18 leu1-32 | Our collection | |
| UFMG-96-A1153a | Wild type | C. Rosa | 8 |
| FY14674 | h90 ade6-216 leu1-32 lys1-131 ura4-D18 crn1::crn1-GFP-HA-Kanr | Y. Hiraoka/YGRCb | |
| NM01 | cdc25-22 bud6-3GFP:kanMX | F. Chang | 18 |
| NM15 | cdc25-22 leu1−::GFP-bgs4:leu1+ | F. Chang | 18 |
| NM39 | cdc25-22 tea1-3GFP:kanMX | F. Chang | 18 |
| NM145 | cdc25-22 CRIB-GFP:ura4+ | F. Chang | 18 |
| YSM120 | h+tea4-GFP-kanMX6 ade6−leu1-32 ura4-D18 | F. Chang | 14 |
| YSM1342 | h−nif1-GFP-kanMX | S. Martin | 13 |
| PT.907 | cdc25-22 GFP-mod5:kanRleu1-32 h+ | Phong Tran | 33 |
| JM414 | pom1-GFP::kanMX6 h− | J. Moseley | 2 |
| JM430 | Mid1-mEGFP::kanMX6 ura4-D18 leu1-32 ade6-M21X h+ | J. Moseley | 26 |
| CA5818 | h−leu1-32 ura4-D18 for3:3GFP(kanMX6) | K. Shiozaki | 31 |
| CA4699 | h−leu1-32 ura4-D18 rga4:GFP(kanMX6) | K. Shiozaki | 31 |
| KGY2628 | h−sid4-GFP-kanR ade6-M210 ura4-D18 leu1-32 | K. Gould | 34 |
| DL04A03 | h+ade6-m216 ura4-D18 leu1-32 tea1::KanMX4 | Bioneer | |
| V2-22-B02 | h+ade6−ura4-D18 leu1-32 lys1::KanMX4 | Bioneer | |
| JA1375 | Strain 556 carrying p02/G12 (leu1-32::htb1-YFP-leu1+) | This study | |
| JA1374 | Strain 556 carrying p18/E01 (leu1-32::spo20-YFP-leu1+) | This study | |
| JA1530 | ura4-D18 lys1::KanMX4 (selected from crossing strains A1153 and V2-22-B02) | This study | |
| JA1568 | leu1-32 lys1::KanMX4 (selected from crossing strains A1153 and V2-22-B02) | This study | |
| JA1551 | leu1-32::htb1-YFP-leu1+ (selected from crossing strains JA1568 and JA1375) | This study | |
| JA1577 | leu1-32::spo20-YFP-leu1+ (selected from crossing strains JA1568 and JA1374) | This study | |
| JA1523 | crn1::crn1-GFP-HA-Kanr (selected from crossing strains A1153 and FY14674) | This study | |
| JA1543 | CRIB-GFP:ura4+ (selected from crossing strains JA1530 and NM145) | This study | |
| JA1545 | leu1−::GFP-bgs4:leu1+ (selected from crossing strains JA1551 and NM15) | This study | |
| JA1476 | bud6-3GFP:kanMX (selected from crossing strains A1153 and NM01) | This study | |
| JA1529 | for3:3GFP(kanMX6) (selected from crossing strains A1153 and CA5818) | This study | |
| JA1489 | tea1-3GFP:kanMX (selected from crossing strains A1153 and NM39) | This study | |
| JA1542 | tea4-GFP-kanMX6 (selected from crossing strains A1153 and YSM120) | This study | |
| JA1578 | GFP-mod5:kanR (selected from crossing strains A1153 and PT.907) | This study | |
| JA1500 | pom1-GFP::kanMX6 (selected from crossing strains A1153 and JM414) | This study | |
| JA1501 | Mid1-mEGFP::kanMX6 (selected from crossing strains A1153 and JM430) | This study | |
| JA1527 | rga4:GFP(kanMX6) (selected from crossing strains A1153 and CA4699) | This study | |
| JA1556 | nif1-GFP-kanMX (selected from crossing strains A1153 and YSM1342) | This study | |
| JA1567 | tea1::KanMX4 (selected from crossing strains A1153 and DL04A03) | This study | |
| JA1576 | sid4-GFP-kanR (selected from crossing strains A1153 and KGY2628) | This study | |
| JA1240 | Strain 556 carrying pREP41GFP-c att c CpY1 | N. Bone | |
| JA1582 | Strain 556 carrying pREP41GFP-c att c CpY1 (selected from crossing strains JA1551 and JA1240) | This study | |
| Plasmids | |||
| p02/G12 | pDUAL nmt42-htb1-HA-YFP-ura4+/leu1+ | M. Yoshida/Riken BRCc | 17 |
| p18/E01 | pDUAL nmt42-spo20-HA-YFP-ura4+/leu1+ | M. Yoshida/Riken BRC | 17 |
Strain UFMG-96-A1153 is referred to as A1153 hereafter.
Y. Hiraoka/YGRC, Y. Hiraoka of the Yeast Genetic Resource Centre (YGRC).
M. Yoshida/Riken BRC, M. Yoshida of the Riken BioResource Center (Riken BRC).
Microscopy.
For imaging of invasive filament formation, a compact group of cells was inoculated onto the glass section of 35-mm glass-bottomed dishes (MatTek) and then covered in 5 ml LNB medium that felt cool to the hand. Time-lapse imaging of growing mycelia was performed using a Zeiss LiveCell Axiovert 200 imaging system with an ×63 magnification oil-immersion objective. For time-lapse imaging of single-celled growth, strains were plated onto the bottom of the agar in glass-bottomed dishes. All time-lapse imaging was performed at 30°C in a heated chamber. Cell lengths were measured using SimplePCI (Hamamatsu).
