Abstract
Eukaryotic cells employ a variety of mechanisms to maintain protein quality control and homeostasis. Here we provide evidence that one such mechanism in Saccharomyces cerevisiae involves the regulated release of excess or misfolded proteins to the extracellular space. The overexpression of an epitope-tagged allele of the glycosylphosphatidylinositol (GPI)-linked cell wall protein Utr2/Crh2p (Utr2/Crh2-green fluorescent protein [GFP] or -hemagglutinin [HA]) causes endoplasmic reticulum (ER) stress and the secretion of Crh2-GFP/HA into the extracellular space. Secretion is dependent on two GPI-linked aspartyl proteases (Yps1p/2p) and components of the unfolded protein response (Ire1p and Hac1p) but is independent of ER-associated degradation (ERAD) components such as Hrd1p and Doa10p. Supporting the idea that this process represents a mechanism for protein quality control, the level of Crh2-HA is increased in strains lacking Bst1p, a protein required for the proteasomal degradation of GPI-linked proteins. Furthermore, secretion is dependent on Sec18p, indicating that it requires ER-to-Golgi trafficking, and accordingly, Crh2-HA accumulates in the ER in ire1Δ and bst1Δ mutants by cycloheximide chase experiments. Since a fraction of Utr2/Crh2-GFP properly localizes to the cell wall in an ire1Δ mutant, extracellular secretion appears to occur through a pathway that is distinct from the normal GPI protein-trafficking pathway. Taken together, these data support a model in which the unfolded protein response (UPR)/yapsin-mediated extracellular release of overexpressed Utr2/Crh2-HA or -GFP is an alternative pathway for the removal of excess or misfolded secretory proteins functioning in parallel with proteasome-mediated degradation in S. cerevisiae. This model provides an explanation for the deleterious effects of Yps1/2p on the industrial production of some recombinant proteins in S. cerevisiae.
The identification and processing of misfolded or misprocessed proteins are crucial components of eukaryotic cell homeostasis (30). In addition to their fundamental importance to eukaryotic cell biology, the mechanisms of protein quality control are of keen interest because a variety of human diseases are linked to the accumulation of misfolded proteins (21). The model organism Saccharomyces cerevisiae has played an important role in the delineation of two of the most important mechanisms through which eukaryotic cells manage misfolded proteins (31): endoplasmic reticulum (ER)-associated degradation (ERAD) and the unfolded protein response (UPR).
Through ERAD, proteins resistant to chaperone-mediated refolding are identified, retrotranslocated from the ER, tagged with ubiquitin, and, ultimately, degraded by the 26S proteasome (39). ERAD is constitutively active and, during unstressed vegetative growth, appears sufficient to process the load of misfolded proteins in yeast. Our current understanding of ERAD indicates that there are three different ERAD subtypes based on whether the misfolded lesion is in the luminal region of a protein (ERAD-L), the membrane region (ERAD-M), or the cytosolic region (ERAD-C) (16). ERAD-L is mediated by the Hrd1p complex, while ERAD-C is mediated by the Doa10 complex. ERAD-M is the most recently described subtype and appears to be dependent on Hr1p and Hrd3p but independent of other components of the Hrd1p ERAD-L complex (8).
When the cell encounters conditions that increase levels of unfolded proteins, a second mechanism, called the UPR, is activated to compensate for elevated levels of ER stress (28). The UPR is an ER-to-nucleus signaling pathway that is initiated by ER stress and induces the transcription of a large number of genes (36). In yeast, the UPR is triggered when unfolded proteins are detected by the transmembrane sensor Ire1p. Ire1p contains protein kinase and endoribonuclease activities that are essential to its role in UPR (11, 22). Ire1p oligomerizes in the presence of unfolded proteins and undergoes autophosphorylation, which in turn activates its RNase activity (34). Ire1p RNase activity is specific for the mRNA of the transcription factor Hac1p, its only known substrate. In yeast, HAC1u mRNA (“u” for “uninduced”) is constitutively transcribed but is not translated due to the presence of an inhibitory intron. Activated Ire1p removes the intron from HAC1u, and tRNA ligase rejoins the two exons to generate HAC1i (“i” for “induced”). HAC1i is then efficiently translated, and the resulting Hac1p transcription factor translocates to the nucleus, where it initiates the transcriptional program of the UPR (36). In higher eukaryotes, two additional pathways (PERK and ATF) also mediate the activation of the UPR (28).
Although the UPR and ERAD represent the two best-studied processes by which cells compensate for the development of ER stress, it is clear that more complex and specific mechanisms underlie these general schemes (30). For example, Fujita et al. recently demonstrated that an important class of membrane and yeast cell wall proteins, glycosylphosphatidylinositol (GPI)-linked proteins, is targeted to the proteasome through a pathway distinct from that of traditional ERAD (13). The degradation of a misfolded form of the canonical yeast GPI-linked protein Gas1p is dependent on the inositol-deacetylase Bst1p but is independent of Hrd1p and Doa10p. This alternative pathway of degradation is consistent with the fact that the trafficking of properly folded GPI proteins to the cell surface is also distinct from that of other secretory cargo (9).
In this report, we describe experiments indicating that aberrant or excess GPI-linked proteins are also removed by secretion to the extracellular space in a process that is dependent on both the UPR and two members of the yapsin family of GPI-linked aspartyl proteases, Yps1p and Yps2p (15). The yapsins are the founding members of a family of GPI-linked yeast aspartyl proteases that are present in many fungi, including pathogenic yeasts such as Candida albicans (1) and Candida glabrata (18) and industrially important yeasts such as Pichia pastoris (41). Although the physiological roles of the yapsins are just beginning to be understood, this family of proteases appears to function as secretases that release membrane and cell wall-localized proteins from the cell surface (14, 15, 20, 38). Our results are consistent with this general function and further suggest that the yapsins play a role in protein quality control. Taken together, the experiments described below provide evidence for a novel mechanism by which at least some misfolded or excess GPI proteins are removed by yeast and provide additional insights into the function of the yapsin family of yeast aspartyl proteases.
MATERIALS AND METHODS
Strains, growth conditions, and materials.
Yeast strains used in this study are listed in Table 1 and were derived from either the S288C (BY4741) or W303 (CRY1) genetic background. Single yeast mutants in the BY4741 background were obtained from a yeast deletion collection (Invitrogen, Carlsbad, CA). Yeast strains were grown in yeast-peptone-dextrose (YPD) or synthetic dropout medium prepared according to standard recipes (6). All incubations were performed at 30°C unless noted otherwise. Escherichia coli DH5α cells (Invitrogen, Carlsbad, CA) were used for routine plasmid manipulations. All chemicals were obtained from Sigma (St. Louis, MO) and used as received. Oligonucleotides were synthesized by IDT (Coralville, IA).
Table 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| CRY1 (W303-1A) | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 | R. Fuller Laboratory Collection |
| HKY20 | CRY1 yps1Δ::LEU2 | 19 |
| HKY21 | CRY1 yps2Δ::HIS3 | 19 |
| DKY5 | CRY1 yps3Δ::KAN | 20 |
| DKY9 | CRY1 yps6Δ::KAN | 20 |
| DKY13 | CRY1 yps7Δ::KAN | 20 |
| HKY24 | HKY20 yps2Δ::HIS3 | 19 |
| DKY39 (5ypsΔ) | HKY24 yps3Δ::KAN yps6Δ::KAN yps7Δ::KAN | 20 |
| DKY104 | W303-1A ire1Δ::TRP1 | 35 |
| DKY103 | W303-1A leu2-3,112::LEU2- GAL-CPY*-HA | 35 |
| DKY101 | DKY103 ire1Δ::TRP1 | 35 |
| ESY39 | W303-1A his3-11::HIS3- UPRE-lacZ | 35 |
| BY4741 | MATahis3Δleu2Δ met15Δ ura3Δ | Research Genetics |
| DKY1000 | BY4741 ire1Δ::KAN | Research Genetics |
| DKY147 | BY4741 ire1Δ::NAT | 32 |
| DKY1012 | BY4741 hac1Δ::KAN | Research Genetics |
| DKY1013 | BY4741 bst1Δ::KAN | Research Genetics |
| DKY1002 | BY4741 hrd1Δ::KAN | 32 |
| DKY1003 | BY4741 doa10Δ::KAN | 32 |
| DKY1014 | BY4741 erv25Δ::KAN | 32 |
| DKY1015 | BY4741 emp24Δ::KAN | 32 |
| DKY1016 | BY4741 htm1Δ::KAN | 32 |
| DKY1017 | DKY147 bst1Δ::KAN | This study |
| CWS18-10 | W303-1A sec18-1 | R. Fuller Laboratory Collection |
Yeast strain construction.
