Abstract
The enzyme α-1,6-mannosyltransferase (OCH-1) is required for the synthesis of galactomannans attached to the N-linked oligosaccharides of Neurospora crassa cell wall proteins. The Neurospora crassa och-1 mutant has a tight colonial phenotype and a defective cell wall. A carbohydrate analysis of the och-1 mutant cell wall revealed a 10-fold reduction in the levels of mannose and galactose and a total lack of 1,6-linked mannose residues. Analysis of the integral cell wall protein from wild-type and och-1 mutant cells showed that the mutant cell wall had reduced protein content. The och-1 mutant was found to secrete 18-fold more protein than wild-type cells. Proteomic analysis of the proteins released by the mutant into the growth medium identified seven of the major cell wall proteins. Western blot analysis of ACW-1 and GEL-1 (two glycosylphosphatidylinositol [GPI]-anchored proteins that are covalently integrated into the wild-type cell wall) showed that high levels of these proteins were being released into the medium by the och-1 mutant. High levels of ACW-1 and GEL-1 were also released from the och-1 mutant cell wall by subjecting the wall to boiling in a 1% SDS solution, indicating that these proteins are not being covalently integrated into the mutant cell wall. From these results, we conclude that N-linked mannosylation of cell wall proteins by OCH-1 is required for their efficient covalent incorporation into the cell wall.
The fungal cell wall is an important organelle that protects the cell from various environmental stresses. It is a dynamic structure that interacts with the environment and is modified to accommodate growth, cell division, and development. Fungal cell walls have been shown to contain β-1,3-glucan, α-1,3-glucan, β-1,6-glucan, mixed β-1,3/β-1,4-glucans, chitin, and mannan/galactomannan (6, 19). These polysaccharide polymers constitute 80 to 85% of the cell wall mass, while glycoproteins constitute the remaining 15 to 20% (6). The cell wall glycoproteins are required for vital functions, like structural support, signal transduction, biofilm formation, and cell wall biosynthesis. In the case of pathogenic fungi, the cell wall is critical for the invasion of host tissues (8). Because of their accessibility and the crucial functions they perform, cell wall proteins could be important targets for the development of antifungal therapeutics.
The glucan and chitin cell wall polymers are synthesized by enzyme complexes (glucan synthases and chitin synthases) that are associated with the plasma membrane. Glucan and chitin are vectorially passed into the cell wall space during synthesis and cross-linked together in the cell wall space. The mannan and galactomannan present in the cell wall are found as glycoconjugates on cell wall proteins. Mannosylation of cell wall proteins occurs in the endoplasmic reticulum (ER) and Golgi apparatus at O-linked and N-linked glycosylation sites. In Saccharomyces cerevisiae, mannosylation of N-linked glycosylation is initiated by the addition of an α-1,6-linked mannose residue by Och1p (33). In the filamentous fungus Neurospora crassa, the structure of the galactomannan associated with N-linked sites has not been definitively determined, but N. crassa has most of the enzymes defined in yeast for the mannosylation of N-linked oligosaccharides (14). An analysis of N-linked oligosaccharides from N. crassa glycoproteins showed that the glycoproteins are modified by the addition of short α-1,6-mannans with short α-1,2-mannose branches that are terminated by galactofuranose residues (31, 32). The N. crassa posttranslational modifications appear to differ from those found in S. cerevisiae by having shorter mannan chains and by the presence of terminal galactofuranose residues.
Mannosylation of glycoproteins has been extensively studied in yeast. In S. cerevisiae, OCH1 encodes the α-1,6-mannosyltransferase enzyme that mediates the addition of the initial α-1,6-mannose in the synthesis of long mannans which are attached to the N-linked oligosaccharides (22, 33). Knockout mutants of OCH1 are viable but exhibit a temperature-sensitive growth pattern and are sensitive to cell wall perturbation reagents (34). Mutants for Candida albicans homologs of OCH1 had near-normal growth rates but were much less virulent (3). Mass spectrometry analysis of glycoproteins from the S. cerevisiae och1 and C. albicans och1 mutants showed that the α-1,6-mannose core was absent (3, 33). In Kluyveromyces lactis, the KlOCH1 gene has been shown to be important for cell wall organization and to give a hypersecretion phenotype (37). OCH1 mutants have also been identified in Pichia angusta, Yarrowia lipolytica, Pichia pastoris, and Schizosaccharomyces pombe, and these mutants have cell wall-related phenotypes (2, 9, 17, 38). However, a recent report of OCH1 knockout mutants of Aspergillus fumigatus indicates that these mutants do not have a cell wall-defective phenotype (18).
Mannosylation of cell wall proteins has not been extensively studied in filamentous fungi. We report on the characterization of the N. crassa knockout mutant of the α-1,6-mannosyltransferase, och-1. The mutant was generated by the Neurospora genome knockout project (10). The N. crassa och-1 mutant has a severe growth defect and exhibits a tight colonial phenotype. We demonstrate that the och-1 mutant exhibits a defect in cell wall biosynthesis. A carbohydrate analysis of the mutant cell wall showed a drastic reduction in mannose and galactose content with a compensatory increase in the glucose content. The och-1 cell wall also showed a reduced cell wall protein content as assessed by a Coomassie brilliant blue dye binding assay and by proteomic analysis. Protein secretion assays showed that the mutant releases large amounts of cell wall protein into the growth medium. We demonstrate that the och-1 mutant is defective in covalently cross-linking known cell wall proteins into the cell wall matrix. Our data demonstrate that the N-linked galactomannan, which is built upon the mannose residue added by OCH-1, is required for the incorporation of cell wall proteins into the cell wall matrix.
MATERIALS AND METHODS
Strains and culturing conditions.
N. crassa wild-type strain 74-OR23-IV A and och-1 knockout mutant strains were maintained in Vogel's medium with 2% sucrose at room temperature (12). The och-1 knockout mutant was generated during the Neurospora genome project by replacing the α-1,6-mannosyltransferase gene with a hygromycin resistance cassette (10). The och-1 knockout mutant was obtained from the Fungal Genetics Stock Center (Kansas City, MO). Cloning, sequencing, and transformation experiments were done as described by Colot et al. (10) and by Maniatis et al. (27).
Complementation of och-1 knockout mutation.
Knock-in experiments targeting the insertion of a wild-type copy of och-1 downstream of the his-3 locus were performed using the pBM61 plasmid as described by Margolin et al. (28). The och-1 gene (NCU00609) is 1,194 bases in length from the start site to the stop site, including introns, as given in the Neurospora genome database. For transformation purposes, ∼1,500 bases upstream of the start site and ∼500 bases downstream of the stop site were included as part of the gene. Primers containing added XbaI and XmaI restriction sites (forward primer, ACTATCTAGAGCCCGATTTCATGCTGGAACAG; reverse primer, CTTACCCGGGTTCGGTGACGATTTCGGACC) were used in amplifying the gene and inserting it as an XbaI/XmaI fragment into pBM61. A plasmid carrying the och-1 gene was sequenced to ensure that it contained no mutations and was used to transform an och-1 his-3 mutant as described by Margolin et al. (28).
Cell wall stress tests.