Fluorescence imaging was performed using a laser scanning Zeiss LSM 510 confocal microscope with an ×100 magnification oil-immersion objective. Fluorescent maximum-intensity stacks were made by taking images from 10 slices spaced 0.5 μm apart and then assembling the image sequence into a maximum-intensity stack using ImageJ (NIH). The intensity levels of Tea1-3GFP (GFP stands for green fluorescent protein) at the tips were measured by analysis of a 50- by 50-pixel section of the tip from a maximum-intensity stack in ImageJ. Time course imaging of Tea1-3GFP delivery to the cell tip was performed from a maximum-intensity stack taken at 10-s intervals over a 480-s period. Staining of invasive filaments with FM 4-64 was performed by removing a section of agar containing invasive filaments and then placing the block of agar onto a 20-μl droplet of water containing 0.8 mM FM 4-64 (Invitrogen).
All images were further processed with Zeiss LSM Image Browser, SimplePCI, and Adobe Photoshop CS2 9.0. For movies (see the supplemental material), the images were converted into Avi files using SimplePCI.
Molecular genetics.
Small-scale DNA isolation from S. pombe cells was done as described previously (19). PCR amplification was performed with KOD polymerase, buffer, and protocol supplied by Novagen (Darmstadt, Germany). The oligonucleotides were 21 nucleotides in length and were supplied by Eurofins MWG (Ebersberg, Germany). DNA sequencing was also performed by Eurofins MWG-Biotech. Transformation of strains with plasmids (Table 1) was performed as described previously (9).
RESULTS
Identification of a strain of S. pombe that exhibits more efficient invasive growth.
Nearly all current research with S. pombe uses strains related to strain 972, originally described by Leupold (11). Most of our previous work on S. pombe invasive growth also used this strain (1, 5). However, with this strain, invasion is initiated by only a small number of cells and then extends directly underneath a large mass of cells on the surface. This has so far prevented observation of the process by microscopy in real time; previous images were from sections cut from the agar, in which further filamentous growth has stopped. We therefore investigated other isolates of S. pombe, with the aim of finding a strain more amenable to microscopy.
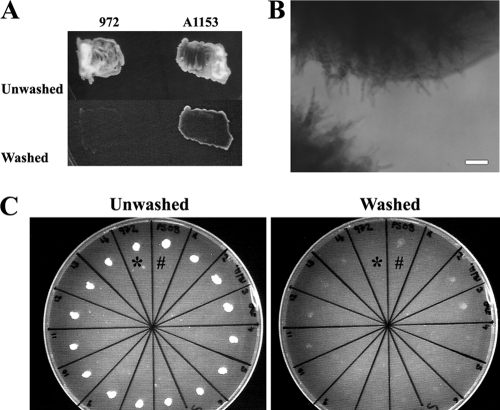
We found that S. pombe strain UFMG-96-A1153 (8) (referred to as A1153 hereafter) exhibited more efficient invasion than strain 972 (Fig. 1A). A notable property of this growth was the emergence of invasive filaments extending beyond the immediate area underneath the surface cells (Fig. 1B). Using glass-bottomed dishes, growing invasive filaments could thus be observed by microscopy.
Fig. 1.
Strain A1153 invades more efficiently than strain 972. (A) Strains 972 and A1153 were plated on LNB medium at high density, and after 4 days, the surface cells were removed by washing. (B) Strain A1153 forms invasive filaments that extend beyond the immediate area of the surface colony. Bar, 50 μm. (C) Mating of strain A1153 with strain 972. Strains 972 (*), A1153 (#), and 14 progeny of a cross between the two (remaining segments) were incubated for 2 days. Strain A1153 had invaded more strongly than strain 972, while the progeny from the cross showed various levels of invasion intermediate between the two parental strains.
To investigate the relationship between A1153 and 972 derivatives, the strains were mated, and hybrid progeny were identified on appropriate selective media. Recombinants were readily recovered but at a low efficiency. This may be due to the fact that strain A1153 is homothallic (not shown), and perhaps some alleles are incompatible between the strains. The recombinants displayed a range of efficiencies of invasion that varied between the levels shown by the parental strains (Fig. 1C). This suggested that multiple loci are likely to contribute to the difference in efficiency between the strains. However, the low efficiency of recombinant recovery prevented a rigorous tetrad analysis. Nevertheless, the strains are clearly sufficiently close to allow mating, which therefore facilitates the introduction of markers from the 972 background into one which allowed real-time observation of filament formation.
Using available primers, 8 regions of the A1153 genome were amplified. Sequence analysis of a total of 5,863 bp of coding and noncoding regions identified 26 differences from the reference 972 sequence (see Tables S1 and S2 in the supplemental material) (http://www.genedb.org/genedb/pombe/), a divergence of 0.44%. Using the same regions from another sequenced strain, NCYC132 (http://www.broadinstitute.org/annotation/genome/schizosaccharomyces_group/MultiHome.html), there were 42 differences between it and strain 972 (0.71%) and 38 differences between strains NCYC132 and A1153 (0.65%). Thus, strain A1153 appears to be no more divergent from the standard strain 972 than is the only other S. pombe strain whose complete genome is currently available.
Both the single-celled and mycelial morphologies of strain A1153 are comparable to those of strain 972.
