Abstract
Entamoeba histolytica, the protist that causes amebic dysentery and liver abscess, has a truncated Asn-linked glycan (N-glycan) precursor composed of seven sugars (Man5GlcNAc2). Here, we show that glycoproteins with unmodified N-glycans are aggregated and capped on the surface of E. histolytica trophozoites by the antiretroviral lectin cyanovirin-N and then replenished from large intracellular pools. Cyanovirin-N cocaps the Gal/GalNAc adherence lectin, as well as glycoproteins containing O-phosphodiester-linked glycans recognized by an anti-proteophosphoglycan monoclonal antibody. Cyanovirin-N inhibits phagocytosis by E. histolytica trophozoites of mucin-coated beads, a surrogate assay for amebic virulence. For technical reasons, we used the plant lectin concanavalin A rather than cyanovirin-N to enrich secreted and membrane proteins for mass spectrometric identification. E. histolytica glycoproteins with occupied N-glycan sites include Gal/GalNAc lectins, proteases, and 17 previously hypothetical proteins. The latter glycoproteins, as well as 50 previously hypothetical proteins enriched by concanavalin A, may be vaccine targets as they are abundant and unique. In summary, the antiretroviral lectin cyanovirin-N binds to well-known and novel targets on the surface of E. histolytica that are rapidly replenished from large intracellular pools.
Entamoeba histolytica causes amebic dysentery and liver abscess in the developing world (10, 20, 29). We are interested in E. histolytica glycoproteins containing Asn-linked glycans (N-glycans) for numerous reasons. E. histolytica makes an N-glycan precursor that contains 7 sugars (Man5GlcNAc2-PP-dolichol) rather than 14 sugars (Glc3Man9GlcNAc2-PP-dolichol) made by most animals, plants, and fungi (21, 31, 44). E. histolytica N-glycans are used for quality control of glycoprotein folding in the endoplasmic reticulum (ER) lumen, and there is positive selection for sites of N-linked glycosylation in secreted and membrane proteins of E. histolytica (5, 11, 53).
Unprocessed Man5GlcNAc2, by far the most abundant E. histolytica N-glycan, is present on the plasma membrane and vesicular membranes (31). The antiretroviral lectin cyanovirin-N, which is specific for α-1,2-linked mannose present on unprocessed N-glycans, binds E. histolytica N-glycans and forms aggregates or caps on the surface of E. histolytica trophozoites (1, 25, 31, 44, 45). E. histolytica glycoproteins are also capped by the plant lectin concanavalin A (ConA), which has a broader carbohydrate specificity (mannose and glucose) than cyanovirin-N (3, 16, 18, 19). Heavy subunits of the Gal/GalNAc lectin, the most important E. histolytica vaccine candidate, have 7 to 10 potential sites for N-linked glycosylation (32, 39, 43). Inhibition of N-glycan synthesis results in Gal/GalNAc lectins that are unable to bind to sugars on host epithelial cells.
Carbohydrates appear to be an important target on the surface of E. histolytica as anti-proteophosphoglycan (PPG) monoclonal antibodies bind to O-phosphodiester-linked glycans and protect animal models from amebic infection (6, 33, 35, 40, 48). Lectin affinity columns are a powerful method for enriching unique parasite glycoproteins that may be identified by mass spectrometry (MS) of tryptic fragments (17, 55). For example, we recently used the plant lectin wheat germ agglutinin to dramatically enrich glycoproteins with short N-glycans of Giardia (42).
The goal of the present studies was to explore further the interaction of the antiretroviral lectin cyanovirin-N with E. histolytica trophozoites in vitro. Questions asked included the following: Are E. histolytica glycoproteins with N-glycans replenished on the plasma membrane after capping with cyanovirin-N? What is the effect of cyanovirin-N capping on other amebic virulence factors and/or vaccine candidates (e.g., the Gal/GalNAc lectin and PPG)? Is capping by cyanovirin-N mediated by actin, as described for capping by the Gal/GalNAc lectin and ConA? What is the effect of the cyanovirin-N on amebic phagocytosis of mucin-coated beads, a surrogate assay for virulence? Which trophozoite glycoproteins are potential targets of cyanovirin-N (identified by mass spectrometry of lectin-enriched E. histolytica proteins)? Are any of them potential vaccine candidates?
MATERIALS AND METHODS
Fluorescence microscopy.
Logarithmic-phase trophozoites of the genome project HM1 strain of E. histolytica were chilled to release adherent organisms, concentrated by low-speed centrifugation, and washed in chilled phosphate-buffered saline (PBS) (29). For surface labeling, trophozoites were incubated for 30 min at 4°C in cyanovirin-N labeled with either Alexa Fluor 488 (green) or Alexa Fluor 585 (red) (1, 31). Cyanovirin-N-labeled trophozoites were washed three times in PBS and then fixed for 10 min at 4°C in 2% paraformaldehyde in 100 mM phosphate, pH 7.4. For capping experiments, trophozoites labeled with cyanovirin-N were warmed to 37°C for 15 min prior to fixation.
To determine whether N-glycans are replenished on the surface of trophozoites capped with cyanovirin-N, we treated capped and fixed organisms with PBS containing 2% bovine serum albumin (BSA) to quench free aldehydes and then labeled them with cyanovirin-N conjugated to a different Alexa Fluor dye. To demonstrate actin fibrils, we permeabilized capped and fixed organisms with 0.1% Triton X-100 and then stained them with 0.1 mg/ml phalloidin conjugated to Alexa Fluor 480 for 1 h at 4°C (18). To determine whether cyanovirin-N cocaps other E. histolytica antigens, we incubated capped and fixed E. histolytica with an Alexa Fluor-labeled mouse monoclonal antibody to the Gal/GalNAc lectin (a generous gift of William Petri) (32, 39). Alternatively, capped and fixed E. histolytica organisms were incubated with an Alexa Fluor-labeled mouse monoclonal antibody to the E. histolytica PPG (a generous gift of Michael Duchêne) (33).
For internal labeling with cyanovirin-N, we fixed E. histolytica trophozoites for 10 min at 4°C, and Triton X-100 was added to a final concentration of 0.1% for 1 min. Cells were gently pelleted by centrifugation, washed with PBS-2% BSA, and then incubated with cyanovirin-N, as described above. Similar methods were performed for labeling the surface and interior of E. histolytica with anti-Gal/GalNAc antibodies and for determining whether Gal/GalNAc lectins are replenished on the parasite surface after capping.