The och-1 mutant and wild-type control were subjected to various cell wall stress conditions. The strains were grown on slants of Vogel's medium with 2% sucrose agar or in Vogel's liquid medium with 2% sucrose containing a cell wall stress reagent. The growth of the mutant was observed at 30°C over a period of 72 h and compared to the growth of the wild type. Cell wall stress reagents included 10% NaCl (salt stress), 2 M glycerol (hyperosmolarity), 0.05 mM hydrogen peroxide (oxidative stress), 0.01% SDS (detergent), 10 mg/ml calcofluor white (CFW) (chitin inhibitor), and 10 μg/ml caspofungin (glucan inhibitor). Caspofungin acetate was obtained as a kind gift from Merck Research Laboratories (Rahway, NJ). The optimum concentration of the cell wall stress reagents for testing the mutant was determined by subjecting the wild-type strain to a range of concentrations and identifying a concentration which was slightly below the concentration at which the wild-type cells were unable to grow. Apart from subjecting the mutant to cell wall stress reagents, it was also subjected to growth at 18°C and 37°C to test for a temperature-sensitive growth phenotype.
Cell wall preparations for N. crassa.
Cell walls were prepared as described previously (26). Seven- to 10-day-old conidia from wild-type and och-1 strains were used to inoculate 1 liter of Vogel's liquid medium containing 2% sucrose and cultured at 25°C in a shaking incubator for 12 to 14 h. Vegetative hyphae were harvested by filtration through a filter paper placed on a Buchner funnel. The harvested vegetative hypha cultures were then frozen with liquid nitrogen in a mortar and ground to a fine powder with a pestle while intermittently adding liquid nitrogen. The ground hyphae were then resuspended in an extraction buffer consisting of phosphate-buffered saline (PBS) (27).
To purify cell walls, disrupted hypha preparations were first subjected to a centrifugation step (4,000 × g for 5 min) to separate the cell walls from the cytosolic proteins. The crude cell walls were washed twice with PBS and resuspended in PBS containing 1% SDS. The resuspended cell wall pellets were boiled in the SDS-containing extraction buffer for 15 min, allowed to cool, and collected by centrifugation (10,000 × g for 5 min). The supernatant was collected as “SDS-solubilized fraction” and was used to examine the proteins that are noncovalently associated with the cell wall in Western blot analyses. The cell wall pellet was washed twice with ice-cold PBS and once with distilled water. The centrifugation step for these washes was carried out at 10,000 × g for 5 min. The washed pellet was considered “purified cell wall” and was subjected to deglycosylation using trifluoromethanesulfonic acid (TFMS), which completely solubilized the cell wall. To identify integral cell wall proteins, the TFMS-solubilized cell wall was subjected to nano-liquid chromatography/tandem mass spectrometry (LC/MS-MS) analysis. The purified cell wall fraction was also subjected to an aniline blue binding analysis to assess the levels of β-1,3-glucan present and Coomassie brilliant blue dye binding analysis to assess the levels of cell wall protein and sent to the Complex Carbohydrate Research Center (CCRC; Athens, GA) for carbohydrate analyses.
Protein secretion assay.
Wild-type and mutant cells were grown in Vogel's liquid medium with 2% sucrose for 12 to 14 h at room temperature in a shaking incubator. Under sterile conditions, the cells were collected by filtration over a Buchner funnel, resuspended into fresh Vogel's liquid medium with 2% sucrose, and allowed to grow for an additional 2 h on the shaking incubator. The cells were collected again by filtration on a Buchner funnel, and the medium was collected for the analysis of secreted proteins. Trichloroacetic acid (TCA) precipitation was used to collect the secreted proteins from the medium. Acetone and TCA were added to the medium to a final concentration of 50% acetone and 12.5% TCA, and the proteins were allowed to precipitate for 24 h at −20°C. The precipitated proteins were collected by centrifugation, washed twice with −20°C acetone, and lyophilized. Samples of the lyophilized protein were then subjected to SDS-PAGE, Western blot analysis, and TFMS digestion.
Carbohydrate and glycosyl linkage analysis of cell walls.
Glycosyl composition of cell walls was analyzed by combined gas chromatography/mass spectrometry (GC/MS) of the per-O-trimethylsilyl (TMS) derivatives of the monosaccharide methyl glycosides produced from the sample by acidic methanolysis at the CCRC. For glycosyl linkage analysis, the samples were permethylated, depolymerized, reduced, and acetylated, and the partially methylated alditol acetates (PMAAs) were analyzed at the CCRC by GC/MS.
TFMS digestion of cell walls and secreted protein.
To analyze the secreted protein and the proteins covalently linked to the cell walls, the samples were treated with TFMS using a procedure described previously (26). TFMS, anisole, and pyridine were obtained from Sigma-Aldrich Chemical Company (St. Louis, MO). Secreted protein samples and cell wall pellets were lyophilized overnight to ensure complete dryness of the samples. To maintain anhydrous conditions during the TFMS treatment, all glass tubes and syringes used were dried under a vacuum, and the digestion was performed in a chamber being continually purged with liquid nitrogen. Initially, a solution of 16% anisole in TFMS was prepared and 1.25 ml of this mixture was added to 20 mg of the purified cell wall sample or to a sample of secreted protein collected from 500 ml of culture medium. The samples were then purged with liquid nitrogen, quickly covered with Parafilm, and placed in the N2-filled chamber at 4°C. During the course of the digestion, the samples were periodically mixed with a Pasteur pipette, purged with N2 gas, and covered again with Parafilm. The cell wall samples and the samples of secreted protein were completely solubilized in 5 h, as assessed by visual inspection. After the cell walls had been solubilized, 3.75 ml of a solution of pyridine-methanol-H2O (3:1:1) was added in a dropwise fashion to each of the digests, which were continually swirled in a dry ice-ethanol bath. The samples were then left in the dry ice-ethanol bath for 20 min, followed by incubation for another 20 min at −20°C. The samples were then removed and allowed to thaw, and 1 ml of 5% ammonium bicarbonate solution was added to each sample. The released proteins were then precipitated by adding 6 ml of 25% TCA in acetone and incubating at −20°C for 24 h. The precipitated proteins were collected by centrifugation at 10,000 × g, washed three times in ice-cold 100% acetone, and briefly dried. The proteins were then resuspended in PBS with 1% SDS and subjected to a 10-min boiling treatment. Protein concentrations of all samples were determined using the Bio-Rad DC protein assay kit. The amount of protein released from up to 5 mg of starting cell wall material was separated by SDS-PAGE on a 4 to 12% Bis-Tris NuPAGE gel and visualized using the SilverQuest silver staining kit (Invitrogen Life Technologies, Carlsbad, CA) or with Coomassie blue (27).
Nano-LC/MS-MS analysis of peptides.
Samples containing 20 μg of integral cell wall protein released by TFMS digestion or 20 μg of TFMS-digested secreted protein were briefly subjected to SDS-PAGE. The gel was then stained with Coomassie brilliant blue, and the stained part of the gel was excised and sent to Midwest Bio Services (Overland Park, KS) for nano-LC/MS-MS-based identification.
Data analysis (MS-MS database search).
Cell wall proteins were identified by matching the peptide sequences obtained from the MS-MS analysis with the protein sequences from the Neurospora genome available at the Broad Institute (www.broad.mit.edu) using the TurboSEQUEST software. Only those proteins with multiple peptides and/or single peptides with an XC (correlation coefficient) score of >2.5 for 2 ions or >3.0 for 3 ions were accepted as accurate identifications in this analysis.
Western blot analysis of cell wall and secreted protein samples.
Western blot analysis of cell wall protein, secreted protein, SDS-soluble cell wall protein, and cytosolic protein samples was done using ACW-1 antibody as described previously (7). Polyclonal antibody directed against purified GH72-5/GEL-1 was raised in rabbits.