The IRE1 open reading frame was deleted by standard PCR-based one-step gene replacement with a KANMX cassette generated from the deletion collection ire1Δ::KAN strain by using standard procedures (17). Correct integration was determined by PCR with primers flanking the sequence used to generate the knockout cassette. All ire1Δ mutants displayed the expected hypersensitivity to 2-deoxyglucose (2). The KANMX cassette was switched to the NATMX cassette according to a previously reported procedure (37).
Plasmids.
Multicopy plasmids containing CRH1-green fluorescent protein (GFP) (JV89G), CRH2-GFP (pJV40G), or CRH2-hemagglutinin (HA) (pNBC13) expressed from endogenous promoters were generously provided by J. M. Rodriquez-Pena (7, 27). pAR213 containing the CWP1-GFP fusion was a gift from F. Klis (24). prYPS1-lacZ contains the promoter region (665 bp upstream of ATG) fused to lacZ in Yep375R (20).
Microscopy.
All microscopy was performed by using logarithmic-phase cultures in synthetic medium. Cells were harvested and washed with fresh medium, and an aliquot was mixed with 0.8% low-melt agar on the microscope slide to mount. Images were obtained with a Nikon Eclipse 300 epifluorescence microscope with a Cool Snap charge-coupled-device (CCD) camera and processed with PhotoShop software. Vacuole staining was performed by using CellTrackerBlue (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions (26).
β-Galactosidase reporter assay.
Cultures of yeast strains harboring lacZ reporters grown overnight were diluted to an optical density at 600 nm (OD600) of 0.1 in YPD medium and grown for 2 doublings (∼3 h). The cultures were exposed to a stressor for 5 h and harvested, and the pellets were flash-frozen in liquid nitrogen. Cell lysates were prepared and β-galactosidase activity was determined as described previously (6). The specific activity for the extracts is expressed as Miller units (nmol/mg protein/min). Each reported value represents the mean of data from at least two experiments, with independent transformants of the strain performed in duplicate or triplicate. The error is expressed as the standard error of the mean (SEM).
Colony immunoblotting.
Cells from freshly streaked plates (2 days) were patched onto agar plates (YPD or appropriate dropout medium), or a 10-fold dilution series (initial OD600 of 1) of a culture grown overnight was plated onto agar plates by using a pronging tool (VP Scientific, San Diego, CA). The plates were allowed to dry for 30 min, and the cells were overlaid with a nitrocellulose membrane (Amersham). After 24 h of incubation at 30°C, the membrane was removed, washed under a stream of distilled water, and incubated in blocking suspension (5% nonfat milk in 50 mM Tris [pH 7.5], 150 mM NaCl, and 0.05% Tween 20 [TBST]) for 1 h with rocking. The membranes were then incubated at room temperature for 3 h in a fresh block suspension containing anti-HA (1:5,000) (12CA5; Roche), anti-GFP (1:5,000) (Living Colors; Clontech), or anti-Kar2p (1:500) (Santa Clara) antibodies. The TBST-washed membranes were then incubated with a 1:5,000 dilution of horseradish peroxidase-conjugated anti-mouse secondary antibody (Amersham) in block suspension, washed with TBST, and developed for chemiluminescence with an ECL-Plus detection kit (Amersham).
Western blotting.
Cultures grown overnight were diluted to an OD600 of 0.1, grown to logarithmic phase, harvested, and flash-frozen. Medium proteins were precipitated by using trichloroacetic acid. Cell extracts (25 to 50 OD600 equivalents) were prepared by glass bead lysis in a lysis buffer consisting of 150 mM NaCl, 50 mM Tris (pH 7.5), and 5 mM EDTA containing a cocktail of protease inhibitors (Mini-Complete protease inhibitors; Roche) using a BeadBeater (7 cycles of 20 s of vortexing and 60 s of incubation on ice). Extracts were normalized by the protein concentration (Bradford assay; Bio-Rad), fractionated by SDS-PAGE (10% gel), and transferred onto nitrocellulose membranes. The membranes were then blocked overnight at 4°C and processed as described above for the colony immunoblotting.
Cycloheximide chase experiments.
Logarithmic-phase cultures were treated with cycloheximide (50 μg/ml, final concentration), and samples (10 OD600 equivalents) were removed at 0, 30, 60, and 90 min (35). The cells were harvested, flash-frozen, and analyzed by Western blotting as described above. Degradation rates were quantified by densitometry using time point zero as 100%. The means for each time point are based on data from three independent experiments, and error bars are standard deviations.
RESULTS
Utr2/Crh2-GFP is mislocalized in yapsin mutants.
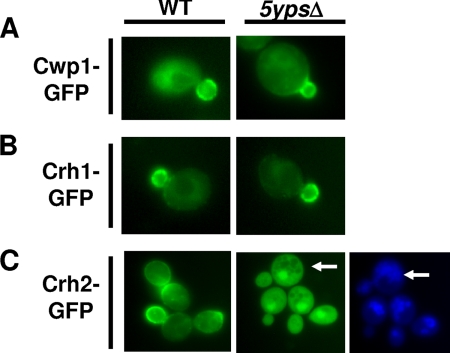
Recently, three substrates have been identified for the S. cerevisiae aspartyl protease Yps1: Msb2p, a signaling mucin (38); Pir4p, a cell wall-linked structural protein (14); and Gas1p, a GPI-linked protein involved in glucan remodeling (14). Since the yapsins play an important, but incompletely understood, role in maintaining yeast cell wall integrity (14, 15, 20), we were interested in determining if they process other cell wall-related GPI proteins. Toward that end, we examined the localization of GFP fusion constructs of the cell wall-related GPI proteins Cwp1p (24), Crh1p (27), and Utr2/Crh2p (27) in the quintuple yapsin mutant 5ypsΔ using alleles expressed from multicopy plasmids under the control of its endogenous promoter. The localization of Cwp1p (Fig. 1A) and Crh1p (Fig. 1B) was unaffected by the deletion of the yapsins and corresponded to previously reported localization patterns (24, 27). Crh2p, on the other hand, clearly accumulated within the cell in a compartment that is distinct from the vacuole (Fig. 1C), with a significant portion of the signal distributed throughout the cytosol (although UTR2 is the Saccharomyces Genome Database [SGD] preferred name for this gene, Crh2p has been more commonly used in recent literature, and we will use this designation for the remainder of the manuscript). The concentrated signal does not colocalize with the nucleus (data not shown) and is suggestive of the class E compartment of endocytosis mutants (25). However, we have not further characterized the specific nature of this body.
Fig. 1.
Crh2-GFP is mislocalized in the 5ypsΔ quintuple yapsin mutant. wt and 5ypsΔ cells expressing the indicated GFP fusion proteins from multicopy plasmids under endogenous promoters were grown to logarithmic phase in synthetic dextrose medium lacking uracil, harvested, and analyzed by fluorescence microscopy as described in Materials and Methods. (A) CWP1-GFP. (B) CRH1-GFP. (C) CRH2-GFP. In C, the cells were also stained with the vacuolar marker CellTracker Blue (Invitrogen) (26). Arrows indicate areas where the GFP and vacuolar stainings do not colocalize.