The morphology of strain A1153 was examined and compared with that of strain 972. Single cells were similar in shape and undertook new-end take-off (NETO) following medial fission (data not shown). However, cell length at division, recorded from time-lapse microscopy on both YES and LNB media, was shorter for strain A1153 than for strain 972 (Table 2). The invasive filaments formed by both strains on LNB medium generally appeared similar in morphology (Fig. 2). The lengths of terminal cells at the filament tips were similar for both strains (Table 2).
Table 2.
Comparison of strains 972 and A1153a
| Cell or filament | Medium | Strain | No. of cells | Mean cell length at septation (μm) (SD) | Mean growth rate (μm/h) (SD) |
|---|---|---|---|---|---|
| Single cells | YES | 972 | 100 | 14.7 (1.21) | 2.35 (0.35) |
| YES | A1153 | 100 | 12.8 (1.09) | 2.07 (0.32) | |
| LNB | 972 | 98 | 15.0 (1.51) | 1.63 (0.28) | |
| LNB | A1153 | 100 | 12.4 (1.08) | 1.19 (0.16) | |
| Filaments | |||||
| From time-lapse microscopy | LNB | A1153 | 50 | 20.0 (1.70) | 1.63 (0.96) |
| From excised blocks | |||||
| LNB | 972 | 49 | 18.5 (1.83) | ||
| LNB | A1153 | 50 | 18.9 (1.54) |
The mean cell lengths at septation and mean growth rates for strains 972 and A1153 were determined on YES and LNB media and compared to the values for tip cells in filaments. The average length of septated tip cells of filaments imaged from excised agar blocks is also shown.
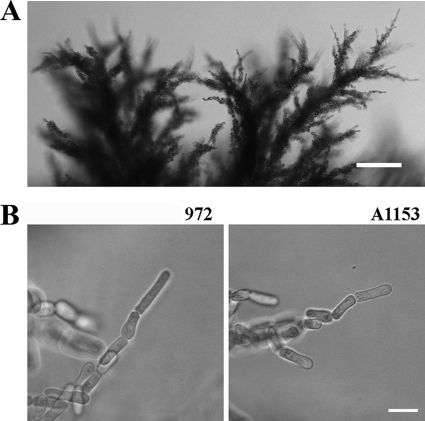
Fig. 2.
The filamentous morphologies of strains A1153 and 972 are similar. (A) Invasive structures formed by strain A1153 after several weeks. The structures are large and complex and resemble those formed by strain 972. Bar, 100 μm. (B) Filaments at single-cell resolution. After 3 days, invasive structures formed by both strains (strains A1153 and 972) were observed in agar slices. The average filament size at division was estimated by measuring the lengths of septated filament tips (Table 2). Bar, 10 μm.
Growth pattern of mycelial development.
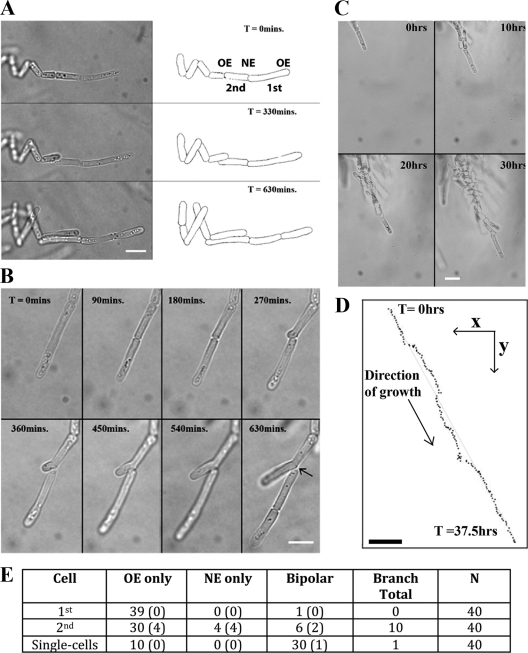
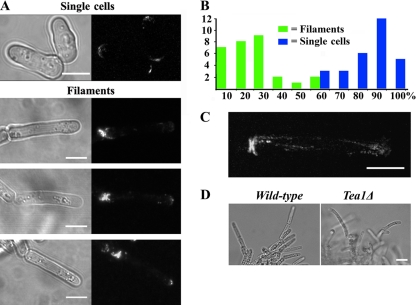
Time-lapse imaging of mycelia of strain A1153 growing in glass-bottomed dishes was performed (see Movie S1 in the supplemental material). Some of the characteristics of filament extension are shown in Fig. 3. The filaments formed outwards from the point of origin and follow a fixed direction of growth. The cells at the tip of the filaments were considerably extended in length (Table 2) and grow monopolarly, apparently without undergoing NETO. Their growth rate slowed before septation, which bisected the cell as normal. The daughters remained in contact at their tips but otherwise appeared to undergo full cell division. Following division, the tip cell continued to grow in a directional, monopolar, and elongated manner and thus continued to extend the filament through the medium. The daughter behind the tip cell sometimes formed a branch point as shown in Fig. 3B, by growing monopolarly from its “new” end, although more frequently, it grew only from its “old” end in the direction away from the filament tip (as in Fig. 3A). The cells further behind the advancing tip appeared more like single cells in growth pattern and dimensions, although as they remained adjacent to each other, growth at the point of contact could give rise to new branches. All of the branching mechanisms contributed to the formation of the larger mycelial structure behind the growing tip (1) (Fig. 3C). The different growth patterns were quantitated in tip cells and the cell immediately behind the tip (Fig. 3E). This confirmed that branching was absent from tip cells and that the second cell from the tip frequently exhibited NETO and hence branch formation.
