Abstract
The first barrier against infection by Candida albicans involves fungal recognition and destruction by phagocytic cells of the innate immune system. It is well established that interactions between different phagocyte receptors and components of the fungal cell wall trigger phagocytosis and subsequent immune responses, but the fungal ligands mediating the initial stage of recognition have not been identified. Here, we describe a novel assay for fungal recognition and uptake by macrophages which monitors this early recognition step independently of other downstream events of phagocytosis. To analyze infection in live macrophages, we validated the neutrality of a codon-optimized red fluorescent protein (yEmRFP) biomarker in C. albicans; growth, hyphal formation, and virulence in infected mice and macrophages were unaffected by yEmRFP production. This permitted a new approach for studying phagocytosis by carrying out competition assays between red and green fluorescent yeast cells to measure the efficiency of yeast uptake by murine macrophages as a function of dimorphism or cell wall defects. These competition experiments demonstrate that, given a choice, macrophages display strong preferences for phagocytosis based on genus, species, and morphology. Candida glabrata and Saccharomyces cerevisiae are taken up by J774 macrophage cells more rapidly than C. albicans, and C. albicans yeast cells are favored over hyphal cells. Significantly, these preferences are mannan dependent. Mutations that affect mannan, but not those that affect glucan or chitin, reduce the uptake of yeast challenged with wild-type competitors by both J774 and primary murine macrophages. These results suggest that mannose side chains or mannosylated proteins are the ligands recognized by murine macrophages prior to fungal uptake.
Candida albicans is an opportunistic fungus that normally resides in the human gut (26) and can cause mucosal infections. When host immune defenses are compromised or when anatomical breaches permit extreme fungal burdens, systemic and often lethal fungal colonization throughout the body can occur. In hospital-acquired bloodstream infections, the rate of mortality, hospital cost, and length of stay associated with disseminated candidiasis now outrank those associated with bacterial infections (37, 43). The most effective host barrier that limits Candida infections is microbial destruction by phagocytic cells of the innate immune system. In a healthy host, phagocytes—macrophages, neutrophils, and dendritic cells—recognize, ingest, and destroy the invading yeast by phagocytosis.
The first step of a fungal infection requires the recognition of yeast by phagocytes. Despite the medical importance of this reaction, it remains poorly understood. As the interface between the yeast and its host, the fungal cell wall is crucial for recognition. The wall is a complex structure consisting of an elastic network of polysaccharides (glucans and chitin) that surrounds the plasma membrane and that in most yeast and fungi contains many different heavily mannosylated proteins (mannan) anchored to the wall in various ways (9, 27–29). Three distinct layers that correspond to these three components can be seen by electron microscopy. The innermost layer is enriched with a small amount of chitin, the outermost layer consists of mannan, and in between these layers are flexible fibrils of β1,3-glucan. Another glucan (β1,6 linked) is relatively minor in amount but is important for maintaining wall structure because it cross-links β1,3-glucan to wall proteins and to chitin (24, 30). Yeast survival relies on the integrity of the cell wall because it shields the yeast from physical stress and osmotic shock. The wall also maintains cell shape, which is a precondition for growth and morphogenesis. The rapid switch between the yeast and hyphal forms that is essential for C. albicans virulence underscores the plasticity of the wall, whose composition, thickness, and structure vary tremendously in response to changes in the environment.
Many phagocytic receptors implicated in fungal recognition have been identified. The interactions between these receptors and fungal wall components activate an array of host defense signaling pathways that promote actin cytoskeletal rearrangements and the membrane remodeling required for phagocytosis, production of toxic metabolites and hydrolytic enzymes within the phagosome that destroy the ingested yeast, and secretion of cytokines that are pro- or anti-inflammatory (for a review, see references 18, 31, and 36). These receptors are members of the C-type lectin receptor and Toll-like receptor families and include proteins that can recognize mannose, glucan, and, possibly, chitin or, possibly, multiligand combinations of these carbohydrates (for reviews, see references 22 and 49). Despite a wealth of information about the signaling cascades elicited by these host receptors, the identity of the fungal cell wall ligands that mediate the initial recognition event during host-fungal interactions remains unclear, in large part because good model systems for studying host-fungal interactions in the context of the live infective environment have been unavailable. Most current assays of fungal recognition rely on indirect readouts, for instance, virulence or cytokine production, which cannot distinguish the initial step of fungal recognition from other downstream events of phagocytosis. In addition, different experimental systems for studying fungal phagocytosis use different cell types that may display unique interactions with C. albicans and vice versa. Thus, there are conflicts in the literature about the contributions of fungal cell wall components to host recognition and phagocytosis.