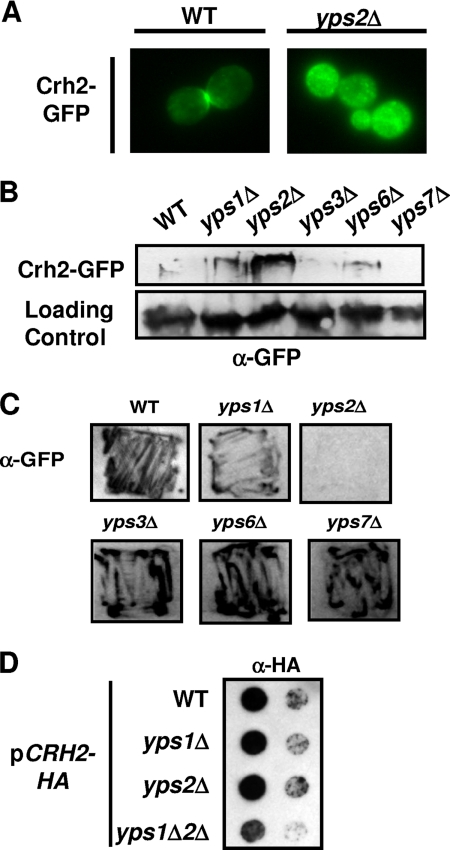
Of the five yapsin single mutants, only yps2Δ showed intracellular accumulation, while the remaining mutants displayed a localization indistinguishable from that of wild-type (wt) cells (Fig. 2A and data not shown). The punctate structures marked by Crh2-GFP resemble those described previously for the Golgi apparatus (12); however, we have not further characterized the intracellular compartment represented by these structures, and thus, the identity of these structures remains speculative. Consistent with this result, Western blot analysis showed increased amounts of intracellular Crh2-GFP in the yps2Δ mutant compared to the wild type (Fig. 2B).
Fig. 2.
Yps1p and Yps2p mediate extracellular secretion of Crh2-GFP/HA. (A) Deletion of YPS2 is sufficient to cause intracellular accumulation of Crh2-GFP. (B) The indicated strains expressing Crh2-GFP were grown to logarithmic phase, harvested, and analyzed by immunoblotting as described in Materials and Methods. A high-molecular-mass protein (>250 kDa) accumulates in yps2Δ and, to a lesser extent, yps1Δ and yps6Δ mutants. (C) Crh2-GFP is secreted to the extracellular medium in a Yps2p-dependent fashion. The indicated strains were patched on agar plates, overlain with a nitrocellulose membrane, and incubated overnight at 30°C. The washed membrane was then probed with anti-GFP antibodies as described in Materials and Methods. (D) Crh2-HA secretion is dependent on both Yps1p and Yps2p.
Since the yapsins were shown previously to mediate the release of cell surface proteins (14, 38), we hypothesized that the intracellular accumulation of Crh2p in yps2Δ mutants may be due to a decreased extracellular release. To test this hypothesis, we used a colony immunoblot assay to determine if Crh2-GFP was secreted to the extracellular space and to assess the effect of the yapsins on secretion. As shown in Fig. 2C, Crh2-GFP is released to the extracellular space. In addition, a null mutation of YPS2 blocks this secretion, supporting our hypothesis that the accumulation of Crh2-GFP in a yps2Δ mutant is due to decreased extracellular secretion. Interestingly, neither Cwp1-GFP nor Crh1-GFP was secreted to the extracellular space in either wild-type cells or yapsin mutants (L. DiDone and D. J. Krysan, unpublished data), suggesting that the yapsins may not process all GPI-linked proteins.
Although the secretion of Crh2-GFP into the growth medium could be part of its normal turnover, there are a number of caveats that must be considered when interpreting this result. First, cell wall proteins are thought to turn over relatively slowly, and the extent of the secretion seems inconsistent with that notion. Second, the Crh2-GFP fusion used in these experiments necessarily contains the GFP moiety within the coding region of the protein because the N and C termini of GPI-linked proteins are removed during processing. This raises the possibility that the processing or folding of the protein could be altered by the presence of the GFP moiety. Indeed, Cabib et al. reported previously that Crh2-GFP appears to be unstable at 37°C (7; DiDone and Krysan, unpublished). To further characterize the secretion of Crh2-GFP, we next examined the secretion of a CRH2 allele containing the smaller HA epitope tag in the same position (7). As shown in Fig. 2D, Crh2-HA was still secreted into the medium, but its secretion was now dependent on both Yps1p and Yps2p. We also examined the level of secretion of the protein expressed from centromeric plasmids and found it to be dramatically reduced for both GFP and HA alleles (data not shown). This finding suggests that the secretion could be due to the fact that the overexpression of CRH2-HA/GFP saturates the capacity of the trafficking pathway; however, we could barely detect the protein in cellular extracts and therefore cannot rule out the possibility that small amounts of CRH2-HA/GFP are secreted from low-copy-number plasmids as well.
Overexpression of CRH2-HA/GFP induces ER stress.
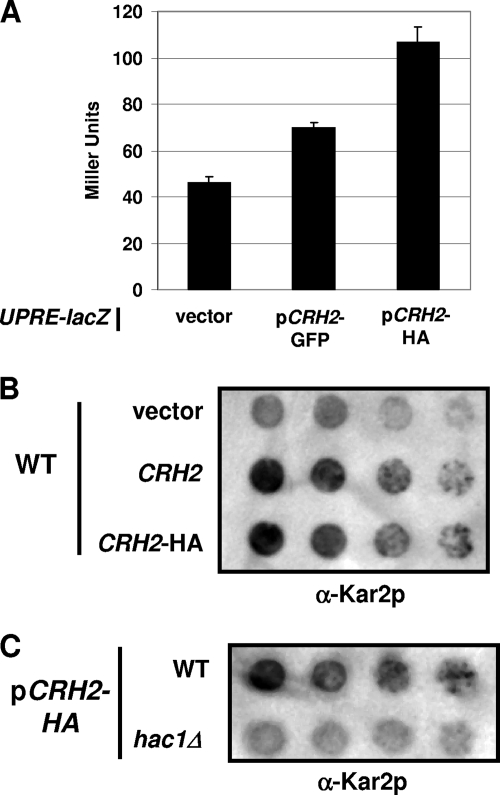
To further explore the possibility that the secretion of Crh2-HA/GFP may be a result of misfolding or saturation of the GPI-trafficking capacity of the cell, we introduced the constructs into a strain containing a UPR reporter in the form of the UPR response element (UPRE) fused to the lacZ gene at the HIS3 locus (35). As shown in Fig. 3A, the expression of both GFP- and HA-tagged alleles activates the UPR reporter compared to the reporter strain containing an empty vector; somewhat surprisingly, the HA allele induced 2-fold-more activity than did the GFP allele. As a comparison, the extensively characterized misfolded allele of carboxypeptidase Y (CPY*) induces a similar level of UPR reporter activity when expressed from its endogenous promoter on a multicopy plasmid (33).
Fig. 3.
Crh2-GFP/HA induces ER stress. (A) CRH2-GFP, CRH2-HA, and the empty vector were expressed in a strain containing a chromosomally integrated fusion of the unfolded response element (UPRE) with the lacZ reporter. Extracts were prepared from logarithmic-phase cells cultivated at 30°C and assayed for β-galactosidase activity as described in Materials and Methods. Graph bars represent the mean Miller units (nmol/mg protein/min) from three independent experiments performed in duplicate, and the error bars indicate the standard errors of the means. (B) A 10-fold dilution series of wild-type cells harboring the indicated plasmids was spotted onto agar plates, incubated overnight, and assayed by colony immunoblotting using anti-Kar2p antibodies as described in Materials and Methods. (C) WT and hac1Δ cells containing the CRH2-HA plasmid were assayed for Kar2p secretion as described in B.
Another consequence of ER stress is the secretion of the ER-resident protein Kar2p (3) into the growth medium. Therefore, we tested whether CRH2-HA expression induced Kar2p secretion. Indeed, Kar2p is released into the growth medium, as determined by colony immunoblotting (Fig. 3B). To assess whether the presence of the epitope tag alone was responsible for the ER stress generated by the construct, we also expressed the untagged allele in the same vector. Since Kar2p secretion was induced by the overexpression of both tagged and untagged alleles of CRH2, it appears that a significant portion of the ER stress generated by these constructs is due to increased levels of CRH2 expression through the multicopy vector; however, we cannot rule out the possibility that the presence of the tag contributes additional stress by inducing a fraction of the protein to be misfolded.