Fig. 3.
Time-lapse microscopy of S. pombe filamentous growth. (A) Montage of time-lapse images with cell outlines traced on the right. Filament tip cells are elongated (Table 2), grow monopolarly throughout the cell cycle, and undergo complete cell division, and no structures that attach adjacent cells are obvious. “1st” and “2nd” refer to the cell at the tip of the filament and the cell directly behind it, and “OE” and “NE” refer to the old and new ends, respectively. (B) Branch formation. Following division, the daughter behind the filament tip can grow monopolarly from its “new” end. More frequently, these daughters grow monopolarly from the “old” end as in panel A. (C) Selected time points from the time-lapse microscopy in Movie S1 in the supplemental material. (D) The position of the filament at each 15-min time point was plotted from Movie S1 in the supplemental material. Through 37.5 h of growth, encompassing nine cell cycles, the filament maintains a stable direction of growth. The straight guidance line represents a distance of 79.5 μm. Bar, 10 μm. (E) Quantitation of the patterns of filamentous growth. From time-lapse microscopy of growing filaments, the 1st and 2nd cells were scored as growing monopolarly from the new or old ends or bipolarly from both. The number of branched cells (branched cells shown in panel B) that were found in each class is shown in parentheses. In some cases, adjacent cells separated to allow them to grow past each other (middle image in panel A). This was classed as bipolar growth without branching. Single cells growing on LNB medium were analyzed in parallel. Time (T) is shown in minutes or hours.
The striking directionality of tip growth is illustrated in Fig. 3C and D and in Movie S1 in the supplemental material. Over a period of 37 h, representing nine cell divisions, the tip maintained an accurate overall direction, with apparent minor deviations from this direction being subsequently corrected.
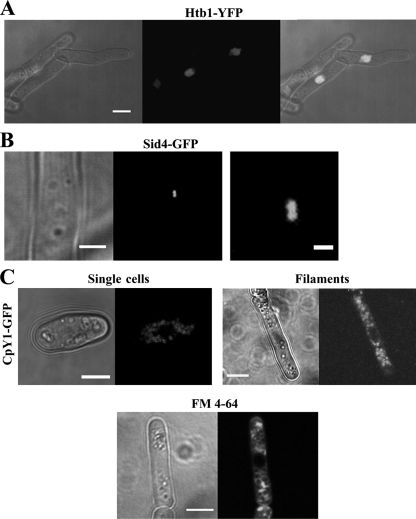
To observe different organelles during mycelial growth, strain A1153 was crossed with strains derived from strain 972 carrying integrated fluorescent markers and efficiently invading progeny selected in each case. Htb1, the equivalent of histone H2B (17), was used to identify nuclei. This showed that all the cells in the filaments were mononucleate (Fig. 4A). To monitor cell cycle progression in the filaments, the spindle pole body (SPB) protein Sid4 (34) was used. This showed duplicated SPBs (Fig. 4B) in all 21 filaments imaged. Since SPB duplication occurs after G1 (4, 35), this indicates that G1, as in single-celled growth, is likely to be relatively short.
Fig. 4.
Cellular organelles in filamentous cells. (A) Filamentous cells stained with nuclear marker Htb1-YFP (YFP stands for yellow fluorescent protein) (strain JA1551). The right panel shows the transmission and fluorescence microscopy images superimposed. The extended filaments are mononucleate. Bar, 5 μm. (B) Filamentous cells stained with Sid4-GFP, a marker for the spindle pole body (SPB) (strain JA1576). The right panel is a higher magnification of the duplicated SPB seen in the left panel. Bars, 2 μm (left panel) and 0.5 μm (right panel). (C) Vacuole morphology in filamentous cells. Filamentous cells were stained with vacuole lumen marker CpY-GFP (strain JA1582) (top) or with vital dye FM 4-64 (bottom), which labels endocytic structures and vacuole membranes. The size of vacuoles appears more irregular in filaments, and they are absent from a region near the tip. All images are single planes. Bars, 5 μm.
Fungal vacuoles can show striking changes during mycelial growth, for example in the fission yeast Schizosaccharomyces japonicus, in which they fuse and congregate at the nongrowing end of the cell at the start of hyphal formation (28). Vacuoles were visualized using both carboxypeptidase Y1 (CpY1) and the endocytic vital dye FM 4-64, which also labels endocytic structures. The vacuoles in filaments were more variable in size than in single cells and appeared to be excluded from a small region at the growing tip (Fig. 4C).
Distribution of polarity-controlling proteins during filamentous growth.
Since the overall pattern of pattern of growth and polarity was clearly altered in filaments, we investigated the localizations of proteins known to control these processes in single cells. First, four proteins related to actin or directly involved in growth, all of which had previously been shown to localize to growing tips, were investigated.