The nuclei of E. histolytica cells labeled with cyanovirin-N or the anti-Gal/GalNAc antibody were stained with 0.1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI), SlowFade antifade solution (Invitrogen) was added, and organisms were visualized with a DeltaVision deconvoluting microscope (Applied Precision, Issaquah, WA) with channels for each fluorochrome. Images were taken at a primary magnification of ×100 and deconvolved using Applied Precision's softWoRx software.
Phagocytosis of mucin-coated spheres.
Assays for E. histolytica phagocytosis of mucin-coated spheres were preformed, as described previously (18). Briefly, E. histolytica trophozoites (105/ml) were incubated with microspheres (107/ml) in culture medium for 15 min at 37°C and then fixed in 2% paraformaldehyde. Phagocytosed beads within 100 cells in each group were counted with a fluorescence microscope. The results were plotted using a modified box and whiskers plot to illustrate the significant shift in phagocytosed beads between the cyanovirin-N-treated and untreated trophozoites. In addition, an analysis of variance (ANOVA) was used to evaluate the statistical significance of the differences in phagocytosis and calculate the P value.
Concanavalin A affinity chromatography of Entamoeba proteins.
Logarithmic-phase E. histolytica trophozoites were harvested on ice, washed in PBS, and sonicated in an ice-water slurry containing 0.1% Triton X-100 and EDTA-free Complete protease inhibitor cocktail (Roche). Insoluble material was removed by centrifugation (at >12,000 × g). Soluble proteins were applied to a ConA-Sepharose column (EY Laboratories, Inc.) (17, 55), and the column was subsequently rinsed with PBS. To avoid collecting proteins that were nonspecifically bound to the ConA resin, we selectively eluted E. histolytica glycoproteins with 50 mM α-methyl mannoside rather than with SDS. Proteins eluted from the ConA column were run on SDS-PAGE gels containing a 4 to 20% gradient of acrylamide (Bio-Rad). In a parallel lane were E. histolytica proteins that were treated twice with 1,000 units of peptide:N-glycanase F (PNGaseF; New England Biolabs) for 9 h at 37°C in NEB G7 phosphate buffer. E. histolytica proteins were transferred to nitrocellulose membranes by electroporation, incubated with horseradish peroxidase (HRP)-conjugated cyanovirin-N, and developed with ECL chemiluminescent substrate (Pierce).
Mass spectrometry.
Mass spectrometry of E. histolytica proteins was performed using two different methods, as two different mass spectrometers were used. For the linear trap quadrupole (LTQ) ProteomeX ion trap mass spectrometer (Thermo Finnigan) present at the Boston University Proteomics Core Facility, E. histolytica peptides were prepared and analyzed using methods that were essentially the same as those used to identify peptides from the E. histolytica cyst wall (54) or from lectin affinity preparations of Giardia glycoproteins (42, 56). In addition, some samples were run on a similar Thermo Finnigan mass spectrometer at the Cancer Center at the Massachusetts Institute of Technology (MIT). Mass spectra were compared to tryptic digests of E. histolytica proteins predicted from whole-genome sequencing using SEQUEST, GPM (The Global Proteome Machine Organization [www.thegpm.org]) open source software, or Mascot software (13, 22, 23).
Two-dimensional protein gels were simulated from mass spectrometry data using GPM, where the position of each protein was determined by its predicted pI and mass, not including posttranslational modifications, and the size of the spot was proportional to the number of observed ions corresponding to that protein. These two-dimensional gels highlighted relative abundances of secreted and plasma membrane proteins (defined by either an N-terminal ER-targeting sequence or a transmembrane helix [TMH]) (24, 36) versus nucleocytoplasmic proteins (defined by the absence of these features). The Excel files in the supplemental material each show the merged results of four mass spectrometric experiments using Mascot software. Proteins previously identified as hypothetical because they showed no homology to other eukaryotic proteins were assigned simple names based upon their topology (e.g., unique nucleocytosolic protein, unique secreted protein, unique type 1 membrane protein, unique glycosylphosphatidylinositol [GPI]-anchored protein, etc.). GPI anchors were predicted using the algorithms of Eisenhaber et al. (15). Where there seemed a good match in the nonredundant (NR) database as demonstrated by a high score with BLASTP (2), we renamed the E. histolytica protein (e.g., “cysteine proteinase” or “disulfide isomerase” rather than “conserved hypothetical protein”).
To identify occupied N-glycan sites, we used a two-dimensional chromatography approach. A peptide mixture from the tryptic digestion of ConA-enriched E. histolytica glycoproteins was treated with PNGaseF to remove N-glycans and to convert Asn to Asp. PNGaseF-treated peptides and an untreated control were separated using strong cationic exchange (SCX) chromatography prior to Nanoflow reversed-phase high-performance liquid chromatography (HPLC)-coupled tandem mass spectrometry (MS/MS). SCX chromatography was performed on a Beckman Coulter ProteomeLab PF2D using a PolySulfoethyl A column. The buffers used were the following; buffer A, 7 mM KH2PO4, pH 2.65, 30% acetonitrile (ACN; vol/vol); buffer B, 7 mM KH2PO4, 350 mM KCl, pH 2.65, 30% ACN (vol/vol); buffer C, 50 mM K2HPO4, 500 mM NaCl, pH 7.5. Peptides were separated using a linear gradient from 0% to 70% of buffer B in 30 min, from 70% to 100% of buffer B in 10 min, and then 100% of buffer B for 6 min. The flow rate used was 0.5 ml/min. Thirteen 2-min fractions were collected. Each fraction was dried to eliminate acetonitrile before LC-MS/MS.