ER-stress-induced Kar2p secretion is dependent upon an intact UPR in yeast (3), and if CRH2-HA-induced Kar2p secretion was the result of ER stress, it should be blocked in a UPR mutant. To test this hypothesis, we expressed CRH2-HA in a hac1Δ mutant and compared Kar2p secretion with that of the wild type. Indeed, Kar2p secretion was reduced in the hac1Δ mutant compared to the wild type (Fig. 3C). This observation further supports the idea that CRH2 expression from a multicopy plasmid induces ER stress. Finally, it is important to note that since the CRH2-HA constructs complement crh2Δ mutants (7) and CRH2-GFP displays proper localization patterns in wild-type cells (27), there appears to be a significant portion of the protein that is functional.
Secretion of Crh2-HA is dependent on Sec18p.
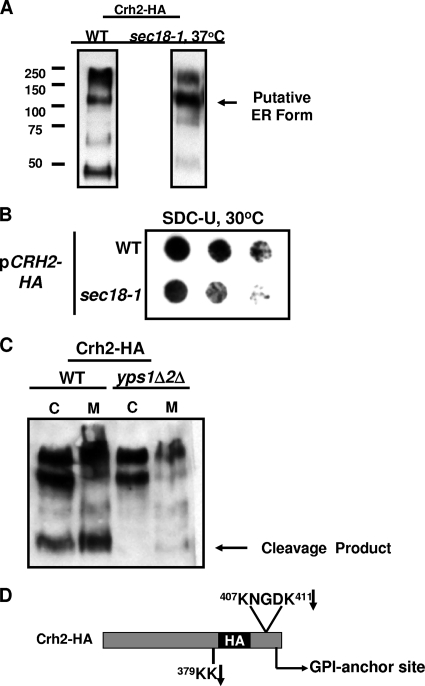
To characterize the intracellular processing of Crh2-HA, we examined cellular extracts from wild-type cells harboring CRH2-HA by Western blotting (Fig. 4A). The presence of multiple molecular species in these blots suggests either that Crh2-HA is undergoing degradation or that immature, partially processed intermediates of Crh2-HA are accumulating. The high-molecular-mass species corresponds to the typical size of a fully mature GPI protein (13). The intermediate-mass band (∼110 kDa) is also larger than the mass predicted from the peptide sequence of Crh2p (50 kDa) and may be a partially processed immature form. The lower-molecular-mass band, on the other hand, is smaller than the mass corresponding to the amino acid sequence and therefore is likely to be a degradation product.
Fig. 4.
Identification of the Chr2-HA ER form and yapsin cleavage products. (A) Western blot analysis of intracellular extracts of WT cells incubated at 30°C and sec18-1 cells after shift to the restrictive temperature of 37°C. (B) Colony immunoblot analysis of Crh2-HA secretion in wt and sec18-1 cells incubated at 30°C and processed as described in the legend of Fig. 1C. (C) wt and yps1Δ yps2Δ cells harboring CRH2-HA were grown to logarithmic phase in synthetic medium. The cells were harvested, and total cell extracts (C) were prepared. Medium proteins (M) were precipitated with trichloroacetic acid-deoxycholate. Samples were analyzed by Western blotting as described in Materials and Methods. Medium proteins corresponding to equivalent numbers of cells were loaded into each lane. Cellular proteins were normalized by protein concentration. (D) Schematic representation of Crh2-HA with the sequences of Yps1/2p cleavage sites that are consistent with the 40- to 50-kDa Western blot band are indicated. The arrows indicate the residue where cleavage is predicted.
To further characterize the multiple molecular species observed in the immunoblot, CRH2-HA was expressed in the sec18-1 strain, a strain containing a temperature-sensitive allele of the ER AAA-ATPase SEC18, and analyzed by immunoblotting. Sec18p is required for ER-to-Golgi transport, and a shift of this strain to the restrictive temperature causes a block in secretory trafficking at the ER (4). As shown in Fig. 4A, the intermediate-molecular-weight species accumulates in the cell, while the higher- and lower-molecular-weight species are depleted. This strongly suggests that the intermediate-molecular-weight species represents the partially processed “ER form” of Crh2-HA and that secretion may be dependent on ER-to-Golgi transport. Consistent with this notion, decreased amounts of Crh2-HA are secreted from sec18-1 cells incubated at the permissive temperature (Fig. 4B). Since the ER forms of GPI proteins are typically not observed at steady state or are present at very low levels (13), the prominence of the ER form of Crh2p in wild-type cells (Fig. 4A) suggests that its overexpression may saturate the GPI protein-trafficking capacity of the cell and thereby cause ER stress.
Finally, the secretory block induced by the loss of Sec18p function also prevents the formation of the lower-molecular-weight species detected in wild-type cells. Since Yps1p and Yps2p are involved in the secretion of Crh2-HA, it seemed likely that this low-molecular-weight species is a yapsin-derived cleavage product. Consistent with this hypothesis, the lower-molecular-weight band is absent when CRH2-HA is expressed in the yps1Δ yps2Δ yapsin double mutant (Fig. 4C). This observation also indicates that the yapsin-mediated processing of secreted Crh2-HA occurs after transport from the ER and is consistent with previous results suggesting that yapsin processing occurs at the cell surface (14, 19, 38).
The HA tag is between residues 383 and 384, near the C terminus. The cleavage fragment is approximately 40 to 45 kb. Based on previous work characterizing the cleavage sites of Yps1p and Yps2p (14, 15), two likely cleavage sites are present in the C-terminal region of Crh2p and are indicated in Fig. 4D. Processing at both sites would lead to the small fragment, while cleavage only at the pair of lysines in the S/T region could account for the larger fragments. These sites correspond closely to the position of Yps1 cleavage sites in Gas1p and Yps1p itself, identified previously by Gagnon-Arsenault et al. (14). There are many other basic residues N terminal to the tag that could also represent sites of cleavage but that would not be apparent, since they would not retain the epitope tag. Therefore, we cannot rule out that other cleavage products are also generated.
Consistent with the colony immunoblot shown in Fig. 2D, there is some Crh2-HA that is secreted into the medium in the yps1Δ yps2Δ mutant, indicating that the results of the immunoblot assay correlate well with Western blotting. In addition, these results suggests that although Yps1 and Yps2p are the predominant proteases involved in the release of Crh2-HA, other members of the yapsins may participate; alternatively, we cannot rule out that some protein is also released nonproteolytically. These observations are consistent with the fact that Gagnon-Arsenault et al. also observed a Yps1p-independent release of yapsin substrates (14).
Secretion of Crh2-HA/GFP is dependent on UPR and independent of ERAD.
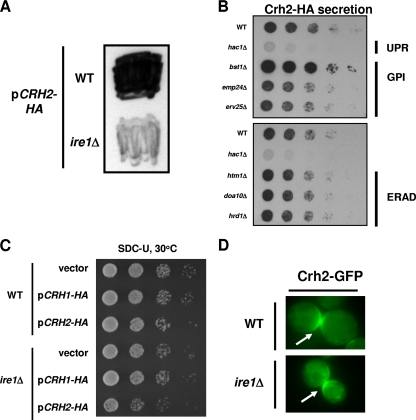
To test the hypothesis that the secretion of Crh2-HA/GFP is a manifestation of a protein quality control process, we expressed CRH2-HA in a strain lacking IRE1, the master regulator of UPR in yeast. As shown in Fig. 5A, secretion was dramatically decreased in the ire1Δ strain compared to an isogenic wild-type strain. To further characterize the genetic requirements for Crh2-HA secretion, we next expressed CRH2-HA in a set of mutants involved in UPR, ERAD, and GPI protein trafficking and examined the effect on secretion by colony immunoblotting (Fig. 5B). Consistent with the result displayed in Fig. 5A, secretion was also dependent on HAC1, the transcription factor for UPR. Secretion was affected little by the deletion of either Emp24p or Erv25p, two proteins involved in both the trafficking of GPI proteins (9) and the Bst1p-mediated degradation of Gas1p (13). Similarly, strains lacking components of ERAD secreted Crh2-HA to the same extent as the wild type. Interestingly, the deletion of BST1 significantly increased the amount of secreted Crh2-HA. Since Bst1p is required for the proteasomal degradation of misfolded Gas1p (13), the increase in Crh2-HA secretion upon its deletion strongly supports our hypothesis that secretion may be an alternative pathway for the removal of GPI proteins when the Bst1p pathway is saturated or absent.