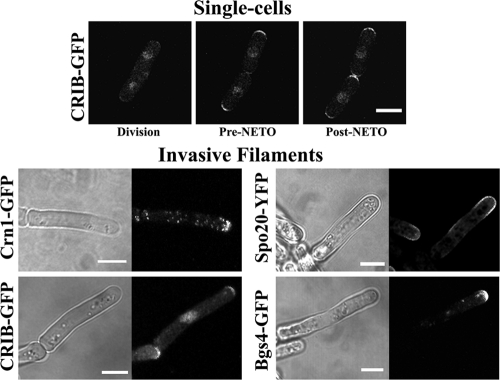
Crn1 (coronin), a marker for F-actin patches (27), the CRIB (Cdc42/Rac interactive binding) domain of Saccharomyces cerevisiae Glc2p that specifically binds the GTP-bound activated form of Cdc42 (24, 31), Spo20, a probable lipid exchange protein thought to be involved in membrane deposition and essential for invasion (5, 22), and Bgs4, a β-glucan synthase involved in cell wall deposition (3), were each observed as GFP fusions within the filaments. All four showed strong enrichment at the growing end of the filament tip (Fig. 5). The same proteins when observed in single cells all exhibited a bipolar distribution following NETO as previously reported (Fig. 5 and data not shown) (3, 22, 27, 31).
Fig. 5.
Four factors required for growth are restricted to the filament tip. The indicated polarity factors are enriched at the growing end of the filaments (strains JA1523, JA1543, JA1577, and JA1545). Single cells are shown at division and before and after new-end take-off (pre-NETO and post-NETO, respectively). Fluorescent images are maximum-intensity stacks; light transmission images are single planes. Bars, 5 μm.
The polarity factor Tea1 in single cells is distributed with roughly equal intensities at both ends throughout the cell cycle (16) (Fig. 6A). In contrast, in the filament tip cells, Tea1 exhibited a strongly monopolar distribution, concentrated at the nongrowing end of the cell (Fig. 6A). This conclusion was supported by quantification of the Tea1-3GFP fluorescence in 29 filament tip cells (Fig. 6B). To investigate whether Tea1 continued to be delivered by microtubules to the growing end in filaments, Tea1-3GFP fluorescence was imaged by time-lapse microscopy (see Movie S2 in the supplemental material). This indicated that Tea1 continued to be delivered to both ends of the tip cell and therefore suggested that its residence time at the growing end is likely to be short.
Fig. 6.
Tea1 abundance at the growing tip is reduced in filamentous cells. (A) Tea1-3GFP distribution in single cells and filaments (strain JA1489). In single cells, Tea1-3GFP is located equally at both tips. In filaments, it is enriched strongly at the nongrowing end. Bars, 5 μm. (B) Quantitation of Tea1-3GFP distribution. Maximum-intensity stacks of Tea1-3GFP fluorescence were collected for both singly growing and filamentous cells of strain JA1489. The graph shows the fluorescence intensity of Tea1-3GFP at the growing end of the filament as a percentage of the intensity at the nongrowing end, plotted as 10% bins. For single cells, the fluorescence from the end with weaker intensity is plotted relative to the intensity from the other end. A total of 29 cells or filaments were examined. (C) Time-lapse imaging of Tea1-3GFP delivery to the tip of a filamentous cell in strain JA1489 (see Movie S2 in the supplemental material). Tea1-3GFP appears to be delivered equally to both ends of the filament. The image shows a maximum-intensity stack taken at 10-s intervals over 480 s. Bar, 5 μm. (D) A strain deleted for Tea1 (strain JA1567) produces filaments that are bent, resulting in aberrant mycelial morphology compared to that of the wild type (see Movie S3 in the supplemental material). Bar, 10 μm.
We previously reported that deletion of tea1 does not reduce the level of invasion but does affect mycelial morphology (5). For a more detailed examination of how tea1 deletion affects mycelial morphology, a tea1-deleted strain was crossed with strain A1153, invasive progeny were selected, and growth of this strain was monitored by time-lapse microscopy (Fig. 6D; see Movie S3 in the supplemental material). The filament tips in this strain grew with a meandering trajectory and lacked the directionality of wild-type cells.
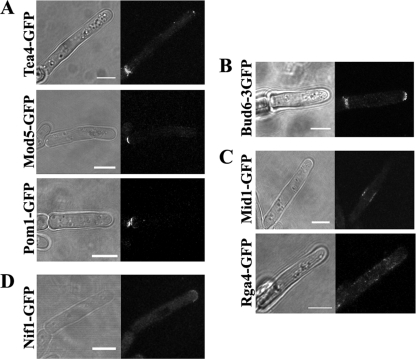
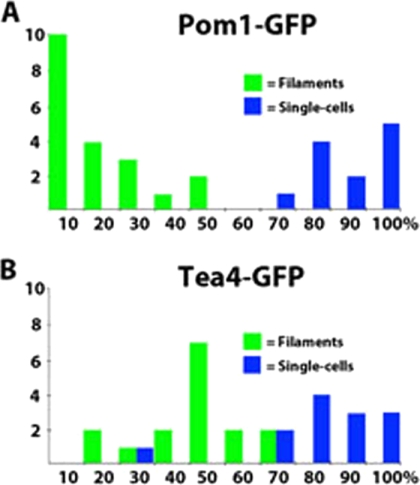
Tea1 either directly or indirectly controls the localization of numerous other polarity factors at the cell tip (12). The localizations of several of these polarity factors were investigated. Tea4 is associated with Tea1 on the microtubule ends and requires it for correct localization at the cell tip (14, 32). Tea4 in filaments showed a similar distribution to Tea1, heavily enriched at the nongrowing end (Fig. 7A and 8). Mod5 is a membrane-anchored protein thought to be required to tether Tea1 to the growing tip, although it also relies on Tea1 for correct localization to the tip (29). In filaments, Mod5, like Tea1, was found to concentrate at the nongrowing end (Fig. 7A). The kinase Pom1 has multiple functions in controlling cell polarity and growth and also requires Tea1 for localization to the cell tip (2). It too showed a strong localization to the nongrowing end of filaments (Fig. 7A and 8). The extent of polarization of both Tea4 and Pom1 was determined as for Tea1 (Fig. 8). This showed that Tea4 polarized to a similar degree as Tea1, consistent with the proteins being in a complex, while Pom1 appeared to show a greater extent of polarized localization. When the imaged strains were grown as single cells, all three proteins, Tea4, Mod5, and Pom1, displayed a bipolar distribution similar to those previously described (data not shown).