Here, we make use of a novel assay to help clarify discrepancies that currently exist in this field. We developed a biologically neutral red fungal fluorescent biomarker that can be stably introduced into most yeast and fungi to monitor C. albicans-host interactions during infection in live cells or animals. This permitted development of quantitative competition assays to measure uptake by macrophages of red fluorescent protein (RFP)- or green fluorescent protein (GFP)-labeled cells as a single isolated event within the complex process of phagocytosis in live cells. We apply this system to address two fundamental questions regarding fungal recognition by murine macrophages. First, do these macrophages display a preference toward yeast forms versus filamentous fungal forms, and second, how do the various fungal cell wall components contribute to this preference during the initial stage of fungal recognition? We demonstrate that, given a choice, J774 macrophages recognize and ingest yeast cells far more rapidly and efficiently than hyphal cells. Importantly, competitive fungal uptake by murine macrophages, both immortalized cell lines and primary cells, is markedly inhibited by reduction of cell wall mannan but not glucan or chitin. This points to a critical role for mannose side chains or mannosylated proteins as key fungal recognition ligands.
MATERIALS AND METHODS
Yeast media, strains, and growth conditions.
C. albicans and Saccharomyces cerevisiae strains were grown in standard rich medium (yeast extract, peptone, adenine, dextrose [YPAD]) supplemented with 50 μg/ml adenine or in synthetic dropout (SD) medium supplemented with the appropriate nutritional requirements (23). Uridine (50 μg/ml) was added to all media used to grow C. albicans except SD (−Ura). The yeast strains used in this study are listed in Table 1. C. albicans strains were derived from strain CAI4 or the related strain BWP17. S. cerevisiae strains were derived from strain SEY6210 or BY4741. Candida glabrata strains were derived from strain BG88b. All comparative analyses used prototrophic C. albicans strains. Uracil, histidine, and arginine prototrophs were made by targeting URA3 to the RP10 locus using the URA3 integrative plasmid CIp10 (34), linearized by digestion with StuI (Table 2); by targeting HIS1 to the his1::hisG locus using the integrative pGEM-HIS plasmid, linearized with Nru1 (50); or by targeting ARG4 to the RP10 locus using the ARG4 integrative plasmid Clp-ARG4 (see below), linearized with StuI. C. albicans strains overproducing yEmRFP at high copy numbers were isolated after transformation with the URA3/ARS-containing plasmid pADH1p-Cherry (25) and performing four sequential selections for uracil prototrophs on SD (−Ura) plates until the colony color turned distinctly pink. Similarly, an isogenic C. albicans strain overproducing GFP (strain SKY43) was isolated using the URA3/ARS GFP-containing plasmid pADH-GFP. C. albicans strains containing a single, integrated copy of yEmRFP were constructed by targeting CIp10-Adh-Cherry (marked with URA3) or CIp-HIS-Adh-Cherry (marked with HIS1) to the RP10 locus. Single or multiple integrations of ADH1p-yEmRFP were confirmed by Southern blotting (data not shown). S. cerevisiae and C. glabrata strains expressing yEmRFP or yEGFP (yeast enhanced green fluorescent protein) were made by transformation with URA3/2μm yEpGAP-Cherry or yEpGAP-yEGFP (12).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| C. albicans | ||
| CAI4 | ura3Δ::λimm434/ura3Δ::λimm434 | 19 |
| BWP17 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 50 |
| SKY35 | CAI4 and RP10::URA3 | This study |
| SKY36 | BWP17 and hisG::HIS1 | This study |
| SKY38 | BWP17 and PADH1-yEmRFP-URA3-ARS | 25 |
| SKY39 | SKY38 and his1::HIS1 | This study |
| SKY40 | CAI4 and PADH1-yEmRFP-URA3-ARS | This study |