LC-MS/MS was performed using a nanoAcquity ultra-performance liquid chromatography (UPLC) capillary system (Waters Corp., Milford, MA), coupled to an LTQ-Orbitrap hybrid mass spectrometer (ThermoFisher Scientific, San Jose, CA) equipped with a TriVersa NanoMate ion source (Advion, Ithaca, NY). Sample concentration and desalting were performed online using a nanoAcquity UPLC trapping column (180 μm by 20 mm; packed with 5-μm, 100-Å-pore-size Symmetry C18 material; Waters Corp.) at a flow rate of 15 μl/min for 1 min. Separation was accomplished on a nanoAcquity UPLC capillary column (100 μm by 100 mm; packed with 1.7-μm,130-Å-pore-size bridged ethyl hybrid [BEH] C18 material; Waters Corp.). A linear gradient of A and B buffers (buffer A, 3% ACN–0.1% formic acid [FA]; buffer B, 97% ACN–0.1% FA) from 7% to 45% buffer B over 124 min was used at a flow rate of 0.5 μl/min to elute peptides into the mass spectrometer. Columns were washed and reequilibrated between LC-MS/MS experiments. Electrospray ionization was carried out at 1.7 kV using the NanoMate, with the LTQ heated capillary set to 150°C.
Mass spectra were acquired in the Orbitrap in the positive-ion mode over the range of m/z 300 to 2,000 at a resolution of 60,000. Mass accuracy after internal calibration was within 4 ppm. Simultaneously, tandem MS spectra were acquired using the LTQ for the five most abundant, multiply charged species in the mass spectrum with signal intensities of >8,000 noise levels. MS/MS collision energies were set at 35%, using helium as the collision gas, and MS/MS spectra were acquired over a range of m/z values dependent on the precursor ion. Dynamic exclusion was set such that MS/MS for each species was acquired a maximum of twice. All spectra were recorded in profile mode for further processing and analysis.
Xcalibur software was used for MS and MS/MS data analysis, while peptide and protein assignments were conducted using Mascot to search against the E. histolytica database employing an error window of 6 ppm on the precursor ions and 0.6 Da on the fragment ions. Table 1 shows occupied N-glycan sites where the predicted Asn was converted to Asp by PNGaseF treatment, resulting in a shift in mass of +1 Da.
Table 1.
E. histolytica glycoproteins with occupied N-glycan sites as shown by PNGaseF treatment and mass spectrometry
| Accession no. | Protein name or description | Mascot score | Sequence coverage (%) | Glycosylation site(s)a |
|---|---|---|---|---|
| EHI_012270 | Gal/GalNAc lectin heavy subunit | 2,478 | 43 | ANLTER |
| EHI_133900 | Gal/GalNAc lectin heavy subunit | 2,007 | 45 | QYNTSCEPK, VSNCTEDLVR |
| EHI_006980 | Gal/GalNAc lectin intermediate subunit | 1,011 | 36 | NVSNDCECNDKHIPTSIDK, DGFYQIENATDGVYCSPCPAK |
| EHI_065330 | Gal/GalNAc lectin intermediate subunit | 959 | 43 | SPYSNCTTCIESNYYPK |
| EHI_077530 | Unique GPI-anchored glycoprotein | 201 | 37 | NETSVAVSEENFK |
| EHI_138750 | Receptor-kinase | 143 | 13 | YDVPYNESLDK, SCQTCNSTIDPK |
| EHI_093710 | Secreted glycoprotein similar to Igl1 | 141 | 34 | VNGSCIQTIDACDTYVNPLFEK, YNFYLPTNGTCER |
| EHI_161040 | Unique secreted protein with PKEDQ repeats | 384 | 42 | ENECNETEEYK |
| EHI_008120 | Unique type 1 membrane glycoprotein | 201 | 18 | SFLLNITK |
| EHI_109920 | Unique basic secreted glycoprotein | 198 | 26 | NVTDYDVHSVR |
| EHI_054530 | Serine carboxypeptidase | 151 | 12 | YFINDTIYSK |
| EHI_151440 | Cysteine proteinase | 138 | 27 | DYPYTATNGTCQYDADK |
| EHI_093970 | Cysteine proteinase | 133 | 16 | FNVTSITTK |
| EHI_188160 | Unique ER glycoprotein | 107 | 23 | KPIAYFTNSTLGLTK |
| EHI_022130 | Unique secreted glycoprotein | 100 | 29 | IPTENR(T), VNASIIESYKK |
| EHI_133530 | Unique secreted glycoprotein | 78 | 31 | VNASIIESLK, ALSDAVSNVLPNTTDK |
| EHI_194850 | Unique secreted glycoprotein | 65 | 8 | INVIDAFEGDTNLTIPK |
| EHI_178470 | Unique type 1 transmembrane glycoprotein | 62 | 9 | TNYSFPSPPSVSEYSDFK |
| EHI_096530 | Asp-rich type 1 membrane glycoprotein | 62 | 8 | YISSVNCSATENYETK |
| EHI_024530 | Unique type 1 membrane glycoprotein | 52 | 23 | EIVNVTGQIGTLITR |
| EHI_098000 | Unique type 1 membrane glycoprotein | 51 | 17 | VSSGNVSYR |
| EHI_180770 | Unique type 1 membrane glycoprotein | 50 | 15 | YIQGDDNVINR |
| EHI_048140 | Unique type 2 membrane glycoprotein | 50 | 28 | VSNTTDSGNATSTK |
| EHI_185260 | Unique type 1 membrane glycoprotein | 50 | 5 | FGYASLFADPTNCSVR |
| EHI_107300 | Unique GPI-anchored glycoprotein | 50 | 23 | IYCDPVEGPIIFNFNNSK |
Boldface indicates where the predicted Asn was converted to Asp by PNGaseF treatment.
MS data.
Mass spectrometric data have been deposited in AmoebaDB (4).
RESULTS
E. histolytica glycoproteins are capped by the antiretroviral lectin cyanovirin-N and then replenished from large intracellular pools.