Fig. 7.
Distribution of seven other growth and polarity factors in filamentous cells. (A) Tea4-GFP, Mod5-GFP, and Pom1-GFP (from strains JA1542, JA1578, and JA1500, respectively) are enriched strongly only at the nongrowing ends of filamentous cells. (B) Bud6-3GFP (strain JA1476) is localized at both ends of the filament as in single cells. (C) Mid1-GFP and Rga4-GFP (from strains JA1501 and JA1527, respectively) exhibit a pattern of localization in filamentous cells similar to that found in single cells. (D) Nif1-GFP (strain JA1556) is enriched strongly at the growing end of the filamentous cell. Bars, 5 μm.
Fig. 8.
Quantitation of Pom1-GFP (A) and Tea4-GFP (B) distribution, measured as for Tea1 (Fig. 6). Tea4 shows a degree of polarization similar to that for Tea1, while Pom1 appears to be more stringently polarized.
Tea1 and Pom1 are also known to control the localization of proteins at the nongrowing end. The localization of several of these proteins was investigated. Bud6 is thought to be an activator of the For3 formin, releasing it from its self-inhibition (15). Both proteins require Tea1 for correct localization to the nongrowing tip before NETO (6, 7). Bud6 showed a bipolar distribution in filaments, with similar concentrations at both ends of the cell (Fig. 7B). Attempts to image For3, which is essential for invasion (5), were not successful, as introduction of the For3-3GFP marker severely affected filament morphology (data not shown). Rga4 is a GTPase-activating protein (GAP) for Cdc42 and is localized to the cell sides and excluded from both cell ends in single cells; it interacts with, and its localization is regulated by, Pom1 (31). Rga4 in filaments was also excluded from both cell ends (Fig. 7C). Mid1 in single cells is localized as a medial broad band around the nucleus, but in the absence of Pom1, it becomes distributed over the entire nongrowing end (25). Mid1 in filaments was still localized as a medial broad band around the nucleus (Fig. 7C). These results indicate that Pom1 and Tea1 appear to function at the nongrowing end of the tip cell similarly to their inferred roles at the “old” ends of single cells.
Finally, Nif1, an inhibitor of Cdr1 kinase, localizes to the cell poles in a microtubule-independent but actin-dependent manner (13, 36). Nif1 and Pom1 are often enriched at opposite ends of the cell, raising the possibility that Pom1 may act to exclude Nif1 (13). Since Pom1 in filament cells was enriched only at the nongrowing ends, the localization of Nif1 was determined. It was found to be enriched only at the growing end of the filament (Fig. 7D).
DISCUSSION
S. pombe is a long-established model for the organization of polarized unicellular growth. The discovery of an alternative multicellular form, which invades the growth medium, raises the question of how the mechanisms controlling single-celled growth are reorganized to produce different cell shapes and structures. We have described here the characteristics of invasive filamentous growth in S. pombe and associated this behavior with changes in the localizations of a range of key factors known to control orderly polarized growth in this organism.
To record the growth of filaments by microscopy, it was necessary to use an isolate, A1153, which invades more efficiently than the standard laboratory strain 972, or recombinants between the two strains. At the sequence level, the two strains appear to differ by no more than might be typical for geographically separate isolates. In addition, progeny from mating the strains, when grown as single cells, show a growth pattern and localizations of a range of markers that are indistinguishable from those previously reported in strains derived from strain 972. Clearly, it would be of interest to define the specific genetic changes that contribute to more efficient invasion. However, at a minimum, several genes are involved (Fig. 1), making genetic analysis difficult. In principle, it may eventually be possible to pursue these differences by a combination of serial backcrossing and genome sequencing of the progeny.
Concerning the “normal” behavior of S. pombe, it should be noted that the large majority of S. pombe isolates are from, or related to, artificial fermentations. Strain A1153 was isolated from an artisanal production of cachaca in Brazil (8). Although this is described as spontaneous fermentation, in fact, part of each brew is kept and inoculated into the subsequent batch. This, or any other repetitive liquid culturing, might select against the propensity to attach to and invade solid substrates. It might be of interest to investigate strains that are genuinely “wild” and not exposed to human selection. However, it is not clear whether such strains of S. pombe have yet been identified.
Cells at the tips of filaments are significantly elongated but mononuclear, while the extent of elongation diminishes away from the filament tip. There is apparently complete separation between adjacent cells, and hence, they can be described as pseudohyphae rather than true hyphae. Judging by the duplication of the spindle pole body (SPB), the tip cells appear to be predominantly in S or G2, but they do not undergo new-end take-off (NETO), which resumes only in cells behind the tip. The tip grows with strikingly accurate directionality over prolonged periods of time and numbers of divisions, despite the lack of any obvious overall rigid structure to define the direction. It might be possible for the larger mycelial organization to play a role in this directionality, but perhaps it is more likely that external gradients within the medium are required (see below).