| SKY41 | BWP17 and PADH1-FLAG-yEmRFP-URA3-ARS | This study |
| SKY42 | BWP17 and RP10::URA3 PADH1-FLAG-yEmRFP | This study |
| SKY43 | BWP17 and PADH1-yEGFP-URA3-ARS | This study |
| SKY51 | BWP17 and RP10::URA3 PADH1-yEmRFP | This study |
| SKY52 | CAI4 and RP10::URA3 PADH1-yEmRFP | This study |
| GSC1(+/−/−) | CAI4 and gsc1Δ::hisG/gsc1Δ::hisG-URA3-hisG/GSC1 | 33 |
| CAP1-3121 | CAI4 and pmt1Δ::hisG/pmt1Δ::hisG | 33 |
| CAP1-31 | CAI4 and PMT1/pmt1Δ::hisG | 46 |
| P2-22 | CAI4 and PMT2/pmt2Δ::hisG | 46 |
| CAP4-21 | CAI4 and PMT4/pmt4Δ::hisG | 46 |
| CAP4-2162 | CAI4 and pmt4Δ::hisG/pmt4Δ::hisG | 46 |
| NGY205 | CAI4 and och1ΔΔ::hisG/och1Δ::hisG | 3 |
| SKY54 | CAI4 and pmt1Δ::hisG/pmt1Δ::hisG PADH1-yEmRFP-URA3-ARS | This study |
| SKY55 | CAI4 and PMT1/pmt1Δ::hisG PADH1-yEmRFP-URA3-ARS | This study |
| SKY56 | CAI4 and PMT2/pmt2Δ::hisG PADH1-yEmRFP-URA3-ARS | This study |
| SKY57 | CAI4 and PMT4/pmt4Δ::hisG PADH1-yEmRFP-URA3-ARS | This study |
| SKY58 | CAI4 and pmt4Δ::hisG/pmt4Δ::hisG PADH1-yEmRFP-URA3-ARS | This study |
| SKY53 | CAI4 and och1Δ::hisG/och1Δ::hisG PADH1-yEmRFP-URA3-ARS | This study |
| S. cerevisiae | ||
| SEY6210 | MATα ura3-52 his3-Δ200 leu2-3,112 trp1-Δ901 lys2-801 suc2-Δ9 | 41 |
| JPY12 | SEY6210 and mnn10Δ::LEU2 | 13 |
| VMY2 | SEY6210 and hoc1Δ::LEU2 | 35 |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| YLR342W | BY4742 and fks1Δ::Kanr | Open Biosystems |
| YBR023C | BY4742 and chs3Δ::Kanr | Open Biosystems |
| YNL322C | BY4742 and kre1Δ::Kanr | Open Biosystems |
| YPR159W | BY4742 and kre6Δ::Kanr | Open Biosystems |
| C. glabrata | ||
| BG88b | his3Δ(1 + 631) ura3Δ(−85 + 932)::Tn903 Neor | 12 |
| SKY60 | BG88b and PADH1-yEmRFP-URA3-ARS | This study |
| SKY61 | BG88b and PADH1-yEGFP-URA3-ARS | This study |
Table 2.
Plasmids used in this study
| Name | Relevant features | Reference or source |
|---|---|---|
| CIp10 | CaURA3 integrative plasmid | 34 |
| CIp10-HIS1 | CaHIS1 integrative plasmid | This study |
| CIp-ADH-yEmRFP | PADH1-yEmRFP in CIp10 | 25 |
| CIp-HIS1-ADH-yEmRFP | PADH1-yEmRFP in CIp-HIS1 | This study |
| CIp-ADH-FLAG-yEmRFP | Like CIp-ADH1-yEmRFP but encodes N-FLAG-tagged mRFP | This study |
| pADH-yEmRFP | PADH1-yEmRFP in CaURA3/ARS plasmid | 25 |
| pADH-FLAG-yEmRFP | Like pADH-yEmRFP but encodes FLAG-tagged mRFP | This study |
| pADH-yEGFP | PADH1-yEGFP in CaURA3/ARS plasmid | This study |
| YEpGAP-yEmRFP | 2μm URA3 plasmid containing PADH1-yEmRFP | 25 |
| YEpGAP-yEGFP | 2μm URA3 plasmid containing PADH1-yEGFP | This study |
Plasmids.
Plasmids and their relevant features are listed in Table 2. Standard molecular biology techniques were used for all DNA manipulations, and the sequences of all DNA generated by PCR were verified by DNA sequence analysis. CIp10-ADH-yEmRFP and pADH-yEmRFP contain the C. albicans codon-optimized mRFP gene (mCherry), driven by the C. albicans ADH1 (CaADH1) promoter in an integrative plasmid and an ARS-containing plasmid, respectively (25). CIp-ADHp-FLAG-yEmRFP and pADHp-FLAG-yEmRFP are identical to the CIp-ADH-yEmRFP and pADH-yEmRFP plasmids but encode yEmRFP tagged at the N terminus with the FLAG epitope (DYKDDDK). CIp10-HIS1 is an integrative HIS1-marked plasmid and was targeted to the his1::hisG locus in strain BWP17 by linearization with XbaI. CIp10-HIS1 was constructed by replacing the URA3 gene in CIp10 with a 2.6-kb BamHI/SacI fragment containing C. albicans HIS1 (CaHIS1). CIp10-ARG4 is an integrative ARG4-marked plasmid made by replacing the URA3 gene in CIp10 with a BamHI/SacI fragment containing C. albicans ARG4 (CaARG4) and 1 kb of 5′ and 3′ flanking sequence. This plasmid was targeted to the RP10 locus after linearization with StuI. YEpGAP-GFP is an S. cerevisiae 2μm/URA3 plasmid containing TDH3 promoter-driven yEGFP. It was constructed by replacing the yEmRFP open reading frame in YEpGAP-yEmRFP with yEGFP.