Aniline blue binding assay.
As a measure of the amounts of β-1,3-glucan present in the wild-type and mutant cell walls, we assessed the ability of the purified cell wall to bind aniline blue. The assay was carried out as previously described (7).
Coomassie brilliant blue dye binding assay.
To compare the protein contents of the wild-type and mutant cell walls, the ability of the wall to absorb Coomassie brilliant blue dye (Bio-Rad, Hercules, CA) was assessed. Lyophilized purified cell wall material from wild-type and mutant cells was washed with distilled water and resuspended in a 15-ml Corex tube at a concentration of 2 mg/ml in water. The cell wall material was subjected to vortexing, and aliquots of cell wall material were taken with a micropipette outfitted with a large orifice tip into microcentrifuge tubes. Distilled water was added to the tube to give a volume of 0.5 ml, and 0.5 ml of 1× Coomassie dye reagent (Bio-Rad Laboratories, Hercules, CA) was added. The samples were incubated with agitation at room temperature for 30 min. The samples were then centrifuged to pellet the cell wall, and the amount of Coomassie blue remaining in the supernatant was determined at an optical density of 465 nm (OD465). The amount of dye absorbed was determined by comparing the OD465 of the samples with the OD465 of a control sample without cell wall material.
RESULTS
The och-1 mutant exhibits morphological defects.
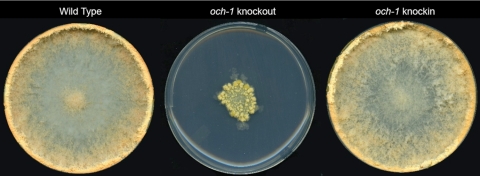
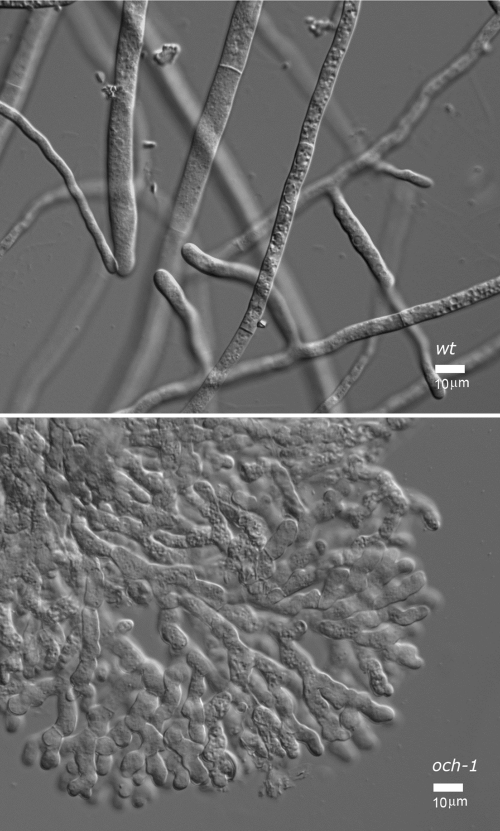
The och-1 mutant was generated by the Neurospora genome knockout project (10) and obtained from the Fungal Genetics Stock Center (Kansas City, MO). It was identified as being an interesting mutant during a screening of the knockout library for morphological mutants. The N. crassa och-1 gene (NCU00609) is a homolog of the OCH1 gene of S. cerevisiae. The gene encodes a 373-amino-acid type II transmembrane protein with a nucleotide sugar binding motif between residues 150 and 210. The S. cerevisiae Och1p has been shown to be localized to the cis-Golgi apparatus (30). The knockout mutant was generated by replacing the gene with a hygromycin resistance cassette as described previously (10). The presence of the hygromycin resistance cassette in the och-1 mutant was verified by PCR amplification of the region between 500 bases upstream of the hygromycin cassette insertion site and 500 bases into the hygromycin resistance cassette. The N. crassa och-1 mutant grew with a tight colonial morphology, as shown in Fig. 1. Microscopic examination of the mutant showed that the hyphae exhibited a tendency to grow in a globular fashion with an abnormal branching pattern (Fig. 2). The mutant was unable to produce normal aerial hyphae and produced abnormal conidia close to the colony surface. The conidia that are produced have a conidial separation phenotype but are viable. The och-1 mutant produces protoperithecia, the female mating structure, and following mating the protoperithecia mature into perithecia with ascospores. The mutant is unable to release the ascospores from the perithecia, but the ascospores can be collected from the perithecia and are viable. The och-1 mutant colonial phenotype was found to cosegregate with the hygromycin resistance gene used to create the knockout mutation, suggesting that the mutant phenotypes are the result of the loss of the och-1 gene.
Fig. 1.
Growth pattern of the wild-type and the och-1 mutant colonies. The wild type, the och-1 knockout mutant, and the och-1 knockout mutant that was transformed with the wild-type copy of the och-1 gene (knock-in) were grown in Vogel's medium with 2% sucrose agar in plates and slants for a period of 6 to 10 days.
Fig. 2.
Differential interference contrast (DIC) microscopy images of the growing edge of the colony from wild-type cells and och-1 mutant cells at 40× resolution (bar represents 10 μm).
To verify that the mutant phenotypes were the result of the loss of the och-1-encoded α-1,6-mannosyltransferase, complementation experiments were performed in which a wild-type copy of och-1 was used to transform the och-1 knockout mutant. Wild-type morphology was restored to the mutant upon transformation with a copy of the och-1 gene, demonstrating that the mutant phenotypes were due to the loss of the OCH-1 α-1,6-mannosyltransferase activity (Fig. 1).
The och-1 mutant is defective in cell wall biosynthesis.
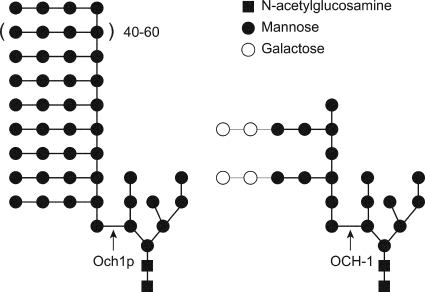
The enzyme encoded by och-1 is a close homolog of the S. cerevisiae Och1p and C. albicans Och1p (α-1,6-mannosyltransferases), which have been found to be required for the synthesis of the long polymannose outer chains on N-linked oligosaccharides attached to cell wall glycoproteins (13, 23). The yeast Och1p α-1,6-mannosyltransferase adds the initial mannose residue to the N-linked oligosaccharides on cell wall and secreted proteins, as shown in Fig. 3 (22, 33). S. cerevisiae and C. albicans och1 mutants have a defect in cell wall biogenesis (3, 11, 21). In order to test whether the N. crassa mutant was affected in cell wall biogenesis, we examined the growth of the och-1 mutant under various cell wall stress conditions. Calcofluor white (CFW) is a known inhibitor of cell wall biosynthesis which functions by binding to chitin and preventing chitin polymerization (8). The och-1 mutant was unable to grow in Vogel's liquid medium with 2% sucrose containing 10 mg/ml CFW, a concentration in which the wild type is able to grow. When tested for growth in other cell wall stress conditions, including 2 M glycerol (hyperosmolarity), 10% NaCl (salt stress), 0.01% SDS (detergent), and 0.05 mM hydrogen peroxide (oxidative stress), the och-1 mutant was unable to grow. Caspofungin acetate (Merck) is a potent antifungal agent that inhibits β-1,3-glucan synthase (5). The och-1 mutant was unable to grow in Vogel's liquid medium with 2% sucrose containing 10 μg/ml caspofungin. The och-1 mutant was also unable to grow at elevated (37°C) temperatures. It has been shown previously that sorbitol can be used for osmotic stabilization of cell wall-defective growth in S. cerevisiae (24, 36). When grown on an agar medium supplemented with sorbitol, the N. crassa och-1 mutant continued to grow in a colonial manner but had an improved growth rate. From these results, we conclude that the och-1 mutant has a severe defect in cell wall biosynthesis.