Of the proteins involved in polarized growth control that we have surveyed, two groups showed notable changes in intracellular distribution compared to single-celled growth. The actin-binding protein Crn1, the lipid exchange protein Spo20, the glycosyl synthase Bgs4, and GTP-bound Cdc42 are all thought to be involved quite directly in the process of growth and are observed at both ends of single cells after NETO. However, all four strongly localized to the growing ends of filaments. This is consistent with the prolonged growth at that end and lack of NETO. In the second group, in contrast, Tea1, which plays a less direct role in organizing the growth pattern and is not absolutely required for growth, accumulated at the nongrowing end, as did two proteins with which Tea1 interacts, Mod5 and Pom1. However, video microscopy indicated that Tea1 continued to be delivered to both ends of the cell. This suggests that all of these proteins continue to play a role in filaments in defining the site and direction of growth but are no longer required when growth at one end is prolonged. Indeed, cells lacking Tea1 can invade quite efficiently, although the resulting structures are clearly disordered (see Movie S3 in the supplemental material), comparable to the effect of deleting the orthologous gene in the filamentous fungus Aspergillus nidulans (30).
The third group of proteins we investigated could be viewed as connecting Tea1 and Pom1 to different parts of the cell cycle. Bud6, which activates the For3 formin, Rga4, a Cdc42 GAP, the anilin Mid1, and the kinase inhibitor Nif1 all localized in filaments similarly to their behavior in single cells. We were unable to localize For3 itself, as a strain expressing For3-3GFP did not form normal filaments. Indeed, this is the only one of the proteins implicated in control of polarity so far found to be essential for invasive growth (5), suggesting that a specific alteration to the behavior or interactions of For3 may be involved in initiating or maintaining filamentous growth.
Studies of the control of single-celled growth of S. pombe have led to the view that the system comprises mutually reinforcing and partially redundant modules rather than a linear hierarchy (12). Thus, the striking changes in localizations that we have observed do not of themselves appear to identify an obvious control point that determines the change in growth pattern. However, they do emphasize that the system must be sufficiently adaptable to produce different growth patterns in response to conditions. The same point applies to another example of monopolar and elongated growth in S. pombe, namely, the formation of shmoos in response to mating pheromone (23). During shmoo development, as in invasive filaments, the abundance of Tea1 at the growing tip is greatly reduced. However, Tea1 protein is not required for normal shmoo formation, and the growth of shmoos does not continue after formation of the initial structure but is halted until fusion with a mating partner occurs. In addition, shmoos are arrested in G1, whereas filamentous cells appear to be mostly in G2.
Another likely parallel between these two states may be their dependence on external factors to determine cell shape. In the case of shmoos, this is pheromone. In filaments, high cell density is essential, while a combination of low levels of nitrogen and high levels of carbon increases the efficiency of the process. Both of these might result in formation of gradients in the medium, either of nutrients or of signals released from the cells, which could trigger invasion or contribute to the linearity of filament growth. Clearly, a major challenge now is to identify the components of these signaling processes and how they interact with the machinery controlling polarity to produce the growth patterns we have described.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following for strains and plasmids: Fred Chang, Kazuhiro Shiozaki, Phong Tran, James Moseley, Sophie Martin, Kathleen Gould, Hiraoka Yashusi, Minoru Yoshida, Bioneer, The Yeast Genetic Resource Centre, Riken BRC, and the late Neil Bone. We are grateful to Iain Hagan for advice. The genome sequence of S. pombe NCYC132 was generated by the Broad Institute.
This work was supported by a studentship from the BBSRC to J. Dodgson and Medical Research Council grant no. G0400062.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 24 September 2010.
REFERENCES
- 1.Amoah-Buahin E., Bone N., Armstrong J. 2005. Hyphal growth in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 4:1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahler J., Pringle J. R. 1998. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 12:1356–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortes J. C. G., Konomi M., Martins I. M., Munoz J., Moreno M. B., Osumi M., Duran A., Ribas J. C. 2007. The (1,3)beta-D-glucan synthase subunit Bgs1p is responsible for the fission yeast primary septum formation. Mol. Microbiol. 65:201–217 [DOI] [PubMed] [Google Scholar]
- 4.Ding R., West R. R., Morphew M., Oakley B. R., McIntosh J. R. 1997. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell 8:1461–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodgson J., Avula H., Hoe K. L., Kim D. U., Park H. O., Hayles J., Armstrong J. 2009. Functional genomics of adhesion, invasion, and mycelial formation in Schizosaccharomyces pombe. Eukaryot. Cell 8:1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feierbach B., Verde F., Chang F. 2004. Regulation of a formin complex by the microtubule plus end protein tea1p. J. Cell Biol. 165:697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glynn J. M., Lustig R. J., Berlin A., Chang F. 2001. Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr. Biol. 11:836–845 [DOI] [PubMed] [Google Scholar]