Protein analysis.
Protein extracts were prepared from mid-logarithmic-phase yeast cultures grown to an optical density at 600 nm (OD600) of ∼2 OD600 units/ml. Five to 10 OD600 units was harvested, washed in phosphate-buffered saline (PBS), and lysed by glass bead beating in PBS containing 0.5% Triton X-100 and protease inhibitors, as described previously (10). Protein concentrations of lysates were determined by the Bradford method. Samples (20 μg) were fractionated by SDS-PAGE (12%) and immunoblotted, as described previously (2). yEmRFP protein levels were quantitated by immunoblotting with anti-FLAG antibodies (Sigma) diluted 1: 2,000 to detect FLAG-tagged yEmRFP expressed at a single copy or multiple copies (Fig. 1C). Primary antibodies were detected by chemiluminescence with horseradish peroxidase-conjugated secondary anti-IgG antibodies (GE/Amersham Biosciences).
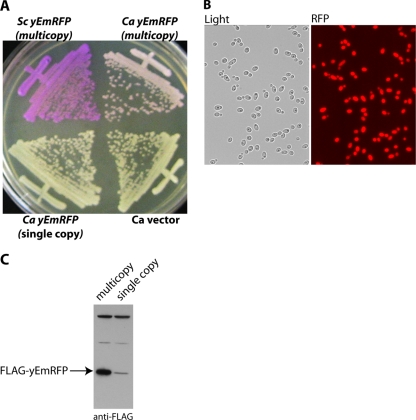
Fig. 1.
High-level production of mRFP in C. albicans. (A) Yeast colony color turns from cream to pink when C. albicans (Ca) overproduces yEmRFP. CAI4 was transformed with pADH-yEmRFP and subjected to multiple rounds of selective and then nonselective growth. Note the colony color phenotype of this strain (strain SKY40) and one that contains a single integrated copy of PADH1-yEmRFP (strain SKY52) after growth on SD (−Ura) medium for 2 days at 30°C. An S. cerevisiae (Sc) strain expressing hundreds of copies of yEmRFP on 2μm plasmids is shown for comparison. (B) Fluorescence microscopy of C. albicans expressing multiple copies of yEmRFP. After multiple rounds of selective growth, almost all of the cells in the population (98 to 99%) remain brightly fluorescent after >10 generations on nonselective YPAD. (C) Elevated yEmRFP production in C. albicans containing multiple, stable copies of yEmRFP. Protein extracts were prepared from C. albicans strains with a single integrated copy of PADH1-FLAG-yEmRFP (strain SKY42) or multiple stable copies (strain SKY41), fractionated by 12% SDS-PAGE, and analyzed by Western blotting with anti-FLAG antibodies.
Analysis of hyphal induction.
Yeast cells were seeded on glass coverslips in 24-well plates to analyze hyphal induction of individual cells, as described previously (40). Briefly, overnight yeast cultures were diluted to 0.5 OD600 units/ml, and 5 μl was spread on each coverslip. These were placed in a 12-well plate with 1 ml of YPAD plus 20% bovine calf serum, and the plate was incubated at 37°C for the required durations. Coverslips were washed with PBS, mounted cell side down on a glass slide, and viewed by microscopy.
Mouse infection assays.
Overnight yeast cultures of C. albicans (strain SKY35) and an isogenic prototrophic variant expressing yEmRFP (strain SKY40) were washed twice and resuspended in sterile water (107 cells per ml). Mice (BALB/c) were injected with 100 μl of yeast (106 cells) in the lateral tail vein. Survival experiments were carried out in groups of two to four mice each. Infected mice were monitored for well-being twice daily. When they were considered moribund (typically, between days 2 and 4 postinfection), the mice were humanely euthanized. All experimental procedures were carried out according to the NIH guidelines for the ethical treatment of animals.
To compare the fungal burdens, kidneys from control and infected mice were dissected, weighed, and homogenized mechanically in PBS (1 kidney per 5 ml PBS). The homogenate was serially diluted and plated on YPAD to determine fungal genotype, phenotype, cell morphology, and fungal load (expressed as the numbers of CFU per gram of kidney tissue). The relative proportions of yeast and hyphal cells in the homogenate were estimated by fluorescence microscopy after the yeast were stained with calcofluor white (CW; Sigma/Aldrich) to a final concentration of 1 μg/ml.