Fig. 3.
Representations of the N-linked oligosaccharides from S. cerevisiae (left structure) and N. crassa (right structure). In S. cerevisiae, Och1p adds the initial α-1,6-mannose of a long α-1,6-mannose chain to the N-linked oligosaccharide (GlcNac 2/mannose 8 structure). Side branches containing 2 α-1,2-mannoses and a terminal α-1,3-mannose are added to the mannoses in the α-1,6-chain. In N. crassa, OCH-1 adds the initial α-1,6-mannose of a short chain of α-1,6-mannoses to the N-linked oligosaccharide (GlcNac 2/mannose 8 structure). Approximately 30% of the mannoses in the chain have side branches containing two α-1,2-mannoses and two galactofuranoses.
The och-1 mutant has altered cell wall carbohydrate content.
To characterize the cell wall defect of the och-1 mutant, we examined the cell wall carbohydrate composition. In S. cerevisiae, Och1p is known to add an initial mannose residue to the N-linked oligosaccharide, which is then further modified to generate a large branched mannan structure. The yeast Och1p specifically recognizes the N-linked oligosaccharide (N-acetylglucosamine 2; mannose 8 structure) as a substrate for mannosylation, as shown in Fig. 3 (33). In the S. cerevisiae och1 mutant, the high-molecular-mass mannans are lost from secreted glycoproteins (34). The N. crassa cell wall does not contain the large mannans characteristic of S. cerevisiae. As shown in Fig. 3, the available data suggest that N-linked oligosaccharides are modified by shorter α-1,6-mannans that have α-1,2-mannan side chains capped by β-1,5-galactofuranose residues (20, 32, 33). If the N. crassa och-1 encoded the α-1,6-mannosyltransferase that mediates the addition of the initial mannose residue for the galactomannan, then we would expect to see a reduction in galactomannan in the mutant cell wall. A carbohydrate analysis of neutral sugars in the lyophilized cell walls from the och-1 mutant and the wild type was performed by GC/MS. The analysis showed that the mutant cell wall had an ∼90% reduction in mannose and an ∼75% reduction in galactose (Table 1). This large reduction in cell wall mannose and galactose suggests that the modification of N-linked oligosaccharides is defective in the och-1 mutant. We saw an increase in the glucose levels of the cell wall carbohydrate in the mutant, from 87.1% to 98.5% (Table 1), which suggests that the mutant may increase glucan synthesis as part of a compensatory mechanism in response to a weakened cell wall. These results indicate that the och-1 mutant has an altered cell wall composition.
Table 1.
Glycosyl composition analysis of the cell wall and linkage analysis of cell wall carbohydrates
| Glycosyl residue or linkage type | % molecular weight of total cell wall ± SD |
|
|---|---|---|
| Wild type | och-1 mutant | |
| Residues | ||
| Mannose (Man) | 7.5 ± 0.42 | 0.8 ± 0.07 |
| Galactose (Gal) | 5.2 ± 0.63 | 0.7 ± 0.42 |
| Glucose (Glc) | 87.1 ± 0.0 | 98.5 ± 0.35 |
| Linkage types | ||
| Terminally linked mannopyranosyl residue (t-Man) | 3.4 | 1.4 |
| 2-Linked mannopyranosyl residue (2-Man) | 5.4 | 0.6 |
| 6-Linked mannopyranosyl residue (6-Man) | 2.0 | 0.0 |
| 2,3-Linked mannopyranosyl residue (2,3-Man) | 1.7 | 0.2 |
| 2,6-Linked mannopyranosyl residue (2,6-Man) | 2.3 | 1.0 |
| Terminally linked galactofuranosyl residue (t-Galf) | 6.8 | 2.0 |
| Terminally linked galactopyranosyl residue (t-Gal) | 1.6 | 1.3 |
| 3-Linked galactopyranosyl residue (3-Gal) | 0.0 | 4.3 |
| 3-Linked glucopyranosyl residue (3-Glc) | 40.1 | 55.2 |
| 4-Linked glucopyranosyl residue (4-Glc) | 18.9 | 16.5 |
| 6-Linked glucopyranosyl residue (6-Glc) | 0.0 | 0.0 |
To further characterize the carbohydrates of the mutant cell wall, samples of mutant and wild-type cell walls were subjected to a glycosyl linkage analysis. The major sugars identified in both samples were 1,3-linked glucose and 1,4-linked glu- cose (Table 1). In agreement with the carbohydrate analysis, the mutant had an increase in the levels of 1,3-linked glucose (glucan), and the various types of mannose linkages and the terminal galactose linkages were dramatically reduced. The increased 1,3-glucan could be due to an increase in α-1,3-glucan, β-1,3-glucan, or both. An aniline blue binding experiment shows that the levels of β-1,3-glucan are elevated in the och-1 mutant (see Fig. S1 in the supplemental material). More importantly, the mutant cell wall was devoid of 1,6-linked mannose, providing verification that the och-1 gene encodes an α-1,6-mannosyltransferase. The major reduction in the levels of 1,6-linked mannose, 1,2-linked mannose, and terminal galactofuranose strongly suggests that in the absence of OCH-1 activity, the synthesis of N-linked galactomannan is compromised. The remaining levels of mannose and galactose present in the mutant cell wall could be the results of O-linked galactomannan synthesis and are consistent with a loss of N-linked mannosylation.
The och-1 mutant has an altered cell wall protein composition.
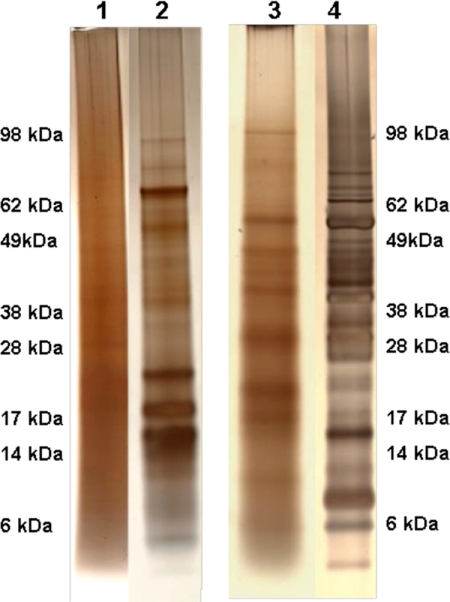
Because the galactomannan component of the cell wall is part of the glycosyl modification of the cell wall glycoproteins, we were interested in learning how the loss of mannosylation affects the protein composition of the cell wall. As one way of asking if the protein composition of the cell wall was affected in the och-1 mutant, a proteomic analysis of integral cell wall proteins released by TFMS, from cell walls of vegetative hyphae, was performed using nano-LC/MS-MS as previously described (26). TFMS not only cleaves the cell wall carbohydrate matrix to release the cell wall glycoproteins but also deglycosylates the glycoproteins to enhance their identification. SDS-PAGE analysis of cell wall proteins released by TFMS from 5 mg of lyophilized cell wall from och-1 mutant and wild-type cells (Fig. 4, lanes 1 and 3) showed that the mutant cell wall seemed to contain fewer identifiable protein bands and that the amount of protein in the bands was smaller than that in the wild-type cell wall. A nano-LC/MS-MS analysis of the protein extracts from the mutant cell wall identified only four proteins with N-terminal signal sequences (Table 2), and these proteins were identified by a limited number of peptides (see Table S1 in the supplemental material). A parallel analysis of the wild-type cell wall identified 15 proteins. We have carried out an extensive proteomic analysis of the wild-type cell and generally identify 15 to 20 cell wall proteins in an analysis (26). Three of the proteins identified in the mutant cell wall, ACW-9 (NCU06185), NCW-2 (NCU01752), and β-glucosidase (NCU09326), were proteins that we routinely identify in the wild-type cell wall. The other protein we identified in the mutant cell wall was a glycosylphosphatidylinositol (GPI)-anchored cell wall protein (NCU00175). The GPI-anchored cell wall protein (NCU00175) was identified as a cell wall protein for the first time in our analysis of the mutant cell wall. This cell wall protein may have been produced as part of the compensatory mechanism in response to a weakened cell wall by the mutant. What is striking about the proteomic analysis is the fact that most of the cell wall proteins we routinely identify in the wild-type cell wall were not identified in the mutant, suggesting that the och-1 mutant has an altered cell wall protein composition.