Fig. 5.
Crh2-HA secretion is dependent on the unfolded protein response and independent of ERAD components. (A) The indicated strains expressing CRH2-HA were analyzed by colony immunoblotting as described in the legend of Fig. 2. (B) CRH2-HA was expressed in strains with mutations in the UPR (hac1Δ), GPI trafficking (emp24Δ and erv25Δ), and ERAD (htm1Δ, doa10Δ, and hrd1Δ) and analyzed by colony immunoblotting. (C) The indicated strains were spotted onto plates of synthetic dextrose medium lacking uracil as a 10-fold dilution series and incubated for 2 days at 30°C. (D) The localization of Crh2-GFP in wt and ire1Δ cells was analyzed as described in the legend of Fig. 1. The arrows indicate the expected budneck/septum localization in both strains (24).
The expression of misfolded proteins in UPR and ERAD mutants unable to properly process such species frequently causes growth defects (35). We therefore expressed CRH2-HA in the wt and an ire1Δ mutant and examined their growths at 30°C. Consistent with our hypothesis, the overexpression of CRH2-HA in the ire1Δ mutant caused a mild growth defect (Fig. 5C). The overexpression of CRH1-HA, which does not lead to secretion, does not affect the growth of an ire1Δ mutant, indicating that UPR-mediated secretion allows the cell to compensate for the toxicity of CRH2-HA.
Since Crh2-HA/GFP secretion is dependent on UPR and is increased in mutants lacking components of the pathway responsible for the degradation of misfolded GPI proteins (e.g., Bst1p), it could represent an alternative pathway by which misfolded GPI proteins are cleared from the cell. If that is the case, then blocking secretion should not affect the localization of that portion of the total cellular Crh2-GFP that is properly folded because secretion and cell wall localization would occur through separate pathways. To test this possibility, we examined the localization of Crh2-GFP in the ire1Δ mutant (Fig. 5D), a strain that does not secrete Crh2-HA. Consistent with our hypothesis, Crh2-GFP localized to the septum of large-budded cells, as observed for wild-type cells (27). These data indicate that a fraction of the Crh2-HA/GFP is properly folded and is trafficked to its appropriate location in the cell wall. A separate fraction of misfolded or excess protein appears to be secreted from the cell through a distinct, UPR-dependent trafficking pathway.
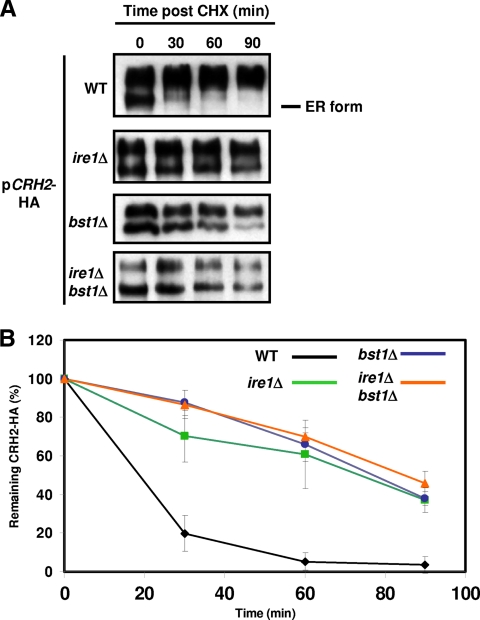
The ER form of Crh2-HA is stabilized in ire1Δ and bst1Δ mutants.
Since protein quality control mechanisms function primarily in the ER, impaired processing of misfolded proteins leads to an accumulation of the ER forms of those proteins (1). We therefore performed cycloheximide chase experiments to characterize the effects of ire1Δ and bst1Δ mutants on the trafficking of overexpressed Crh2-HA. As shown in Fig. 6A, the decay of the ER form of Crh2-HA is significantly delayed in both bst1Δ and ire1Δ mutants as well as in an ire1Δ bst1Δ mutant. Quantification of these data further emphasizes the delayed disappearance of the ER form of the protein in these mutants (Fig. 6B). Although the rate of turnover for the ER forms of Crh2-HA in the ire1Δ bst1Δ double mutant was similar to that shown by either single mutant, the double mutant showed an increased ratio of the ER to mature forms, suggesting that they both contribute to independent mechanisms for the removal of misfolded GPI proteins (data not shown) and corroborating the synthetic growth defect of the ire1Δ bst1Δ mutant in the presence of overexpressed CRH2-HA (Fig. 5C).
Fig. 6.
Decay of the ER form of Crh2-HA is delayed in ire1Δ and bst1Δ mutants. (A) The indicated strains were grown to logarithmic phase and treated with 50 μg/ml cycloheximide, and 10 OD600 cell equivalents were harvested and frozen at the indicated time points. Extracts of each sample were analyzed by Western blotting. (B) Western blots generated from three independent cycloheximide chase experiments for each strain were analyzed by densitometry to determine the amount of ER Crh2-HA remaining at each time point. The data were normalized by using time zero as 100%. Error bars indicate standard deviations.
YPS1 expression is regulated by Ire1p during ER stress, and the yps2Δ mutant genetically interacts with the ire1Δ mutant.
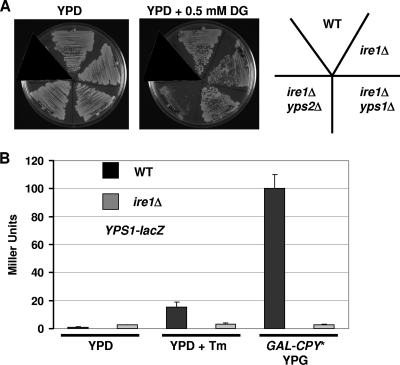
Previously reported results from our laboratory as well as from others indicate that yapsins process at least three types of cell surface-localized proteins (14, 38). Therefore, it is unlikely that Yps1p and Yps2p function only to remove excess or misfolded proteins. However, the yapsins could process proteins from the cell surface within the context of a variety of secretory and protein-trafficking pathways. To further explore the role of the yapsins in protein quality control, we examined the genetic interactions between YPS1/2 and IRE1 by constructing a corresponding set of double mutants and comparing their growth to the growth of single strains under ER stress. The growth of each strain depicted in Fig. 7A was similar to that of the wild type at 30°C on rich medium, and each single mutant grew similarly to the wild type at the indicated concentrations of 2-deoxyglucose (data not shown), a well-characterized inducer of ER stress (2). At concentrations of 2-deoxyglucose minimally toxic to the ire1Δ mutant, the yps2Δ ire1Δ mutant showed a clear synthetic defect (Fig. 7A), while the growth of the yps1Δ ire1Δ mutant was identical to that of the wild type at this as well as at higher concentrations of 2-deoxyglucose (data not shown). These data suggest that YPS2 may function in parallel with IRE1, while the epistatic relationship of IRE1 with YPS1 indicates that these two genes may function in the same pathway. Indeed, transcriptional profiling of UPR has shown that YPS1 expression is induced by ER stress, while YPS2 expression changed little (35).
Fig. 7.
Genetic interactions of YPS1 and YPS2 with the unfolded protein response. (A) The indicated strains were streaked onto plates containing YPD and YPD plus 2-deoxyglucose (DG) and incubated at 30°C for 2 days. (B) WT, ire1Δ, GAL-CPY*, and ire1Δ GAL-CPY* strains harboring a plasmid with the promoter of YPS1 fused to lacZ were grown to the logarithmic phase and treated with tunicamycin to induce ER stress or galactose to induce the expression of the misfolded protein CPY*. After 5 h of exposure to the stress condition, the cells were harvested, and the β-galactosidase activity of cellular extracts was assayed as described in the legend of Fig. 3.