Bone marrow macrophage isolation and culture conditions.
Bone marrow-derived macrophages were isolated from BALB/c mice, as described previously (8, 39). Bone marrow cells from two femurs were suspended in culture medium (Dulbecco's modified Eagle medium [DMEM] with a high glucose concentration [4,500 μg/ml] supplemented with 20% heat-inactivated fetal bovine serum [HyClone], 30% L-cell-conditioned medium, 2 mM l-glutamine, and 1 mM sodium pyruvate) and seeded into 100-mm-diameter petri dishes (Nunc) at 4 × 106 cells/plate. After 3 days of incubation at 37°C in the presence of 5% CO2, the medium was removed and replaced with fresh medium. After an additional 2 days of incubation, macrophages that selectively adhered to the dishes were washed in cold PBS, harvested, and transferred to 24-well plates (1.5 × 105 cells/well) with 1 ml of medium. After an additional 24-h incubation, fungal competition assays (see below) were performed.
J774 cell culture conditions.
The murine BALB/c macrophage-like J774 cells were maintained in DMEM–10% horse serum (Gibco) and incubated at 37°C in the presence of 5% CO2. Preconfluent cultures were diluted and plated on coverslips in 24-well plates (104 cells/well). After 24 h incubation, the medium was replaced with fresh CO2-independent DMEM–10% heat-inactivated serum. Phagocytosis and macrophage morphology were unaffected by growth in CO2-independent or nonbuffered DMEM. Therefore, cells were maintained in CO2-independent medium after yeast addition to minimize the pH fluctuations caused by removing plates from the incubator during microscopy.
Macrophage phagocytosis assay.
Fresh overnight yeast cultures were grown in YPAD–50 μg/ml uridine. Before addition to macrophages, yeast cells were harvested by centrifugation, washed once in PBS, and resuspended in PBS. The yeast cell concentration was estimated by measurement of the OD600 (∼2 × 107 yeast cells/OD600 unit) and then measured precisely with a hemacytometer to adjust the final concentration to 107 yeast cells/ml PBS. Yeast cells were added to J774 or primary macrophages at multiplicities of infection (MOIs) that ranged from 1 to 10. At various times after addition of the yeast cells, CW was added (final concentration, 1 μg/ml) directly to the culture medium for ∼10 s before the cells were viewed (no washing step was required). CW was prepared in H2O (1 mg/ml), titrated with 1 M NaOH to clear the turbidity, and filter sterilized. To stain the surface of macrophages, fluorescein isothiocyanate (FITC)-conjugated Pisum sativum agglutinin (PSA-FITC; Sigma) was added to the cells (1:2,000 dilution). CW/PSA-FITC-stained samples were viewed immediately, without washing, by removing the coverslips, mounting them on a glass slide, and sealing them with nail polish. We used a Zeiss Axioskop fluorescence microscope, a high-performance Dage-MTI charge-coupled-device camera, and Scion Image software for capture of single black-and-white images. Fluorescent signals were sufficiently bright so that no further image processing, other than colorization, was required. Images were colorized using the AdobePhotoshop CS program, and all images in a given figure were processed together.
Time-lapse photography.
J774 cells were precultured in DMEM–10% horse serum in 35-mm glass-bottom petri dishes (In Vitro Science) with 10 mm/0.13-μm-thick glass bottom inserts. After an overnight incubation, the plates were transferred to a Zeiss Axioscop inverted microscope equipped with a heated stage and Plexiglas CO2 chamber. Throughout the experiment, cultures were maintained at 37°C in 5% CO2. Five minutes after the addition of yeast, time-lapse images (×20 or ×63 magnification) were captured with an AxioCam HR camera, using AxioVision, release 5, software (Zeiss) to configure image acquisition. Images were acquired every 1.0 min by dual channels set for exposure times of 10 to 30 ms (differential interference contrast) and 3.5 s (Texas Red) during a 2- to 4-h period. Merged Nomarski and fluorescent images as Zeiss (.zvi) formatted files were normalized for brightness and contrast using AxioVision software and were subsequently exported as Quicktime movies that were not further processed. At the completion of each time-lapse experiment, CW was added to samples for 5 min to obtain a single merged UV/red fluorescence/Nomarski image that was used to determine the ratio of yeast cells that were internal or external of macrophages (data not shown).
Phagocytosis competition assay.