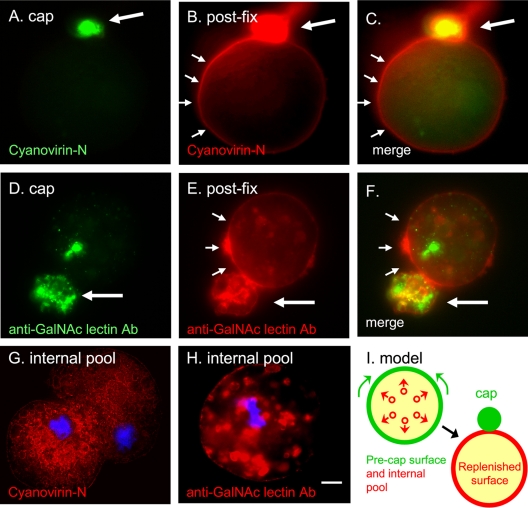
Cyanovirin-N, which labels α-1,2-linked mannose residues in unprocessed N-glycans, evenly stains the surface of E. histolytica trophozoites either kept at 4°C to prevent capping or fixed prior to labeling (see Fig. S1 in the supplemental material). Glycoproteins containing N-glycans are capped on the surface of E. histolytica trophozoites when cyanovirin-N-labeled trophozoites are warmed to 37°C (Fig. 1A and C, large arrows). Subsequent labeling of fixed parasites with cyanovirin-N conjugated to a different Alexa Fluor dye shows that many glycoproteins containing N-glycans are replenished on the surface of E. histolytica trophozoites away from the cap (Fig. 1B and C, small arrows). Similarly, Gal/GalNAc lectins that are capped by a monoclonal antibody are replenished on the surface of E. histolytica away from the cap (Fig. 1D to F; see also Fig. S1 in the supplemental material).
Fig. 1.
Capped cyanovirin-N-binding glycoproteins and Gal/GalNAc lectins are rapidly replenished on the E. histolytica surface from large intracellular pools. (A) Cyanovirin-N forms a tight green cap (large arrow) on the surface of trophozoites warmed for 15 min prior to fixation. (B) The same organism was fixed and then labeled with red cyanovirin-N. Cyanovirin-N binding sites that are replenished on the E. histolytica surface are marked with small arrows. (D) Similar results were obtained with a monoclonal antibody to the Gal/GalNAc lectin, which forms a tight cap on trophozoites prior to fixation (green). (E) After fixation, the Gal/GalNAc lectin that is replenished on the parasite surface is shown with a red anti-Gal/GalNAc antibody. Merged panels are as indicated. (G) Cyanovirin-N (red) binds to a large interior pool of glycoproteins in a fixed and permeabilized ameba. (H) Similar results were obtained with antibodies to the Gal/GalNAc lectin. In panels G and H nuclei are stained with DAPI (blue). Bar, 5 μm. (I) A model for the redistribution of glycoproteins binding cyanovirin-N or anti-GalNAc antibodies from large intracellular pools during capping. See Fig. S1 in the supplemental material for images of cyanovirin-N and anti-Gal/GalNAc antibody labeling of uncapped parasites. Ab, antibody.
The source of the new glycoproteins with N-glycans on the E. histolytica surface after capping is likely the large intracellular pool of glycoproteins containing N-glycans. These glycoproteins are clearly visible on cyanovirin-N labeling of fixed and permeabilized E. histolytica trophozoites (Fig. 1G). Cyanovirin-N binds in a reticular or membrane pattern to glycoproteins of permeabilized E. histolytica. In contrast, the anti-Gal/GalNAc lectin monoclonal antibody labels the membranes and the contents of numerous secretory vesicles throughout the E. histolytica trophozoite (Fig. 1H). A model for the replenishment of capped E. histolytica surface glycoproteins from large intracellular pools is shown in Fig. 1I and discussed below.
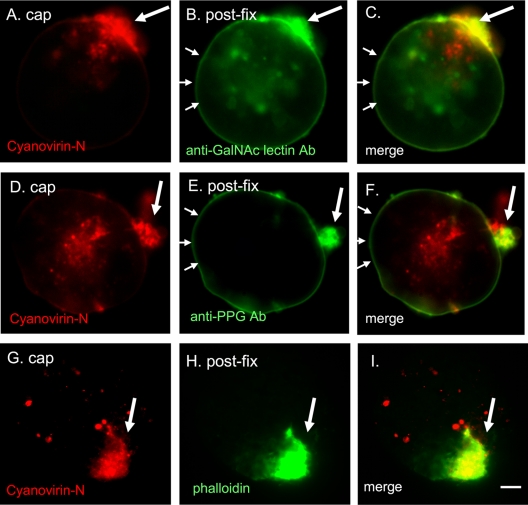
Cyanovirin-N caps the Gal/GalNAc lectin and glycoproteins recognized by an anti-proteophosphoglycan antibody.
Consistent with the presence of 7 to 10 N-glycan sites on each heavy subunit of the Gal/GalNAc lectin, cyanovirin-N cocaps the Gal/GalNAc lectin (Fig. 2A to C). The presence of anti-Gal/GalNAc lectin antibody labeling in areas away from the cap (Fig. 2B and C) is consistent with spontaneous replenishment of the Gal/GalNAc lectin from the large intracellular pools concurrent with the capping event. Cyanovirin-N also cocaps glycoproteins recognized by the anti-PPG antibodies (Fig. 2D to F). There is binding of the anti-PPG in areas away from the cap (Fig. 2E), consistent with replenishment of the PPG from large intracellular pools (data not shown).
Fig. 2.
Cyanovirin-N cocaps and partially depletes the Gal/GalNAc lectin and PPG. E. histolytica trophozoites were capped with cyanovirin-N (red), fixed, and then labeled with monoclonal antibodies to the Gal/GalNAc lectin (green) or to PPG (green). In each case, cyanovirin-N is present in a relatively tight cap (single large arrow), while the Gal/GalNAc lectin and PPG are each present in the cap and along the surface of the protist (series of small arrows). A cyanovirin-N-induced cap on the surface of an E. histolytica trophozoite (G) colocalizes with phalloidin (H) that binds filamentous actin. Merged panels are as indicated. Bar, 5 μm. Ab, antibody.
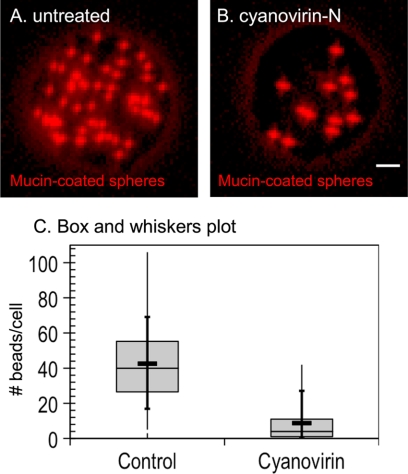
Cyanovirin-N inhibits phagocytosis by Entamoeba trophozoites of mucin-coated beads.