Fig. 4.
Proteomic analysis of cell wall proteins and secreted proteins. Lane 1, och-1 mutant cell wall proteins from 2 mg of cell wall; lane 2, och-1 mutant secreted proteins (20 μg); lane 3, wild-type cell wall proteins from 2 mg of cell wall; lane 4, wild-type secreted proteins (20 μg).
Table 2.
Cell wall proteins identified by nano-LC/MS-MS in the TFMS-deglycosylated cell wall fraction of vegetative hyphae
| Cell wall fraction | Protein | Locus tag |
|---|---|---|
| Wild type | ACW-1/CCG-15 | NCU08936 |
| ACW-2 | NCU00957 | |
| ACW-3 | NCU05667 | |
| ACW-5 | NCU07776 | |
| ACW-6 | NCU03530 | |
| ACW-7 | NCU09133 | |
| ACW-8 | NCU07277 | |
| ACW-9 | NCU06185 | |
| GH17-3/β-glucanase | NCU09175 | |
| GH16-1/mixed linked glucanase | NCU01353 | |
| GH16-7/glycoside hydrolase | NCU05974 | |
| GH72-4/GEL-2 | NCU07253 | |
| β-Glucosidase | NCU09326 | |
| NCW-1 | NCU05137 | |
| NCW-2 | NCU01752 | |
| och-1 mutant | ACW-9 | NCU06185 |
| β-Glucosidase | NCU09326 | |
| NCW-2 | NCU01752 | |
| GPI-anchored cell wall protein | NCU00175 |
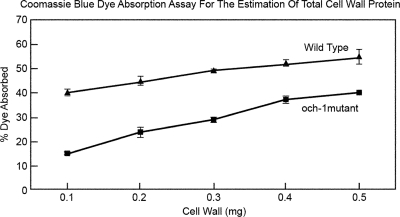
To determine if the och-1 mutant cell wall had a reduced level of protein incorporated into the cell wall matrix, we developed a Coomassie blue dye binding assay to examine the protein levels within wild-type and mutant cell walls. The assay consists of incubating increasing amounts of purified lyophilized cell wall with Coomassie blue dye and allowing the cell wall proteins to bind the dye. Following dye absorption, the cell walls were collected by centrifugation and the amount of unbound dye was determined. Although the assay is not linear with increasing cell wall samples, it is clear that the mutant cell wall does not absorb as much dye as the wild-type cell wall (Fig. 5). Based on the dye binding assay (Fig. 5), SDS-PAGE analysis (Fig. 4), and proteomic analysis (Table 2), we conclude that the mutant cell wall is deficient in protein.
Fig. 5.
Coomassie brilliant blue dye assay of cell wall protein. Wild-type and och-1 mutant cell walls were incubated in the presence of Coomassie blue, and the amounts of Coomassie blue absorbed by the cell wall were determined.
Large amounts of cell wall protein are released into the medium by the och-1 mutant.
With a reduced level of protein being incorporated into the cell wall, we considered whether cell wall proteins were being released into the growth medium. To assess whether cell wall proteins were being lost to the medium, we examined the amount of protein being released into the medium during a secretion assay. We grew the wild type and mutant in a liquid medium, collected the cells, and transferred them into fresh medium for a 2-h period of time. The cells and the medium were collected, and TCA was used to precipitate proteins from the growth medium. We also prepared cellular extracts from the collected cells. Protein assays of the cellular extracts and TCA-precipitated protein from the growth medium showed that wild-type cells secreted 0.5 μg of protein into the medium per mg of cellular protein. The och-1 mutant secreted 9.1 μg of protein per mg of cellular protein, an 18-fold increase in the amount of secreted protein. Clearly, the mutant cell released a larger amount of protein into the medium. This increase in protein released into the medium could be due to an increased secretion of cell wall protein, cell lysis, or a combination of the two.
To identify what types of proteins were being released into the medium, we carried out a proteomic analysis of the och-1 mutant secretome and compared it with the wild-type secretome. Secreted proteins are typically glycoproteins, and to remove the carbohydrates and maximize the identification of these proteins in SDS-PAGE and by proteomic analysis, we treated secreted protein samples from mutant and wild-type cells with TFMS (26). Examination of the secretome from och-1 mutant cultures by gel electrophoresis showed that there were more than 20 proteins being secreted (Fig. 4, lane 2). A nano-LC/MS-MS analysis of the secretome proteins from the och-1 mutant is given in Table 3. The mutant secreted 7 proteins that we normally see in the wild-type cell wall fraction (26): ACW-1 (NCU08936), ACW-12 (NCU08171), GH17-3/β-glucanase (NCU09175), GH72-4/GEL-2 (NCU07253), NCW-1 (NCU05137), NCW-5 (NCU00716), and GH16-5/cell wall glucanase (NCU05686). Most of these proteins were identified by the presence of a number of peptide fragments (see Table S2 in the supplemental material).
Table 3.
Cell wall proteins identified by nano-LC/MS-MS in the TFMS-deglycosylated protein released into the growth medium by vegetative hyphae
| Culture medium | Protein | Locus tag |
|---|---|---|
| Wild type | ACW-1/CCG-15 | NCU08936 |
| ACW-2 | NCU00957 | |
| ACW-3 | NCU05667 | |
| ACW-7 | NCU09133 | |
| ACW-12 | NCU08171 | |
| GH17-3/β-glucanase | NCU09175 | |
| GH16-7/glycoside hydrolase | NCU05974 | |
| GH72-5/GEL-1 | NCU08909 | |
| NCW-1 | NCU05137 | |
| GLA-1/glucamylase | NCU01517 | |
| Predicted secreted protein | NCU00399 | |
| CAT-3/catalase | NCU00355 | |
| Predicted secreted protein | NCU00265 | |
| och-1 mutant | ACW-1/CCG-15 | NCU08936 |
| ACW-12 | NCU08171 | |
| GH17-3/β-glucanase | NCU09175 | |
| GH72-4/GEL-2 | NCU07253 | |
| NCW-1 | NCU05137 | |
| NCW-5 | NCU00716 | |
| GLA-1/glucamylase | NCU01517 | |
| Predicted secreted protein | NCU00399 | |
| Predicted secreted protein | NCU08720 | |
| Endoglucanase | NCU05404 | |
| GH16-5/cell wall glucanase | NCU05686 | |
| CCG-14 | NCU07787 | |
| Exoglucanase | NCU03646 |
The och-1 mutant releases elevated levels of ACW-1 and GEL-1 into the medium.