Competing yeast strains expressing yEGFP or yEmRFP were grown overnight in YPAD, harvested, washed in PBS, and resuspended in PBS to 107 cells/ml. An equal number of each competitor strain was mixed and added to macrophages grown in 24-well plates at MOIs ranging from 1 to 10. Aliquots of input yeast mixtures were serially diluted and plated on YPAD and incubated overnight at 37°C, and the numbers of red and green fluorescent colonies were counted to confirm the 1:1 input ratio of competitor strains (data not shown). At various times after coincubation, CW was added and yeast cells (red, green, or untagged) were scored as being inside or outside macrophages on the basis of their susceptibility to CW staining (blue). The fractions of internalized red (Ri) and green (Gi) yeast cells were calculated by subtracting the number of external (CW+) cells from the total (t) number of cells per field: Ri = Rt − RCW+and Gi = Gt − GCW+.
For each experiment, at least 300 yeast cells were scored, and each experiment was repeated at least three times. The fractions of cells internalized (I) by macrophages for each competitor strain were calculated by IR = (Ri/Rt) and IG = Gi/Gt for red and green cells, respectively.
The percent internalization ratio (PIR) for each competitor was calculated as the fraction of red or green competitor cells internalized as a function of the total number of competing cells, where PIRR + PIRG = 100: PIRR = IR/(IR + IG) (100) and PIRG = IG/(IR +IG) (100) for red and green cells, respectively.
RESULTS
Characterization of yEmRFP-expressing C. albicans.
We previously reported that an ARS/URA3-containing plasmid harboring PADH1-yEmRFP transformed into C. albicans does not replicate episomally but instead behaves as a stable integrant (25). High-level production of this codon-optimized yEmRFP leads to brilliant red fluorescence and an alteration of colony color: S. cerevisiae colonies are bright purple and C. albicans are pink. During the course of characterizing yEmRFP expression in C. albicans transformants, we noticed that after multiple rounds of selection for uracil prototrophs there was a concomitant intensification of both colony color (Fig. 1A) and cell fluorescence phenotypes (Fig. 1B). These results suggested the occurrence of a stable plasmid-dependent chromosomal integration and amplification of the yEmRFP gene. Both Southern and Western blotting experiments supported this idea. Compared to an isogenic strain with a single integrated copy of yEmRFP, we observed an ∼5-fold increased yEmRFP gene copy number (data not shown) and a greater than 20 fold increased RFP level in yEmRFP-overproducing strains subjected to multiple rounds of selection. The plasmid-dependent integration and gene amplification were not dependent on yEmRFP, since replacement of yEmRFP with the yeast-enhanced GFP gene (11) in this plasmid led to C. albicans transformants with an increased green fluorescence phenotype that was also stably inherited (data not shown). Thus, transformation and multiple rounds of selection with these ARS-containing plasmids allow the construction of stable C. albicans strains that overproduce fluorescent proteins.
C. albicans cell division, hyphal growth, and virulence are unaffected by yEmRFP overexpression.
The bright fluorescence conferred by yEmRFP expression suggested that it may be a useful tool for monitoring fungal infection and pathogenesis in living cells. To be a useful biomarker, yEmRFP must be biologically neutral. To test this idea, we measured the effect of yEmRFP overproduction on growth, virulence, and pathogenesis. The effect of yEmRFP expression on vegetative growth was determined by measuring the optical density of isogenic yeast strains inoculated in rich medium at 30°C over 24 to 36 h. Isogenic strains that express yEmRFP (strain SKY40) or that do not (strain SKY35) displayed similar growth curves, with doubling times of ∼1.5 h during the logarithmic phase of growth and entry into the stationary phase at an OD600 of ∼30. These experiments demonstrated that yEmRFP expression did not significantly affect vegetative growth.
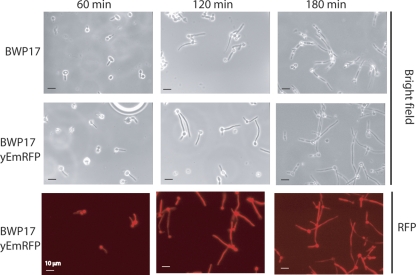
The rate of hyphal extension and hyphal length were measured with yeast cultures seeded on glass coverslips to allow observations of single cells induced to form hyphae by addition of 20% bovine calf serum and a shift to 37°C. No obvious differences in the rate of formation or length of the hyphae were observed as a result of RFP expression (Fig. 2). Germ tube formation was evident after ∼20 min of induction, and filaments continued to extend at similar rates in both strains, unaffected by RFP overproduction. Importantly, during hyphal extension there was no decrease of RFP fluorescence, whose distribution was uniform throughout the cytoplasm of the cell body and along the length of the filament (Fig. 2). Together these results suggested that mRFP is constitutively expressed throughout hyphal formation and that these red fluorescent cells retain their full dimorphic capability.