To confirm this observation and test the hypothesis that Ire1p regulates YPS1 expression, we utilized our previously developed YPS1-lacZ reporter (20) and asked if ER stress induces YPS1 expression. Indeed, treatment with tunicamycin, a glycosylation inhibitor that causes severe ER stress (2), increased YPS1 expression 15-fold (Fig. 7B). Similarly, galactose-induced expression of CPY*, an extensively characterized misfolded protein that induces UPR activation (35), induced ∼100-fold-higher levels of expression of YPS1. Under both sets of ER stress conditions, the increase in YPS1 expression was completely abolished by the deletion of IRE1. Taken together, these data support the idea that the yapsins play a role in the UPR and further support a model by which yapsin proteolysis represents a novel means by which some misfolded or excess GPI proteins are removed from the cell.
DISCUSSION
Crh2p is a cell wall-localized GPI-linked protein that is involved in the cross-linking of chitin to 1,6-β-glucan (7). The laboratories of Arroyo and Cabib have extensively studied this protein (7, 27) and, in the course of that work, utilized two fusion alleles derived from Crh2p containing HA and GFP fused within the C-terminal region of the protein. While both HA and GFP show identical localizations to the cell wall of the budneck (25), the Crh2-HA fusion is stable at 38°C, whereas the Crh2-GFP construct is not (7). Consistent with the instability of the GFP construct, it failed to complement a crh2Δ mutant, while the overexpression of the HA allele did complement the strain (7). Therefore, it appears that the insertion of the GFP moiety within this region affects both the stability and the function of Crh2p, possibly by interfering with folding or processing. However, note that at least a fraction of the total protein produced by this construct is functional, since both the GFP and HA alleles show similar localizations, and the overexpression of the HA-tagged allele complements a crh2Δ mutant.
Based on those previously reported observations, it is therefore not surprising that the overexpression of Crh2-HA/GFP induces ER stress, as our data clearly indicate. However, the ER stress generated by these constructs is not entirely the result of the internal epitope tags, because the overexpression of an untagged allele also induces ER stress (Fig. 3B). The tag plays a role in the amount of ER stress, since the HA allele triggers 2-fold-higher levels of UPR activation than does the GFP allele (Fig. 3A). It is, however, somewhat counterintuitive that the apparently less stable GFP-tagged allele generates less ER stress than does the more stable HA allele. The Western blots of the GFP allele show extensive degradation at the steady state in wild-type cells, and many more bands are present than those observed with the corresponding HA construct (D. J. Krysan, unpublished results). Therefore, we suspect that the intrinsic instability of Crh2-GFP may result in a more rapid clearance of the protein from the ER and, therefore, less activation of the UPR. The more stable Crh2-HA protein, on the other hand, may require a more extensive activation of the UPR to efficiently remove it from the ER.
Although overexpressed Crh2-HA/GFP certainly leads to ER stress, it is important to consider whether these constructs represent an appropriate model substrate for the study of mechanisms of GPI-linked protein quality control. Prior to this study, only one other putatively misfolded GPI-linked protein has been studied: a point mutant of Gas1p (13). Gas1p is a GPI-linked protein that is mainly plasma membrane localized and, like Crh2p, is thought to be involved in the remodeling of cell wall polysaccharides. Fujita et al. previously constructed a point mutant of Gas1p that shows an accumulation of the ER form at steady state and undergoes proteasome-mediated degradation (13). Their studies of the processing of misfolded Gas1p revealed that its proteasome-mediated degradation was independent of the key ERAD proteins Hrd1p and Doa10p and dependent on the inositol deacetylase Bst1p.
If overexpressed Crh2-HA/GFP is an appropriate model substrate for studies of the processing of aberrant or excess GPI proteins, then its processing should share features with those observed for misfolded Gas1p. Similarly to misfolded Gas1p (13), overexpressed Crh2-HA leads to high levels of its ER form, and the deletion of Bst1p significantly delays the decay of the ER form of Crh2-HA in cycloheximide chase experiments. Taken together, the observations that overexpressed Crh2-HA/GFP induces ER stress and is processed in part by pathways previously shown to be involved in the processing of misfolded GPI proteins strongly suggest that this represents an informative system for the study of GPI protein quality control.
In addition to being processed through the Bst1p-dependent proteasome-mediated degradative pathway, we observed that significant amounts of both Crh2-HA and Crh2-GFP are secreted to the extracellular space. The secretion of Crh2-HA/GFP is dependent on the UPR and is increased in strains lacking Bst1p, an inositol deacetylase involved in the proteasome-mediated degradation of GPI proteins (13). Based on these and other observations described above, we propose that the secretion of Crh2-HA/GFP represents a previously unrecognized pathway by which excess or aberrant secretory proteins are removed from the cell when other degradative pathways are saturated or inoperative. It is important that the overexpression of the extensively studied ERAD and UPR substrate CPY* does not lead to secretion (D. Ng, personal communication), suggesting that only some ER-stress-inducing secretory proteins are processed through this process.
To our knowledge, the ER-stress-induced release of secretory pathway proteins to the extracellular space has been described only once previously. Specifically, Belden and Barlowe showed that ER stress induced either by the mutation of proteins in the p24 family of ER transport proteins or by the treatment of cells with the reducing agent β-mercaptoethanol causes the release of the ER-resident chaperone Kar2p/BiP to the extracellular medium (3). This release is completely blocked when the UPR is disabled through the deletion of IRE1, leading those authors to propose that Kar2p secretion is a “hallmark of UPR” (3). Our results indicate that the high-level expression of a GPI-linked cargo protein that induces UPR activation leads to both Kar2p secretion and secretion of the cargo protein, suggesting that this UPR-mediated secretory pathway may not represent simply a nonspecific release of secretory pathway proteins to the extracellular space.
At this point, we cannot completely exclude the possibility that the release of Kar2p/Crh2p is due to stress-induced ER and secretory pathway dysfunction. However, a number of observations from this work and from the work of Belden and Barlowe (3) support the notion that this secretory pathway is not an indirect consequence of ER dysfunction. First, we have shown that the secretion of Crh2-HA is dependent on ER-to-Golgi transport via Sec18p, indicating that components of the trafficking machinery are required for the process. Second, Belden and Barlowe found previously that under conditions in which Kar2p is released, there is very little change in the pattern or quantity of proteins secreted into the growth medium compared to wild-type cells (3). This finding suggests that Kar2p secretion is not due to generalized defects in secretory pathway function leading to a nonspecific release of proteins into the extracellular space. Third, the UPR is an important mechanism by which ER and secretory pathway function is maintained (23, 36). Therefore, if the release of Crh2-HA was the result of ER-stress-mediated secretory pathway dysfunction, then the additional ER stress generated by the loss of the UPR through the deletion of Ire1p would be expected to increase secretion. Hence, the decrease in Crh2-HA and Kar2p secretion observed for ire1Δ mutants is inconsistent with a nonspecific breakdown in secretory pathway function but is consistent with the idea that secretion represents a UPR-mediated pathway by which aberrant proteins are removed by secretion to the extracellular space.
Although more work will be required to fully characterize the secretory pathway proteins involved in the delivery of Crh2-HA/GFP to the extracellular space, our initial results suggest that this pathway is distinct from the pathway by which normal GPI proteins are delivered to the cell surface. First, ire1Δ mutants, which do not secrete Crh2-HA/GFP, still localize a significant portion of the protein to the cell wall, indicating that secretion is not required for appropriate localization and supporting the idea that secretion is distinct from normal GPI protein trafficking. Second, the deletion of two proteins involved in GPI protein trafficking, Emp24p and Erv25p, does not block secretion; indeed, the deletion of either protein leads to Kar2p secretion (3), suggesting that the loss of these proteins may actually trigger the pathway. Third, Western blots of strains with decreased secretion show amounts of fully mature Crh2-HA that are similar to those in wild-type cells (Fig. 4C). The pathway does, however, require Sec18p, a protein required for ER-to-Golgi transport. Thus, the pathway by which Crh2-GFP/HA is secreted to the extracellular space must utilize at least some components of the machinery involved in the trafficking of normal proteins. Since mutants that affect the proteasome-mediated processing of aberrant GPI proteins increase Crh2-HA secretion, we suspect that this pathway represents a mechanism that is operative mainly when the standard, proteasome-mediated pathway is saturated by severe ER stress.