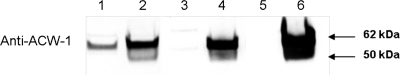
As a third way to follow the release of cell wall proteins into the medium, we chose to look at the secretion of two well-known cell wall proteins, ACW-1 and GEL-1, in a secretion assay. ACW-1 is a major GPI-anchored cell wall protein in N. crassa (7) and a homolog of the yeast Ecm33p cell wall protein. The release of ACW-1 into the medium was examined in a Western blot assay from the samples of the 2-h secretion assay described above. Figure 6 shows that ACW-1 was easily detected in the cellular extracts of both wild-type and mutant cells and that the levels of ACW-1 in the mutant cells were elevated, suggesting that ACW-1 expression is upregulated as part of the compensatory response to a weakened cell wall. C. albicans och1 mutants have been shown to activate the cell wall integrity pathway in response to a weakened cell wall (3). Examination of the secreted proteins shows that small amounts of ACW-1 were released into the medium by the wild-type cell but that the mutant cell released large amounts of ACW-1 (Fig. 6). The N. crassa GH72-5/GEL-1 gene encodes a GPI-anchored cross-linking enzyme with homology to the S. cerevisiae Gas1p and the A. fumigatus Gel1 β-1,3-glucanosyltransferase (26, 29). Gel1 has been shown to be an important cell wall biosynthetic enzyme (29). Western blot experiments using antibody directed against purified GEL-1 showed that the levels of GEL-1 in the cytosol were elevated in the mutant and that the mutant released easily detectable levels of GEL-1 into the medium, while the levels of GEL-1 released from the wild-type cell were below the level of detection in our system (Fig. 7).
Fig. 6.
Secretion of ACW-1 is elevated in the och-1 mutant. Western blot analysis of protein using antibody directed against ACW-1. Lane 1, wild-type cytosolic fraction (30 μg of cytosolic protein); lane 2, och-1 cytosolic fraction (30 μg of cytosolic protein); lane 3, wild-type secreted protein fraction (normalized to the amount of secreted protein from 30 μg of cytosolic protein); lane 4, och-1 secreted protein fraction (normalized to the amount of secreted protein from 30 μg of cytosolic protein); lane 5, wild-type secreted protein fraction (normalized to the amount of secreted protein from 500 μg of cytosolic protein); lane 6, och-1 secreted protein fraction (normalized to the amount of secreted protein from 500 μg of cytosolic protein).
Fig. 7.
Secretion of GEL-1 is elevated in the och-1 mutant. Western blot analysis using antibody directed against GEL-1. Lane 1, wild-type cytosolic fraction (30 μg of cytosolic protein); lane 2, och-1 cytosolic fraction (30 μg of cytosolic protein); lane 3, wild-type secreted protein fraction (normalized to the amount of secreted protein from 30 μg of cytosolic protein); lane 4, och-1 secreted protein fraction (normalized to the amount of secreted protein from 30 μg of cytosolic protein).
The results of our measurements showing that the mutant released much more protein into the medium than the wild-type cell, the proteomic analysis showing the presence of cell wall proteins in the medium from the mutant cells (Table 3), and the Western blot analyses demonstrating that the mutant released elevated levels of the ACW-1 and GEL-1 cell wall proteins into the medium (Fig. 6 and 7) clearly demonstrate that in the och-1 mutant, the cell wall glycoproteins move through the intracellular secretory pathway and pass through the cell wall without being cross-linked into the cell wall glucan-chitin matrix. This suggests that the proper mannosylation of the cell wall proteins is required for their recognition by cell wall cross-linking enzymes and that, in its absence, the incorporation of the proteins into the cell wall glucan-chitin matrix is compromised.
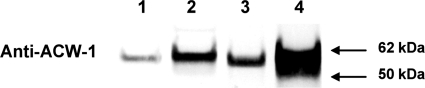
The och-1 mutant is defective in cross-linking ACW-1 into the cell wall.
Once cell wall proteins are released into the cell wall space by secretory vesicles, they become covalently cross-linked into the cell wall glucan-chitin matrix (5, 8). This process is expected to take some time, and cell wall proteins that have been secreted but not yet cross-linked into the wall matrix could be extracted from the cell wall. Our procedure for preparing the purified cell walls used throughout this paper includes a step in which the cell wall fraction is subjected to boiling in a buffer containing 1% SDS to remove proteins that are not covalently attached to the glucan-chitin matrix. If the och-1 mutant is defective in the incorporation of cell wall proteins into the matrix, we expected that boiling the cell wall in 1% SDS would release large amounts of cell wall protein. To directly test whether the och-1 mutant is defective in the incorporation of cell wall proteins, we carried out a Western blot analysis of ACW-1 in the protein fraction released from the cell walls during the treatment step in which the cell wall fraction was boiled in SDS. As shown in Fig. 8, some ACW-1 is released from wild-type cells by the treatment. We ascribe this to the fact that it takes some time for ACW-1 to be covalently incorporated into the wall in wild-type cells. In the wild-type cells, a comparison of the levels of unincorporated, cell wall-associated ACW-1 with the levels of ACW-1 in the cellular extract (representing ACW-1 in transit to the cell wall) shows that the level of unincorporated, cell wall-associated ACW-1 is slightly higher than the level of ACW-1 in transit. Figure 8 also shows that ACW-1 levels are upregulated in the och-1 cells as part of the compensatory mechanism turned on by the weakened state of the mutant cell wall, as mentioned earlier. Most importantly, the och-1 mutant cell wall has large amounts of unincorporated ACW-1, much more than could be explained as arising from the upregulation of ACW-1 expression. Figure 8 shows that the mutant is compromised in its ability to incorporate ACW-1 into the cell wall. A similar analysis for GEL-1 showed that the och-1 mutant cell wall had elevated levels of unincorporated GEL-1 (data not shown). Based on these results, along with the results of the secretion experiments, we conclude that the mannosylation of cell wall proteins by OCH-1 is required for the efficient covalent incorporation of the proteins into the cell wall matrix.
Fig. 8.
The covalent incorporation of ACW-1 into the cell wall is reduced in the och-1 mutant. Western blot analysis using antibody directed against the cell wall protein ACW-1. Lane 1, wild-type cytosolic fraction (30 μg of cytosolic protein); lane 2, och-1 cytosolic fraction (30 μg of cytosolic protein); lane 3, wild-type SDS-soluble cell wall protein fraction (normalized to 30 μg of cytosolic protein); lane 4, och-1 secreted protein fraction (normalized to 30 μg of cytosolic protein).
DISCUSSION
The N. crassa cell wall is composed of polysaccharides and proteins. The wall is known to contain β-1,3-glucan, α-1,3-glucan, chitin, and a number of identified cell wall glycoproteins (5, 7, 26). In the ER, the cell wall glycoproteins are modified by N-linked glycosylation, and many cell wall proteins receive a GPI anchor. Subsequently, galactomannans are added to the N-linked oligosaccharides. As the proteins pass through the secretory pathway, O-linked galactomannans are also added. After being secreted into the cell wall space, integral cell wall proteins are then cross-linked into the cell wall glucan-chitin matrix. The cross-linking of cell wall proteins into the cell walls of filamentous fungi has not been carefully examined.