Fig. 2.
Expression of yEmRFP does not affect the rate or length of hyphal formation. The kinetics and extent of hyphal formation were examined in wild-type strain BWP17 or yEmRFP-overexpressing strain SKY38. Yeast cells were grown to early logarithmic stage (1 to 2 OD600 units/ml), seeded on glass coverslips, and induced to form hyphae by the addition of 20% serum and a temperature shift to 37°C. Coverslips were removed at the indicated times and viewed by light or fluorescence microscopy. It should be noted that these panels do not represent overlapping images from the same field.
To determine if overproduction of yEmRFP in C. albicans affects virulence, BALB/c mice were injected in the tail vein with 1 × 106 yeast cells (see Materials and Methods). In each experiment, which was repeated three times, duplicate mice were infected with yEmRFP-expressing cells (strain SKY40) or an isogenic URA3 prototrophic strain (strain SKY35) and compared to an uninfected control. The results, summarized in Table 3, demonstrated that mice infected with yEmRFP-expressing yeast or nonexpressing strains were equally susceptible to lethal systemic infection. Mice infected with either strain died within 2 to 3 days of infection and had comparable kidney fungal burdens (Table 3). Moreover, all of the yeast colonies recovered from cultured kidney homogenates from mice infected with yEmRFP-expressing yeast were pink, indicating that the red fluorescent phenotype was stably maintained during infection (data not shown). Together, these results demonstrated that production of yEmRFP in C. albicans does not significantly affect its virulence properties in a mouse model of systemic infection.
Table 3.
Mouse infection assay
| Yeast strain | Survival timea (days) | Fungal loadb (CFU/g tissue) |
|---|---|---|
| SKY40 (CAI4 Ura+ yEmRFP) | 2–3 | 8 × 105 |
| SKY35 (CAI4 Ura+) | 2–3 | 5 × 105 |
The survival time is based on averaged results of three separate experiments, using six mice.
Fungal load was determined by counting the number of viable yeast cells recovered from kidney homogenates. Kidneys (n = 6) were weighed prior to mechanical homogenization. CFU represents the number of colonies per gram of tissue and was determined by plating serial dilutions of kidney homogenates on YPAD plates and counting the number of colonies that grew after 2 days of incubation at 30°C.
Overproduction of RFP does not affect fungal uptake or evasion in a macrophage model of infection.
The stable, neutral phenotype of red fluorescent yeast suggested that yEmRFP would be a useful tool to measure fungal recognition and uptake by macrophages. To test this idea, we asked if overproduction of RFP affects phagocytosis of yeast by the murine macrophage cell line J774. The efficiency of yeast uptake, the rate of hyphal formation within the phagosome, and the efficiency of fungal evasion from the macrophage were measured after infection with isogenic yeast strains that do or do not express yEmRFP. Several parameters were varied, including the MOI, macrophage density, and culture conditions (see Materials and Methods). To summarize the results of these experiments, we found no deleterious effect of yEmRFP overproduction on any stage of phagocytosis or macrophage evasion; the kinetics of uptake, the rate of hyphal growth within the macrophage, the time of macrophage perforation, and the morphology of the macrophages during the course of infection were identical whether or not RFP was overproduced (data not shown).
Quantitative fluorescence assay for fungal recognition and uptake by macrophages.
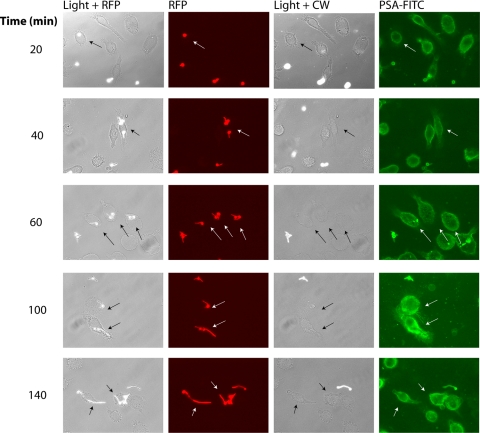
To measure fungal uptake by macrophages, fluorescent yeast cells were added to J774 cells precultured on glass coverslips in multiwell plates (or glass-bottom dishes for time-lapse photography). At various times after the addition of the yeast cells, CW was added to assess the fraction of phagocytosed cells, since this blue fluorescent dye binds only to yeast cells that have not been phagocytosed. Yeast cells that were ingested by macrophages are inaccessible to CW. Therefore, the ratio of CW-negative yeast (inside macrophages) and CW-positive yeast (outside macrophages) provided a means to accurately measure yeast internalization by macrophages (Fig. 3). The outer surface of the macrophage was detected by FITC-labeled PSA, a lectin with high affinity to terminal fucoses α-linked to mannose-containing oligosaccharides. Although fucose is not found on the fungal cell surface, nonspecific FITC-PSA staining of yeast cells outside but not within macrophages was also observed (Fig. 3), probably due to low-affinity PSA binding to cell wall mannans. Nevertheless, FITC-PSA binding enabled a very clear delineation of the macrophage membrane, allowing green (macrophage), red (all yeast), and blue (uningested yeast) to be viewed simultaneously in live cells. The time course of this assay, depicted in Fig. 3 (and see Fig. S1 in the supplemental material for the time-lapse movie), allowed us to distinguish distinct stages of phagocytosis, including (i) yeast cell uptake (at ∼30 min), (ii) germ tube formation and extension of the hyphae within the phagosome (from ∼40 to 120 min), and finally, (iii) macrophage perforation (at ∼180 min).