- 8.Gomes F. C. O., Pataro C., Guerra J. B., Neves M. J., Correa S. R., Moreira E. S. A., Rosa C. A. 2002. Physiological diversity and trehalose accumulation in Schizosaccharomyces pombe strains isolated from spontaneous fermentations during the production of the artisanal Brazilian cachaca. Can. J. Microbiol. 48:399–406 [DOI] [PubMed] [Google Scholar]
- 9.Kanter-Smoler G., Dahlkvist A., Sunnerhagen P. 1994. Improved method for rapid transformation of intact Schizosaccharomyces pombe cells. Biotechniques 16:798–800 [PubMed] [Google Scholar]
- 10.Kumamoto C. A., Vinces M. D. 2005. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 7:1546–1554 [DOI] [PubMed] [Google Scholar]
- 11.Leupold U. 1993. The origins of Schizosaccharomyces pombe genetics, p. 125–128 InHall M. N., Linder P. (ed.), The early days of yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 12.Martin S. G. 2009. Microtubule-dependent cell morphogenesis in the fission yeast. Trends Cell Biol. 19:447–454 [DOI] [PubMed] [Google Scholar]
- 13.Martin S. G., Berthelot-Grosjean M. 2009. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature 459:852–856 [DOI] [PubMed] [Google Scholar]
- 14.Martin S. G., McDonald W. H., Yates J. R., Chang F. 2005. Tea4p links microtubule plus ends with the formin For3p in the establishment of cell polarity. Dev. Cell 8:479–491 [DOI] [PubMed] [Google Scholar]
- 15.Martin S. G., Rincon S. A., Basu R., Perez P., Chang F. 2007. Regulation of the formin for3p by cdc42p and bud6p. Mol. Biol. Cell 18:4155–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mata J., Nurse P. 1997. Tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89:939–949 [DOI] [PubMed] [Google Scholar]
- 17.Matsuyama A., Arai R., Yashiroda Y., Shirai A., Kamata A., Sekido S., Kobayashi Y., Hashimoto A., Hamamoto M., Hiraoka Y., Horinouchi S., Yoshida M. 2006. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24:841–847 [DOI] [PubMed] [Google Scholar]
- 18.Minc N., Bratman S. V., Basu R., Chang F. 2009. Establishing new sites of polarization by microtubules. Curr. Biol. 19:83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreno S., Klar A., Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 [DOI] [PubMed] [Google Scholar]
- 20.Moseley J. B., Mayeux A., Paoletti A., Nurse P. 2009. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature 459:857–860 [DOI] [PubMed] [Google Scholar]
- 21.Nadal M., Garcia-Pedrajas M. D., Gold S. E. 2008. Dimorphism in fungal plant pathogens. FEMS Microbiol. Lett. 284:127–134 [DOI] [PubMed] [Google Scholar]
- 22.Nakase Y., Nakamura T., Hirata A., Routt S. M., Skinner H. B., Bankaitis V. A., Shimoda C. 2001. The Schizosaccharomyces pombe spo20(+) gene encoding a homologue of Saccharomyces cerevisiae Sec14 plays an important role in forespore membrane formation. Mol. Biol. Cell 12:901–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niccoli T., Nurse P. 2002. Different mechanisms of cell polarisation in vegetative and shmooing growth in fission yeast. J. Cell Sci. 115:1651–1662 [DOI] [PubMed] [Google Scholar]
- 24.Ozbudak E. M., Becskei A., van Oudenaarden A. 2005. A system of counteracting feedback loops regulates Cdc42p activity during spontaneous cell polarization. Dev. Cell 9:565–571 [DOI] [PubMed] [Google Scholar]
- 25.Padte N. N., Martin S. G., Howard M., Chang F. 2006. The cell-end factor pom1p inhibits mid1p in specification of the cell division plane in fission yeast. Curr. Biol. 16:2480–2487 [DOI] [PubMed] [Google Scholar]
- 26.Paoletti A., Chang F. 2000. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol. Biol. Cell 11:2757–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelham R. J., Chang F. 2001. Role of actin polymerization and actin cables in actin-patch movement in Schizosaccharomyces pombe. Nat. Cell Biol. 3:235–244 [DOI] [PubMed] [Google Scholar]
- 28.Sipiczki M., Takeo K., Grallert A. 1998. Growth polarity transitions in a dimorphic fission yeast. Microbiology 144:3475–3485 [DOI] [PubMed] [Google Scholar]
- 29.Snaith H. A., Sawin K. E. 2003. Fission yeast mod5p regulates polarized growth through anchoring of tea1p at cell tips. Nature 423:647–651 [DOI] [PubMed] [Google Scholar]
- 30.Takeshita N., Higashitsuji Y., Konzack S., Fischer R. 2008. Apical sterol-rich membranes are essential for localizing cell end markers that determine growth directionality in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 19:339–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatebe H., Nakano K., Maximo R., Shiozaki K. 2008. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr. Biol. 18:322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tatebe H., Shimada K., Uzawa S., Morigasaki S., Shiozaki K. 2005. Wsh(3)/Tea4 is a novel cell-end factor essential for bipolar distribution of Tea1 and protects cell polarity under environmental stress in S. pombe. Curr. Biol. 15:1006–1015 [DOI] [PubMed] [Google Scholar]
- 33.Terenna C. R., Makushok T., Velve-Casquillas G., Baigl D., Chen Y., Bornens M., Paoletti A., Piel M., Tran P. T. 2008. Physical mechanisms redirecting cell polarity and cell shape in fission yeast. Curr. Biol. 18:1748–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomlin G. C., Morrell J. L., Gould K. L. 2002. The spindle pole body protein Cdc11p links Sid4p to the fission yeast septation initiation network. Mol. Biol. Cell 13:1203–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uzawa S., Li F., Jin Y., McDonald K. L., Braunfeld M. B., Agard D. A., Cande W. Z. 2004. Spindle pole body duplication in fission yeast occurs at the G1/S boundary but maturation is blocked until exit from S by an event downstream of cdc10+. Mol. Biol. Cell 15:5219–5230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L., Russell P. 1997. Nif1, a novel mitotic inhibitor in Schizosaccharomyces pombe. EMBO J. 16:1342–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.