Different species of fungi have been found to modify their glycoproteins differently (16). S. cerevisiae has been shown to modify glycoproteins by the addition of a large polymannose structure, while filamentous fungi generate shorter galactomannan structures (14). The posttranslational modifications of glycoproteins can be important targeting determinants, as demonstrated by the tagging of lysosomal protein for import into that organelle by the addition of a posttranslationally added mannose-6-phosphate residue. We demonstrate that in N. crassa the addition of an α-1,6-mannose residue to N-linked oligosaccharides is necessary for their covalent incorporation into the cell wall. This suggests that the incorporation of cell wall proteins into the glucan-chitin matrix may occur between N-linked galactomannan and the matrix.
In this report, we characterize the N. crassa och-1 knockout mutant. The och-1 mutant was identified in a screening of the Neurospora knockout library, and the gene (NCU00609) was found to be a homolog of known α-1,6-mannosyltransferases involved in initiating the addition of mannose to N-linked oligosaccharides. The encoded protein, OCH-1, contains 373 amino acids with a classical DXD sugar binding motif.
Examination of the och-1 mutant showed that it was a morphological mutant with a slow growth rate and a tight colonial phenotype (Fig. 1 and 2). A microscopic examination of the mutant showed that it has an abnormal pattern of hyphal growth and branching. Two interesting aspects of the mutant's phenotype are that when used as a female in mating, it is unable to “throw” the ascospores, and that during the asexual phase of the life cycle, it produces abnormal conidia that do not readily separate from each other. A variety of cell wall stress tests demonstrated that the mutant has a cell wall defect and is compromised in its ability to survive any of the cell wall stress reagents we tested. The altered morphology and growth characteristics of the mutant are likely to be the direct result of the weakened state of the mutant cell wall.
OCH1 mutants have been isolated from a number of different fungi. In S. cerevisiae, C. albicans, K. lactis, P. angusta, Y. lipolytica, S. pombe, and P. pastoris, loss of OCH1 gives rise to morphologies that correlate with alterations in the cell wall (2, 3, 9, 17, 33, 35, 37, 38). In contrast with these yeasts, knockout mutations of the OCH1 genes in the filamentous fungus A. fumigatus do not result in a cell wall-defective phenotype (18). The N. crassa och-1 mutant has a phenotype more in line with the phenotypes seen in S. cerevisiae. There are, however, some important differences between the N. crassa och-1 mutant and the S. cerevisiae mutant. In S. cerevisiae, GPI-anchored cell wall proteins can be cross-linked into the wall through the attachment of β-1,6-glucan to GPI-anchored oligosaccharides (15, 25), and the yeast mutant has not been reported as having a large-scale loss of cell wall protein into the growth medium. In N. crassa, which lacks β-1,6-glucan (4) (Table 1), there is no evidence for the cross-linking of GPI anchors into the cell wall, and the och-1 mutant releases GPI-anchored proteins into the growth medium.
To characterize the carbohydrate polymers in the och-1 mutant cell wall, we examined the carbohydrate composition and the linkages present between the different sugars in wild-type and mutant cell walls. We found that the mutant cell wall had an altered carbohydrate composition, with a major decrease in mannose and galactose residues. This suggests that most of the mannose and galactose present in the N. crassa cell wall is associated with cell wall glycoproteins. Linkage analysis showed that the mutant had a complete lack of 1,6-linked mannose residues. The α-1,6-mannosylation of cell wall proteins has been shown to be associated with N-linked oligosaccharides in S. cerevisiae and C. albicans (13, 14). The carbohydrate analysis also showed a major decrease in mannose residues with 1,2-linkages and galactose residues. The data suggest that OCH-1 is required for the synthesis of N-linked galactomannan (Fig. 3). A comparable cell wall carbohydrate analysis has not been done for the yeast OCH1 mutants, but it has been shown that the cell wall and secreted proteins from the mutants lack the high-mannan structure shown in Fig. 3 (1, 3, 34).
The most important conclusion from our experiments is that the mannosylation of N-linked oligosaccharides is required for the incorporation of cell wall proteins into the cell wall matrix. Using such techniques as SDS-PAGE of TFMS-released cell wall protein, proteomic analysis of cell wall protein, and a Coomassie blue dye binding assay, we found that the och-1 mutant cell wall had less protein than the wild-type cell wall. This provides a partial explanation for the reduced levels of mannose and galactose in the carbohydrate analysis of the och-1 cell wall. Mannose and galactose are incorporated into the wall as oligosaccharides associated with cell wall protein. Proteomic analysis of purified mutant cell walls failed to identify a number of proteins we have routinely found in wild-type cell walls.
We also found that when grown in a liquid medium, the och-1 mutant released large amounts of proteins into the medium. Proteomic analysis of the secreted proteins demonstrated that the growth medium contained a number of proteins that we previously identified as cell wall proteins in wild-type cell walls. Western blot analysis of two well-known cell wall proteins, ACW-1 and GEL-1 (26), demonstrated that large amounts of these cell wall proteins were being released into the medium by the mutant, while the levels of ACW-1 and GEL-1 released from wild-type cells were small or negligible (Fig. 6 and 7). The data indicate that in the absence of OCH-1-directed α-1,6-mannosylation, cell wall proteins are not being covalently incorporated into the cell wall glucan-chitin matrix and are being released into the medium.
We directly tested whether the incorporation of cell wall proteins into the cell wall matrix was compromised in the och-1 mutant and found that the levels of unincorporated, cell wall-associated ACW-1 were elevated in the mutant cell wall. As shown in Fig. 8, the mutant cell wall does contain elevated levels of ACW-1 that have not been covalently linked into the cell wall matrix. We conclude that in the absence of α-1,6-mannosylation, the proteins are not efficiently cross-linked into the wall. Our results support a model for cell wall biogenesis in which the cross-linking of cell wall protein into the wall requires some elements of the N-linked galactomannan, whose synthesis begins with the OCH-1-mediated addition of an α-1,6-linked mannose residue to the N-linked oligosaccharide. The enzymes used to cross-link the proteins into the cell wall may recognize some aspects of the N-linked oligosaccharide-galactomannan structure and mediate the cross-linking of the structure into the glucan-chitin matrix.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by NIH grant R01 078589. The carbohydrate analysis was supported in part by the Department of Energy-funded Center for Plant and Microbial Complex Carbohydrates (DE-FG09-93ER-20097).
We are grateful to David Toczynski for the polyclonal rabbit antibody directed against GH72-5/GEL-1. We thank Alan Siegel and James Stamos for assistance in preparing figures.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 24 September 2010.