Filamentous actin, which is labeled by the fungal toxin phalloidin, is important for amebic motility, capping, and phagocytosis. Actin filaments accumulate in the region of the cyanovirin-N induced cap (Fig. 2G to I), as has been shown for caps by the plant lectin ConA and by the monoclonal antibody to the Gal/GalNAc lectin.
Cyanovirin-N inhibits phagocytosis of mucin-coated beads by E. histolytica trophozoites, a surrogate assay for amebic virulence (Fig. 3A to C). While untreated E. histolytica trophozoites phagocytose 43 ± 22 (mean ± standard deviation [SD]) mucin-coated beads, cyanovirin-N-treated trophozoites phagocytose 9 ± 11 (mean ± SD) beads (P ≪ 0.005). The inhibition of phagocytosis by cyanovirin-N is comparable to that caused by overexpression of a dominant negative p21rac mutant that interferes with localization of actin filaments during phagocytosis (18).
Fig. 3.
Cyanovirin-N inhibits phagocytosis of mucin-coated spheres by E. histolytica trophozoites. Fluorescence micrograph of a control E. histolytica trophozoite (A) that phagocytoses many mucin-coated spheres (red). In contrast, a representative cyanovirin-N-treated E. histolytica trophozoite (B) phagocytoses many fewer red spheres. Bar, 5 μm. (C) Phagocytosis of mucin-coated beads, as illustrated by a modified box and whiskers plot, in which the means are marked with a heavy horizontal line and the medians are marked by a light horizontal line. The lower and upper medians are the edges of each gray box, while the vertical bar shows the minimum and maximum values and the 10th and 90th percentiles (crosses).
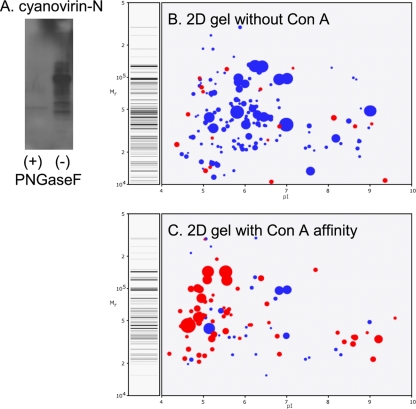
E. histolytica membrane and secreted proteins are dramatically enriched by affinity chromatography with ConA.
Lectin affinity chromatography was performed with ConA-Sepharose because glycoproteins can be eluted with excess α-methyl mannoside. In contrast, glycoproteins bound to cyanovirin-N-–epharose may only be eluted with SDS that introduces nonspecifically bound contaminants. Western blotting showed that cyanovirin-N conjugated to horseradish peroxidase binds to E. histolytica glycoproteins that were enriched by ConA affinity chromatography (Fig. 4A). In contrast, cyanovirin-N no longer binds to Entamoeba glycoproteins treated with PNGaseF to remove N-glycans (Fig. 4A). These results confirm that cyanovirin-N is binding only to E. histolytica N-glycans.
Fig. 4.
E. histolytica secreted and membrane proteins are markedly enriched by a ConA affinity column. (A) Cyanovirin-N binding to a Western blot of ConA-enriched E. histolytica proteins is removed by prior treatment with PNGaseF. Computer-derived, two-dimensional (2D) protein gels show mass spectrometry data from a representative experiment where unfractionated E. histolytica trophozoite proteins (B) and trophozoite proteins after ConA affinity (C) were identified. The size of each spot is proportional to the peptide coverage of the protein. Secreted proteins, which are markedly enriched after ConA, are shown in red. Nucleocytosolic proteins, which are abundant in unfractionated proteins, are shown in blue.
In the absence of ConA affinity chromatography, the vast majority (87%) of 302 E. histolytica proteins identified by mass spectrometry of tryptic peptides are nucleocytosolic (Fig. 4B and C, labeled blue). For example, when proteins are listed by their Mascot score, there are 41 nucleocytosolic proteins before the first secreted protein, a cysteine proteinase (see Excel file S1 in the supplemental material). While they are not the focus of the present study, nucleocytosolic proteins (many of which have greater than 50% peptide coverage) include enzymes involved in fermentation, glycolysis, and protein synthesis as well as chaperones and cytoskeletal proteins.
Following ConA affinity enrichment, the majority of E. histolytica proteins identified (52%) were membrane or secreted, as shown by the presence of N-terminal signals and/or transmembrane helices (Fig. 4B and C, labeled red). For example, 25 of the 30 proteins with the highest Mascot scores are secreted or membrane proteins rather than nucleocytosolic proteins (see Excel file S2 in the supplemental material). These glycoproteins (many of which have greater than 50% peptide coverage) include well-characterized virulence factors such as all three subunits of the Gal/GalNAc adherence lectin, as well as lysosomal proteases and phosphatases (see Excel file S2) (7, 10, 29, 32, 39). ER chaperones, protein disulfide isomerases, peptidyl-prolyl cis-trans isomerases, and calreticulin are all abundant. Of particular interest for discovery of potential vaccine candidates are 27 unique type 1 membrane proteins and six unique GPI-anchored proteins (see the FASTA file in the supplemental material). In the absence of information with regard to the location of any of these unique proteins, in the Excel files in the supplemental material the proteins with TMHs were arbitrarily assigned to the plasma membrane while proteins with an N-terminal signal peptide and no TMHs were assigned to the lysosome.
E. histolytica glycoproteins with occupied N-glycan sites include numerous proteins implicated in amebic pathogenesis.
ConA-enriched glycoproteins were treated with PNGaseF, and peptides in which the predicted Asn was converted to Asp were identified by a shift in mass of +0.984 Da (Table 1). These modified peptides (32 total), which represent occupied N-glycan sites, are absent from E. histolytica proteins that have not been treated with PNGaseF. Glycoproteins (26 total) with occupied N-glycan sites include numerous well-characterized virulence factors and/or vaccine candidates (heavy and intermediate subunits of the Gal/GalNAc lectin, serine and cysteine peptidases, and a receptor kinase) (Table 1) (8, 10, 32). Other glycoproteins with occupied N-glycan sites include 17 unique proteins that are secreted, membrane associated, or GPI anchored. Because some of these unique E. histolytica proteins with occupied N-glycan sites are both short and abundant (e.g., EHI_077530 is 206 amino acids long with 56% peptide coverage and EHI_161040 is 180 amino acids long with 42% peptide coverage), it is likely that they would make good vaccine candidates. A list of unique E. histolytica glycoproteins is shown in the FASTA file in the supplemental material.