An important question regarding the yapsins is, where do they cleave their substrates? Recently, Gagnon-Arsenault et al. reported an elegant study that strongly suggested that yapsin proteolysis occurs at the cell surface and is regulated by the pH of the compartment (14). This finding is consistent with the results of Komano et al. showing that the yapsin-specific cleavage of fluorescent model substrates occurs at the surface of intact cells (19). Yps1p and Yps2p have both been shown to have optimal activity at a low pH, which is present in the extracellular/periplasmic space in yeast. Since the early compartments of the secretory pathway are more basic than those that lead to efficient yapsin proteolysis, cleavage is unlikely to occur within early portions of the pathway, such as the ER (14); indeed, our results show that the trapping of Crh2-HA in the ER blocks proteolysis (Fig. 4C). It is, however, possible that some portion of the proteolysis occurs within the Golgi apparatus. Sievi et al. showed previously the Yps1p-mediated degradation of a mammalian sialyltransferase ectodomain fusion protein (STE6Ne) in the Golgi apparatus of a strain with a temperature-sensitive block in Golgi transport; Yps2p had no effect on the processing of this substrate (33).
Seivi et al. also showed that Yps1p is mislocalized from the plasma membrane to the vacuole in erg6Δ mutants and processes STE6Ne in that acidic compartment (33). Although the cleavage of Crh2-HA could occur in this compartment, the fact that we did not observed an accumulation of uncleaved Crh2-GFP in the vacuole of yapsin mutants (Fig. 1C) argues against this possibility. However, since Crh2-GFP processing and Crh2-HA processing are not identical, we cannot rule out the vacuole as a site of Crh2-HA processing. This model, however, requires that the cleavage products then be transported from the vacuole to the cell surface, a process that is unprecedented to our knowledge.
In addition to the vacuolar targeting model, our observations are consistent with at least three other scenarios for the events leading to the yapsin-mediated release of misfolded/excess proteins such as Crh2-HA. First, the substrates and proteases could traffic in the same vesicles as normal proteins to the cell surface and then undergo proteolysis. In this model, selection between folded and misfolded proteins could result from differential rates between further processing and proteolysis. For example, a cell wall-targeted GPI protein must be cleaved from its anchor and incorporated into the cell wall. If a protein is misfolded or excess protein has saturated the capacity of this process, then its delay may then allow proteolysis to compete, leading to the release of the misfolded, or excess, GPI protein. Second, a similar series of events may occur but within specific vesicles that contain misfolded proteins. An indirect precedence for the packaging of misfolded cargo proteins into distinct vesicles is based on the fact that excess misfolded protein can be trafficked directly to the vacuole when ERAD is saturated, a process that is likely to involve vesicles distinct from those destined for the plasma membrane (35). Last, yapsin cleavage could occur in the Golgi apparatus, as suggested by the results described previously by Seivi et al. (33). Recent work has also revealed that protein quality control surveillance is operative within the Golgi apparatus (40), and thus, it is possible that discrimination between abnormal and normal proteins is made at the Golgi apparatus. After proteolysis, the fragments could then be transported to the surface via either normal vesicles or, alternatively, specific vesicles containing misfolded proteins. The small punctate structures to which Crh2-GFP localizes in the yps2Δ mutant could represent the Golgi apparatus (Fig. 2A) and therefore would provide support for this model. It must be stressed that these models are rather speculative at this point and that additional work will be required to identify the pathways that are involved in the UPR/yapsin-dependent secretion of excess/misfolded proteins.
Although the yapsin family of GPI-linked aspartyl proteases was identified over 15 years ago (15), an understanding of their physiological function is only recently beginning to emerge (1, 14, 15, 18, 20, 38). To date, S. cerevisiae yapsins have been shown to play a role in yeast cell wall integrity (20), the processing of the signaling mucin Msb2p during the transition to filamentous growth (38), and the release of both Gas1p and Pir4p from the cell surface (14). In this work, we have found evidence supporting a role for both Yps1p and Yps2p in protein quality control. YPS1 expression is increased by ER stress in an Ire1p-dependent fashion, a finding that is consistent with data from previously reported transcriptional profiling experiments (36) and is further supported by the epistatic relationship between ire1Δ and yps1Δ mutants during ER stress (Fig. 7A). Yps2p also appears to play a role in protein quality control based on the fact that yps2Δ and ire1Δ mutants display a synthetic genetic interaction during ER stress.
Unlike YPS1, YPS2 expression does not appear to vary significantly during cellular stress (15), and its genetic interaction with Ire1p suggests that its function is independent of the UPR. Furthermore, the surface yapsin activity is dependent almost exclusively on Yps2p under nonstress conditions, while Yps1p is undetectable at the surface unless the cells are exposed to stresses known to increase YPS1 expression levels (15, 19, 20, 33). We therefore propose that Yps2p is constitutively active during unstressed growth and is sufficient to process the basal levels of misfolded or excess protein within the secretory pathway. During ER stress, the UPR is activated and induces YPS1 expression to increase the processing capacity. This model provides a possible explanation for the observation that Yps2p is the only yapsin required for Crh2-GFP processing while both Yps1p and Yps2p process Crh2-HA. Crh2-GFP does not activate the UPR as intensely, as Crh2-HA presumably leads to relatively low levels of Yps1p, and thus, Yps2p carries out the bulk of the processing. Crh2-HA induces the UPR more strongly, and consequently, Yps1p is available to process Crh2-HA. Taken together with previous studies of yapsin function (14, 38) in S. cerevisiae as well as in the closely related pathogenic yeast C. glabrata (18), the yapsins appear to mediate the release of proteins from the surface of yeast cells during a variety of cellular process, including cell wall remodeling, cell surface-initiated signaling, and protein quality control.
A number of groups have found that the presence of yapsins and their homologues adversely affects the production of heterologously expressed, recombinant proteins in S. cerevisiae and Pichia pastoris (5, 10, 29, 41) by causing a large amount of degraded protein. Our data implicating yapsins in secretory pathway quality control provide a compelling physiological explanation for these observations. Yps1p has been found to be particularly deleterious to recombinant protein production, and its presence leads to the degradation of a variety of recombinant proteins (5, 10). The regulation of YPS1 expression by the UPR suggests that heterologous or high-level expression could activate the UPR, leading to increased expression levels of YPS1 and, ultimately, to an increased degradation of the secreted protein. Recent results indicating that Yps2p also contributes to the degradation of some recombinant proteins (10) are also consistent with our findings that Yps1p and Yps2p both have a role in secretory pathway homeostasis.
In summary, we have provided evidence for a previously unrecognized, UPR-mediated pathway by which an ER-stress-inducing GPI-linked protein is secreted from the cell. We propose that this pathway represents a mechanism by which the cell removes excess or aberrant proteins from the cell when other pathways for degradation are saturated or inoperative. In addition, we have identified a new substrate for the yapsins and provided evidence supporting a role for this family of proteins in protein quality control. Studies designed to further characterize the types of proteins secreted to the extracellular space by this pathway as well as the components of the secretory pathway that mediate this secretion are in progress and will be reported in due course.
ACKNOWLEDGMENTS
We thank the following investigators for generously providing strains, plasmids, and/or reagents: J.-M. Rodriguez-Pena (Madrid, Spain), R. Fuller (Michigan), D. Ng (Singapore), F. Klis (Amsterdam, Netherlands), and P. de Groot (Amsterdam, Netherlands). We also thank J.-M. Rodriguez-Pena and D. Ng for helpful discussions.
This work was supported by grants from the National Institutes of Health (grant K08AI06927 to D.J.K.) and Strong Children's Research Center (to D.J.K.).
Footnotes
Published ahead of print on 10 September 2010.