REFERENCES
- 1.Ballou C. E. 1990. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 185:440–479 [DOI] [PubMed] [Google Scholar]
- 2.Barnay-Verdier S., Boisrame A., Beckerick J.-M. 2004. Identification and characterization of two α-1,6-mannosyltransferases, Anl1p and Och1p, in the yeast Yarrowia lipolytica. Microbiology 150:2185–2195 [DOI] [PubMed] [Google Scholar]
- 3.Bates S., Hughes H. B., Munro C. A., Thomas W. P. H., MacCallum D. M., Bertram G., et al. 2006. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 281:90–98 [DOI] [PubMed] [Google Scholar]
- 4.Borkovich K. A., Alex L. A., Yarden O., Freitag M., Turner G. E., Read N. D., et al. 2004. Lessons from the genome sequences of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouffard F. A., Zambias R. A., Dropinski J. F., Balkovec J. M., Hammond M. L., Abrazzo G. K., et al. 1994. Synthesis and antifungal activity of novel cationic pneumocandin B(o) derivatives. J. Med. Chem. 37:222–225 [DOI] [PubMed] [Google Scholar]
- 6.Bowman S. M., Free S. J. 2006. The structure and synthesis of the fungal cell wall. Bioessays 28:799–808 [DOI] [PubMed] [Google Scholar]
- 7.Bowman S. M., Piwowar A., Al Dabbous M., Vierula J., Free S. J. 2006. Mutational analysis of the glycosylphosphatidylinositol (GPI) anchor pathway demonstrates that GPI-anchored proteins are required for cell wall biogenesis and normal hyphal growth in Neurospora crassa. Eukaryot. Cell 5:587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaffin W. L. 2008. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 72:495–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi B.-K., Bobrowicz P., Davidson R. C., Hamilton S. R., Kung D. H., Li H., et al. 2003. Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. Proc. Natl. Acad. Sci. U. S. A. 100:5022–5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., Litvinkova L., et al. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U. S. A. 103:10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen P. J., Schultz J., Horecka J., Stevenson B. J., Jigami Y., Sprague G. F. 2000. Defects in protein glycosylation cause SHO1-dependent activation of STE12 signaling pathway in yeast. Genetics 155:1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis R. H., DeSerres F. J. 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 27:79–143 [Google Scholar]
- 13.Dean N. 1999. Asparagine-linked glycosylation in yeast Golgi. Biochim. Biophys. Acta 1426:309–322 [DOI] [PubMed] [Google Scholar]
- 14.Deshpande N., Wilkins M. R., Packer N., Nevalainen H. 2008. Protein glycosylation pathways in filamentous fungi. Glycobiology 18:626–637 [DOI] [PubMed] [Google Scholar]
- 15.Fujii T., Shimoni H., Iimura Y. 1999. Structure of the glucan-binding sugar chain of Tip1p, a cell wall protein of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1427:133–144 [DOI] [PubMed] [Google Scholar]
- 16.Gemmill T. R., Trimble R. B. 1999. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta 1426:227–237 [DOI] [PubMed] [Google Scholar]
- 17.Kim M. W., Kim E. J., Kim J.-Y., Park J.-S., Oh D.-B., Shimma Y.-I., et al. 2006. Functional characterization of the Hansenula polymorpha HOC1, OCH1, and OCR1 genes as members of the yeast OCH1 mannosyltransferase family involved in protein glycosylation. J. Biol. Chem. 281:6261–6272 [DOI] [PubMed] [Google Scholar]
- 18.Lambou K., Perkhofer S., Fontaine T., Latge J. P. 2010. Comparative functional analysis of the OCH1 mannosyltransferase families in Aspergillus fumigatus and Saccharomyces cerevisiae. Yeast 27:625–636 [DOI] [PubMed] [Google Scholar]
- 19.Latge J.-P. 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 66:279–290 [DOI] [PubMed] [Google Scholar]
- 20.Leal J. A., Jimenez-Barbero J., Gomez-Miranda B., Prieto A., Domenech J., Bernabe M. 1996. Structural investigation of the cell-wall galactomannan from Neurospora crassa and N. sitophila. Carbohydr. Res. 283:215–222 [DOI] [PubMed] [Google Scholar]
- 21.Lee B. N., Elion E. A. 1999. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc. Natl. Acad. Sci. U. S. A. 96:12679–12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehle L., Eiden A., Lehnert K., Haselbeck A., Kopetzki E. 1995. Glycoprotein biosynthesis in Saccharomyces cerevisiae: ngd29, an N-glycosylation mutant allelic to och1 having a defect in the initiation of outer chain formation. FEBS Lett. 370:41–45 [DOI] [PubMed] [Google Scholar]
- 23.Lesage G., Bussey H. 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu C.-F., Montijn R. C., Brown J. L., Klis F., Kurjan J., Bussey H., Lipke P. N. 1995. Glycosylphosphatidylinositol-dependent cross-linking of alpha-agglutinin and beta-1,6-glucan in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 128:333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maddi A., Bowman S. M., Free S. J. 2009. Trifluoromethansulfonic acid-based proteomic analysis of cell wall and secreted proteins of the ascomycetous fungi Neurospora crassa and Candida albicans. Fungal Genet. Biol. 46:768–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis T., Fritsch E. F., Sambrook C. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 28.Margolin B. S., Freitag M., Selker E. U. 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa. Fungal Genet. Newsl. 44:34–36 [Google Scholar]
- 29.Mouyna I., Fontaine T., Vai M., Monod M., Fonzi W. A., Diaquin M., et al. 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275:14882–14889 [DOI] [PubMed] [Google Scholar]
- 30.Munro S. 2001. What can yeast tell us about N-linked glycosylation in the Golgi apparatus? FEBS Lett. 498:223–227 [DOI] [PubMed] [Google Scholar]
- 31.Nakajima T., Yoshida M., Hura N., Matsuda K. 1984. Structure of the cell wall proteogalactomannan from Neurospora crassa. I. Purification of the proteoheteroglycan and characterization of alkali-labile oligosaccharides. J. Biochem. 96:1005–1011 [DOI] [PubMed] [Google Scholar]
- 32.Nakajima T., Yoshida M., Nakamura M., Hura N., Matsuda K. 1984. Structure of the cell wall proteogalactomannan from Neurospora crassa. II. Structural analysis of the polysaccharide part. J. Biochem. 96:1013–1020 [DOI] [PubMed] [Google Scholar]
- 33.Nakanishi-Shindo Y., Nakayama K., Tanaka A., Toda Y., Jigami Y. 1993. Structure of the N-linked oligosaccharides that show the complete loss of α-1,6-polymannose outer chain from och1, och1 mnn1, and mnn1 alg3 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 268:26338–26345 [PubMed] [Google Scholar]
- 34.Nakayama K., Nagasu T., Shimma Y., Kuromitsu J., Jigami Y. 1992. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagines-linked oligosaccharides. EMBO J. 11:2511–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohashi T., Ikeda Y., Tanaka N., Nakakita S., Natsuka S., Giga-Hama Y., Takegawa K. 2009. The och1 mutant of Schizosaccharomyces pombe produces galactosylated core structures of N-linked oligosaccharides. Biosci. Biotechnol. Biochem. 73:4070–4414 [DOI] [PubMed] [Google Scholar]
- 36.Scrimale T., Didone L., de Mesy Bentley K. L., Krysan D. J. 2009. The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 20:164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uccelletti D., Farina F., Rufini S., Magnelli P., Abeijon C., Palleschi C. 2006. The Kluyveromyces lactis α-1,6-mannosyltransferase KlOch1p is required for cell-wall organization and proper functioning of the secretory pathway. FEMS Yeast Res. 6:449–457 [DOI] [PubMed] [Google Scholar]
- 38.Yoko-o T., Tsukahara K., Watanabe T., Hata-Sugi N., Yoshimatsu K., Nagasu T., Jigami Y. 2001. Schizosaccharomyces pombe och1+ encodes α-1,6-mannosyltransferase that is involved in outer chain elongation of N-linked oligosaccharides. FEBS Lett. 489:75–80 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.