DISCUSSION
Capping is more complex than previously supposed.
While actin-mediated capping of amebic proteins has been described (3, 16, 18, 19, 50), this is the first demonstration, to our knowledge, of replenishment of surface antigens from large intracellular pools. This process is shown in the model in Fig. 1I, where the precap surface antigens are shown in green, and the precap internal pool of antigens is shown in red. During capping of the green antigens by cyanovirin-N or antibodies to the Gal/GalNAc lectin, the red antigens move from internal pools to cover the parasite surface.
Because replacement occurs so quickly, the effects of cyanovirin-N on amebic phagocytosis in vitro (shown here) and of antibodies to the Gal/GalNAc lectin (32, 39, 43) and to PPG (6, 33, 35, 40, 48) on amebic virulence in vivo are likely not simply based upon clearing the relevant proteins from the parasite surface. Instead, the effects of cyanovirin-N and of antibodies to the Gal/GalNAc lectin or to PPG are likely also mediated by perturbation of the cytoskeleton during capping (3, 16, 18, 19, 50) and/or by signals transduced by various receptors (Gal/GalNAc lectin and/or receptor kinases) (8). Conversely, it does not appear that E. histolytica trophozoites escape anti-Gal/GalNAc lectin or anti-PPG antibodies by capping and removing antigens from their surfaces as both the Gal/GalNAc and PPG are rapidly replenished from large intracellular pools.
While there was no surprise that the E. histolytica Gal/GalNAc lectin has occupied N-glycan sites (32), it was not possible in advance to predict that glycoproteins recognized by anti-PPG antibodies are also capped by cyanovirin-N (6, 33, 35, 40, 48). The latter result suggests that some E. histolytica glycoproteins contain both N-glycans and O-phosphodiester-linked glycans.
E. histolytica glycoproteins include well-characterized virulence factors, as well as numerous unique proteins that may be novel vaccine candidates.
ConA affinity chromatography enabled the identification of >100 E. histolytica secreted and membrane proteins by mass spectrometry. The gel-free mass spectrometric methods used here are easier than cutting proteins from two-dimensional protein gels and result in relatively fewer cytosolic proteins identified than methods in which membranes or lysosomes are isolated (12, 26, 37, 51, 52, 55, 56). However, these other mass spectrometric studies reveal differences between virulent and avirulent strains of Entamoeba and demonstrate accessory proteins (e.g., Rabs) involved in vesicle sorting, endocytosis, and protein secretion.
Gal/GalNAc lectins are among the most abundant E. histolytica glycoproteins identified here, consistent with their prior identification by monoclonal antibodies and their importance in amebic pathogenesis (32, 39, 43). Dozens of unique and abundant E. histolytica glycoproteins identified here by mass spectrometry include new vaccine candidates (type 1 membrane proteins and GPI-anchored proteins) and/or new proteins involved in pathogenesis (secreted proteins). Of course, vaccine candidates and proteins involved in pathogenesis may be overlapping (e.g., the Gal/GalNAc lectin) (39). Recombinant versions of these unique E. histolytica glycoproteins, many of which are relatively small and not too Cys rich, might be used to vaccinate animal models and so add to the relatively short list of amebic vaccine candidates (Gal/GalNAc lectins, serine-rich E. histolytica protein [SREHP], and the 29-kDa protein) (9, 32, 39, 46, 47). Knockdown or knockout methods might be used to test the roles of these proteins in amebic virulence (27, 34).
These results suggest the possibility that E. histolytica N-glycans may be a new target for antiamebic reagents.
Unprocessed Man5GlcNAc2, by far the most abundant E. histolytica N-glycan, is recognized by the antiretroviral lectin cyanovirin-N that has been overexpressed in Lactobacillus (1, 25, 28, 31, 41, 46). Cyanovirin-N-expressing lactobacilli (“yogurt-plus”) might be introduced into the gastrointestinal tract, where the bacteria may have an antiamebic effect. Other bacterial lectins that target high-mannose N-glycans of HIV (e.g., griffithsin and banana lectin [BanLec]) may have even greater efficacy than cyanovirin-N versus E. histolytica (38, 49). Conversely, it may be possible to vaccinate against amebic infection with high-mannose N-glycans present on Saccharomyces mutants (14, 30).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants AI44070 (to J.S.), GM31318 (to P.W.R.), and RR10888 (to C.E.C.). Support for D.M.R. was provided by the Training Program in Host Pathogen Interactions (T32 AI052070).