REFERENCES
- 1.Albrecht A., Felk A., Pichova J., Naglik J. R., Schaller M., de Groot P., Maccalum D., Odds F. C., Klis F., Monod F., Hube B. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular and host pathogen interactions. J. Biol. Chem. 281:688–694 [DOI] [PubMed] [Google Scholar]
- 2.Back H. S., Schroder M., Lee K., Zhang K., Kaufman R. J. 2005. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods 35:395–416 [DOI] [PubMed] [Google Scholar]
- 3.Belden W. J., Barlowe C. 2001. Deletion of yeast p24 genes activates the unfolded protein response. Mol. Biol. Cell 12:957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonifacino J. S., Glick B. S. 2004. The mechanisms of vesicle budding and fusion. Cell 116:153–166 [DOI] [PubMed] [Google Scholar]
- 5.Bourbannais Y., Larouche C., Tremblay G. M. 2000. Production of full length human pre-elafin, an elastase inhibitor, from yeast requires the absence of a functional yapsin 1 (Yps1p) endoprotease. Protein Expr. Purif. 20:485–491 [DOI] [PubMed] [Google Scholar]
- 6.Burke D. J., Dawson D., Stearns T. 2000. Methods in yeast genetics. CSHL Press, Woodbury, NY [Google Scholar]
- 7.Cabib E., Blanco N., Grau C., Rodriquez-Pena J. M., Arroyo J. 2007. Crh1p and Crh2p are required for the cross-linking of chitin to β(1-6)glucan in the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 63:921–935 [DOI] [PubMed] [Google Scholar]
- 8.Carvalho P., Goder V., Rapoport T. A. 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126:361–373 [DOI] [PubMed] [Google Scholar]
- 9.Castillion G. A., Watanable R., Taylor M., Schwabe T. M. E., Riezman H. 2009. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic 10:186–200 [DOI] [PubMed] [Google Scholar]
- 10.Cho E. Y., Cheon S. A., Kim H., Choo J., Lee D.-J., Ryu H. M., Rhee S. K., Chung B.-H., Kim J.-Y., Kang H. A. 25June2010, posting date Multiple yapsin-deficient mutant strains for high level production of intact recombinant proteins in Saccharomyces cerevisiae. J. Biotechnol. doi:10.1016/j.jbiotec.2010.06014 [DOI] [PubMed] [Google Scholar]
- 11.Cox J. S., Shamu C. E., Walter P. 1993. The transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73:1197–1206 [DOI] [PubMed] [Google Scholar]
- 12.Franzusoff A., Redding K., Crosby J., Fuller R. S., Schekman R. 1991. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J. Cell Biol. 112:27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita M., Yoko-O T., Jigami Y. 2006. Inositol deacetylation by Bst1p is required for the quality control of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell 17:834–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gagnon-Arsenault I., Parise L., Tremblay J., Bourbonnais Y. 2008. Activation mechanism, functional role, and shedding of glycosylphosphatidylinositol-anchored Yps1p at the Saccharomyces cerevisiae cell surface. Mol. Microbiol. 69:982–983 [DOI] [PubMed] [Google Scholar]
- 15.Gagnon-Arsenault I., Tremblay J., Bourbonnais Y. 2006. Fungal yapsins and cell wall: a unique family of aspartic peptidases for a distinctive cellular function. FEMS Yeast Res. 6:966–978 [DOI] [PubMed] [Google Scholar]
- 16.Ismail N., Ng D. T. W. 2006. Have you HRD? Understanding ERAD is DOAble! Cell 126:237–239 [DOI] [PubMed] [Google Scholar]
- 17.Johnston M., Riles L., Hegemann J. H. 2002. Gene disruption. Methods Enzymol. 350:290–315 [DOI] [PubMed] [Google Scholar]
- 18.Kaur R., Ma B., Cormack B. P. 2007. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence in Candida glabrata. Proc. Natl. Acad. Sci. U. S. A. 104:7628–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komano H., Seeger M., Gandy S., Wang G. T., Krafft G. A., Fuller R. S. 1998. Involvement of cell surface glycosyl-phosphatidylinositol-linked aspartyl proteases in α-secretase-type cleavage and ectodomain solubilization of human Alzheimer β-amyloid precursor protein in yeast. J. Biol. Chem. 273:31648–31651 [DOI] [PubMed] [Google Scholar]
- 20.Krysan D. J., Ting E. L., Abeijon C., Kroos L., Fuller R. S. 2005. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot. Cell 4:1364–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J. H., Walter P., Yen T. S. 2008. Endoplasmic reticulum stress in disease pathogenesis. Annu. Rev. Pathol. 3:399–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mori K., Ma W., Gething M. J., Sambrook J. 1993. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74:3028–3039 [DOI] [PubMed] [Google Scholar]
- 23.Ng D. T. W., Spear E. D., Walter P. 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 150:77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ram A. F. J., van den Ende H., Klis F. M. 1998. Green fluorescent protein-cell wall fusion proteins are covalently incorporated into the cell wall of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 162:249–255 [DOI] [PubMed] [Google Scholar]
- 25.Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts C. J., Raymond C. K., Yamashiro C. T., Stevens T. H. 1991. Methods for studying the yeast vacuole. Methods Enzymol. 194:644–661 [DOI] [PubMed] [Google Scholar]
- 27.Rodriquez-Pena J. M., Cid V. J., Arroyo J., Nombela C. 2000. A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol. Cell. Biol. 20:3245–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ron D., Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8:519–529 [DOI] [PubMed] [Google Scholar]
- 29.Rourke I. J., Johnson A. H., Din N., Peterson J. G., Rehfeld J. F. 1997. Heterologous expression of human cholecystokinin in Saccharomyces cerevisiae. Evidence for a lysine specific endopeptidase in the yeast secretory pathway. J. Biol. Chem. 272:9720–9727 [DOI] [PubMed] [Google Scholar]
- 30.Sayeed A., Ng D. T. W. 2005. Search and destroy: ER quality control and ER-associated protein degradation. Crit. Rev. Biochem. Mol. Biol. 40:75–91 [DOI] [PubMed] [Google Scholar]
- 31.Schafer A., Wolf D. H. 2005. Yeast genomics in the elucidation of endoplasmic reticulum quality control and associated protein degradation (ERQD). Methods Enzymol. 339:459–468 [DOI] [PubMed] [Google Scholar]
- 32.Scrimale T. S., DiDone L., de Mesy Bentley K. L., Krysan D. J. 2009. The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 20:164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shamu C. E., Walter P. 1996. Oligomerization and phosphorylation of Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 15:3028–3039 [PMC free article] [PubMed] [Google Scholar]
- 34.Sievi E., Sunito T., Makarow M. 2001. Proteolytic function of GPI-anchored plasma membrane protease Yps1p in the yeast vacuole and Golgi. Traffic 2:896–907 [DOI] [PubMed] [Google Scholar]
- 35.Spear E. D., Ng D. T. W. 2003. Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol. Biol. Cell 14:2756–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. 2000. Functional and genomic analysis reveals an essential coordination between unfolded protein response and ER-associated degradation. Cell 101:249–258 [DOI] [PubMed] [Google Scholar]
- 37.Tong A. H., Boone C. 2006. Synthetic genetic analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313:171–192 [DOI] [PubMed] [Google Scholar]
- 38.Vadaie N., Dionne H., Akajagbor D. S., Nickerson S. R., Krysan D. J., Cullen P. J. 2008. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J. Cell Biol. 181:1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vembar S. S., Brodsky J. L. 2008. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9:944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S., Ng D. T. W. 2010. Evasion of endoplasmic reticulum surveillance makes Wsc1p an obligate substrate of Golgi quality control. Mol. Biol. Cell 21:1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao X. Q., Zhao H. L., Xue C., Zhang W., Xiong X. H., Wang Z. W., Li X. Y., Liu Z. M. 2009. Degradation of HAS-AX15(R13K) when expressed in Pichia pastoris can be reduced vie the disruption of the YPS1 gene in yeast. J. Biotechnol. 139:131–136 [DOI] [PubMed] [Google Scholar]