We thank Richard Cook of MIT for some mass spectrometry data.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Adams E. W., Ratner D. M., Bokesch H. R., McMahon J. B., O'Keefe B. R., Seeberger P. H. 2004. Oligosaccharide and glycoprotein microarrays as tools in HIV glycobiology; glycan-dependent gp120/protein interactions. Chem. Biol. 11:875–881 [DOI] [PubMed] [Google Scholar]
- 2.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arhets P., Gounon P., Sansonetti P., Guillen N. 1995. Myosin II is involved in capping and uroid formation in the human pathogen Entamoeba histolytica. Infect. Immun. 63:4358–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aurrecoechea C., Brestelli J., Brunk B. P., Fischer S., Gajria B., Gao X., et al. 2010. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 38:D415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee S., Vishwanath P., Cui J., Kelleher D. J., Gilmore R., Robbins P. W., Samuelson J. 2007. Evolution of quality control of protein-folding in the ER lumen. Proc. Natl. Acad. Sci. U. S. A. 104:11676–11681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya A., Arya R., Clark C. G., Ackers J. P. 2000. Absence of lipophosphoglycan-like glycoconjugates in Entamoeba dispar. Parasitology 120:31–35 [DOI] [PubMed] [Google Scholar]
- 7.Bruchhaus I., Loftus B. J., Hall N., Tannich E. 2003. The intestinal protozoan parasite Entamoeba histolytica contains 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot. Cell 2:501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buss S. N., Hamano S., Vidrich A., Evans C., Zhang Y., Crasta O. R., Sobral B. W., Gilchrist C. A., Petri W. A., Jr 2010. Members of the Entamoeba histolytica transmembrane kinase family play non-redundant roles in growth and phagocytosis. Int. J. Parasitol. 40:833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhry O. A, Petri W. A., Jr 2005. Vaccine prospects for amebiasis. Expert Rev. Vaccines 4:657–668 [DOI] [PubMed] [Google Scholar]
- 10.Clark C. G., Alsmark U. C., Tazreiter M., Saito-Nakano Y., Ali V., Marion S., Weber C., Mukherjee C., et al. 2007. Structure and content of the Entamoeba histolytica genome. Adv. Parasitol. 65:51–190 [DOI] [PubMed] [Google Scholar]
- 11.Cui J., Smith T., Robbins P. W., Samuelson J. 2009. Darwinian selection for sites of Asn-linked glycosylation in phylogenetically disparate eukaryotes and viruses. Proc. Natl. Acad. Sci. U. S. A. 106:13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis P. H., Chen M., Zhang X., Clark C. G., Townsend R. R., Stanley S. L., Jr 2009. Proteomic comparison of Entamoeba histolytica and Entamoeba dispar and the role of E. histolytica alcohol dehydrogenase 3 in virulence. PLoS Negl. Trop. Dis. 3:e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan D. T., Craig R., Link A. J. 2005. Parallel tandem: a program for parallel processing of tandem mass spectra using PVM or MPI and X! Tandem. J. Proteome Res. 4:1842–1847 [DOI] [PubMed] [Google Scholar]
- 14.Dunlop, Bonomelli D. C. C., Mansab F., Vasiljevic S., Doores K. J., Wormald M. R., Palma A. S., Feizi T., Harvey D. J., Dwek R. A., Crispin M., Scanlan C. N. 2010. Polysaccharide mimicry of the epitope of the broadly neutralizing anti-HIV antibody, 2G12, induces enhanced antibody responses to self oligomannose glycans. Glycobiology 20:812–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenhaber F., Eisenhaber B., Kubina W., Maurer-Stroh S., Neuberger G., Schneider G., Wildpaner M. 2003. Prediction of lipid posttranslational modifications and localization signals from protein sequences: big-Pi, NMT and PTS1. Nucleic Acids Res. 31:3631–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espinosa-Cantellano M., Martinez-Palomo A. 1994. Entamoeba histolytica: mechanism of surface receptor capping. Exp. Parasitol. 79:424–435 [DOI] [PubMed] [Google Scholar]
- 17.Ghosh D., Krokhin O., Antonovici M., Ens W., Standing K. G., Beavis R. C., Wilkins J. A. 2004. Lectin affinity as an approach to the proteomic analysis of membrane glycoproteins. J. Proteome Res. 3:841–850 [DOI] [PubMed] [Google Scholar]
- 18.Ghosh S. K., Samuelson J. 1997. Involvement of p21racA, phosphoinositide 3-kinase, and vacuolar ATPase in phagocytosis of bacteria and erythrocytes by Entamoeba histolytica: suggestive evidence for coincidental evolution of amebic invasiveness. Infect. Immun. 65:4243–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guillen N. 1996. Role of signaling and cytoskeletal rearrangements in the pathogenesis of Entamoeba histolytica. Trends Microbiol. 4:191–197 [DOI] [PubMed] [Google Scholar]
- 20.Haque R., Huston C. D., Hughes M., Houpt E., Petri W. A., Jr 2003. Amebiasis. N. Engl. J. Med. 348:1565–1573 [DOI] [PubMed] [Google Scholar]
- 21.Helenius A., Aebi M. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73:1019–1049 [DOI] [PubMed] [Google Scholar]
- 22.Higdon R., Kolker N., Picone A., van Belle G., Kolker E. 2004. LIP index for peptide classification using MS/MS and SEQUEST search via logistic regression. OMICS 8:357–369 [DOI] [PubMed] [Google Scholar]
- 23.Koenig T., Menze B. H., Kirchner M., et al. 2008. Robust prediction of the MASCOT score for an improved quality assessment in mass spectrometric proteomics. J. Proteome Res. 7:3708–3717 [DOI] [PubMed] [Google Scholar]
- 24.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 25.Kwong P. D., Doyle M. L., Casper D. J., Cicala C., Leavitt S. A., Majeed S., Steenbeke T. D., Venturi M., et al. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 26.Leitsch D., Wilson I. B., Paschinger K., Duchêne M. 2006. Comparison of the proteome profiles of Entamoeba histolytica and its close but non-pathogenic relative Entamoeba dispar. Wien. Klin. Wochenschr. 118:37–41 [DOI] [PubMed] [Google Scholar]
- 27.Linford A. S., Moreno H., Good K. R., Zhang H., Singh U., Petri W. A., Jr 2009. Short hairpin RNA-mediated knockdown of protein expression in Entamoeba histolytica. BMC Microbiol. 9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X., Lagenaur L. A., Simpson D. A., Essenmacher K. P., Frazier-Parker C. L., Liu Y., Tsai D., Rao S. S., Hamer D. H., Parks T. P., Lee P. P., Xu Q. 2006. Engineered vaginal lactobacillus strain for mucosal delivery of the human immunodeficiency virus inhibitor cyanovirin-N. Antimicrob. Agents Chemother. 50:3250–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loftus B., Anderson I., Davies R., Alsmark U. C., Samuelson J., Amedeo P., Roncaglia P., Berriman M., et al. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865–868 [DOI] [PubMed] [Google Scholar]
- 30.Luallen R. J., Agrawal-Gamse C., Fu H., Smith D. F., Doms R. W., Geng Y. 2010. Antibodies against Manα1,2-Manα1,2-Man oligosaccharide structures recognize envelope glycoproteins from HIV-1 and SIV strains. Glycobiology 20:280–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magnelli P., Cipollo J. F., Ratner D. M., Cui J., Kelleher D., Gilmore R., Costello C. E., Robbins P. W., Samuelson J. 2008. Unique Asn-linked oligosaccharides of the human pathogen Entamoeba histolytica. J. Biol. Chem. 283:18355–18364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann B. J., Torian B. E., Vedvick T. S., Petri W. A., Jr 1991. Sequence of a cysteine-rich galactose-specific lectin of Entamoeba histolytica. Proc. Natl. Acad. Sci. U. S. A. 88:3248–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marinets A., Zhang T., Guillen N., Gounon P., Bohle B., Vollmann U., Scheiner O., Wiedermann G., Stanley S. L., Duchêne M. 1997. Protection against invasive amebiasis by a single monoclonal antibody directed against a lipophosphoglycan antigen localized on the surface of Entamoeba histolytica. J. Exp. Med. 186:1557–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirelman D., Anbar M., Bracha R. 2008. Epigenetic transcriptional gene silencing in Entamoeba histolytica. IUBMB Life 60:598–604 [DOI] [PubMed] [Google Scholar]
- 35.Moody-Haupt S., Patterson J. H., Mirelman D., McConville M. J. 2000. The major surface antigens of Entamoeba histolytica trophozoites are GPI-anchored proteophosphoglycans. J. Mol. Biol. 297:409–420 [DOI] [PubMed] [Google Scholar]
- 36.Nielsen H., Brunak S., von Heijne G. 1999. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 12:3–9 [DOI] [PubMed] [Google Scholar]
- 37.Okada M., Huston C. D., Oue M., Mann B. J., Petri W. A., Jr., Kita K., Nozaki T. 2006. Kinetics and strain variation of phagosome proteins of Entamoeba histolytica by proteomic analysis. Mol. Biochem. Parasitol. 145:171–183 [DOI] [PubMed] [Google Scholar]
- 38.O'Keefe B. R., Vojdani F., Buffa V., Shattock R. J., Montefiori D. C., Bakke J., Mirsalis J., d'Andrea A. L., Hume S. D., Bratcher B., Saucedo C. J., McMahon J. B., Pogue G. P., Palmer K. E. 2009. Scaleable manufacture of HIV-1 entry inhibitor griffithsin and validation of its safety and efficacy as a topical microbicide component. Proc. Natl. Acad. Sci. U. S. A. 106:6099–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petri W. A., Jr., Haque R., Mann B. J. 2002. The bittersweet interface of parasite and host: lectin-carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu. Rev. Microbiol. 56:39–64 [DOI] [PubMed] [Google Scholar]
- 40.Prasad R., Tola M., Sharma M. P., Bhattacharya A. 1992. Recognition of Entamoeba histolytica lipophosphoglycan by a strain-specific monoclonal antibody and human immune sera. Mol. Biochem. Parasitol. 56:279–287 [DOI] [PubMed] [Google Scholar]
- 41.Pusch O., Boden D., Hannify S., Lee F., Tucker L. D., Boyd M. R., Wells J. M., Ramratnam B. 2005. Bioengineering lactic acid bacteria to secrete the HIV-1 virucide cyanovirin. J. Acquir. Immune Defic. Syndr. 40:512–520 [DOI] [PubMed] [Google Scholar]
- 42.Ratner D. M., Cui J., Steffen M., Moore L. L., Robbins P. W., Samuelson J. 2008. Changes in the N-glycome (glycoproteins with Asn-linked glycans) of Giardia lamblia with differentiation from trophozoites to cysts. Eukaryot. Cell 7:1930–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ravdin J. I., John J. E., Johnston L. I., Innes D. J., Guerrant R. L. 1985. Adherence of Entamoeba histolytica trophozoites to rat and human colonic mucosa. Infect. Immun. 48:293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samuelson J., Banerjee S., Magnelli P., Cui J., Kelleher D. J., Gilmore R., Robbins P. W. 2005. The diversity of protist and fungal dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc. Natl. Acad. Sci. U. S. A. 102:1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scanlan C. N., Pantophlet R., Wormald M. R., Ollmann Saphire E., Stanfield R., Wilson I. A., Katinger H., Dwek R. A., Rudd P. M., Burton D. R. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanley S. L., Jr 2006. Vaccines for amoebiasis: barriers and opportunities. Parasitology 133:S81–S86 [DOI] [PubMed] [Google Scholar]
- 47.Stanley S. L., Jr., Becker A., Kunz-Jenkins C., Foster L., Li E. 1990. Cloning and expression of a membrane antigen of Entamoeba histolytica possessing multiple tandem repeats. Proc. Natl. Acad. Sci. U. S. A. 87:4976–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanley S. L., Jr., Huizenga H., Li E. 1992. Isolation and partial characterization of a surface glycoconjugate of Entamoeba histolytica. Mol. Biochem. Parasitol. 50:127–138 [DOI] [PubMed] [Google Scholar]
- 49.Swanson M. D., Winter H. C., Goldstein I. J., Markovitz D. M. 2010. A lectin isolated from bananas is a potent inhibitor of HIV replication. J. Biol. Chem. 285:8646–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tavares P., Sansonetti P., Guillén N. 2000. Cell polarization and adhesion in a motile pathogenic protozoan: role and fate of the Entamoeba histolytica Gal/GalNAc lectin. Microbes Infect. 2:643–649 [DOI] [PubMed] [Google Scholar]
- 51.Teixeira J. E., Huston C. D. 2008. Participation of the serine-rich Entamoeba histolytica protein in amebic phagocytosis of apoptotic host cells. Infect. Immun. 76:959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tolstrup J., Krause E., Tannich E., Bruchhaus I. 2007. Proteomic analysis of Entamoeba histolytica. Parasitology 134:289–298 [DOI] [PubMed] [Google Scholar]
- 53.Trombetta E. S., Parodi A. J. 2003. Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 19:649–676 [DOI] [PubMed] [Google Scholar]
- 54.Van Dellen K. L., Chatterjee A., Ratner D., Magnelli P. E., Cipollo J., Steffen M., Robbins P. W., Samuelson J. 2006. Unique posttranslational modifications of chitin-binding lectins of Entamoeba invadens cyst walls. Eukaryot. Cell 5:836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y., Wu S.-I., Hancock W. S. 2006. Approaches to the study of N-linked glycoproteins in human plasma using lectin affinity chromatography and nano-HPLC coupled to electrospray linear ion trap-Fourier transform mass spectrometry. Glycobiology 16:514–523 [DOI] [PubMed] [Google Scholar]
- 56.Yates J. R., III, Carmack E., Hays L., Link A. J., Eng J. K. 1999. Automated protein identification using microcolumn liquid chromatography-tandem mass spectrometry. Methods Mol. Biol. 112:553–569 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.