Fig. 3.
Phagocytosis of red fluorescent C. albicans by J774 macrophages in real time. Red fluorescent C. albicans cells (strain SKY38) were added to J774 macrophages grown on coverslips in multiwell plates. Yeast cells were added at an MOI of ∼1 yeast cell per macrophage, and at various times after addition of yeast cells, CW (1 μg/ml) and PSA-FITC (20 μg/ml) were added to the culture medium to stain the surface of noningested yeast cells and macrophages, respectively. Cells were viewed by fluorescence microscopy directly, without washing, using RFP, GFP, and 4′,6-diamidino-2-phenylindole filter sets. Arrows denote RFP-positive internalized yeast cells that are inaccessible to staining with CW.
Quantitating fungal recognition and uptake during phagocytosis by competition assays.
The ability to detect live C. albicans cells tagged with green or red fluorescent markers in infected cells suggested that competition assays could be applied to measure the relative contribution of fungal cell wall components to macrophage recognition. Competition assays are powerful because they allow measurement of even subtle phenotypic differences. To confirm that neither RFP nor GFP expression confers a competitive disadvantage during phagocytosis, uptake by J774 cells of RFP-tagged C. albicans challenged with an isogenic untagged competitor was measured in J774 macrophages (Fig. 4 and data not shown). At various times after yeast addition, CW was added and the numbers of red and unlabeled yeasts that were outside macrophages (CW positive) or inside macrophages (CW negative) were counted. The results of this experiment demonstrated that yEmRFP-expressing and non-RFP-expressing yeast cells were internalized by macrophages with an equal efficiency (Fig. 4A). Competition between yEGFP- and yEmRFP-expressing C. albicans yeast cells also showed an ∼1:1 ratio of internalized green and red yeast cells (Fig. 4A). Both yEGFP- and yEmRFP-expressing cells formed hyphae and killed macrophages with the same kinetics (data not shown). Thus, neither GFP nor RFP expression positively or negatively affected recognition and uptake by murine macrophages.
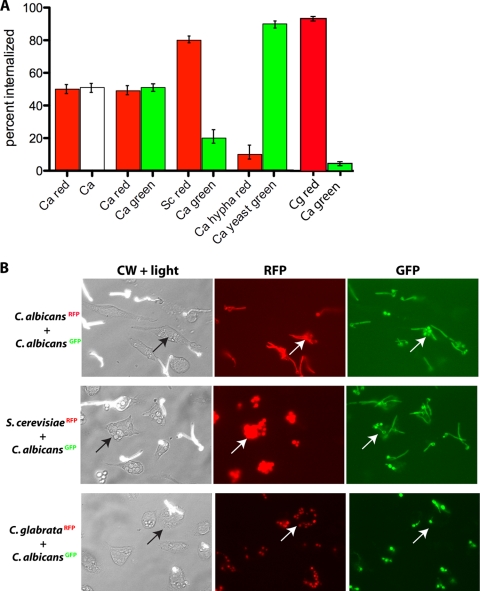
Fig. 4.
Yeast cells are ingested more efficiently than hyphal cells by J774 macrophages. Competition assays were performed with various combinations of S. cerevisiae, C. glabrata, and C. albicans yeast or hyphal cells (preincubated for 20 min with 20% serum at 37°C to induce hyphae). Equal numbers of yeast cells expressing yEmRFP or yEGFP were mixed together and added to J774 macrophages at an MOI of 5. After 40 min, CW was added and red or green cells were scored as being in or out of macrophages. (A) Graphic representation of quantitative competition assays between nontagged C. albicans yeast cells (strain SKY51) and those expressing yEGFP (strain SKY43) that were mixed with RFP-expressing C. albicans yeast cells (strain SKY38), with S. cerevisiae yeast cells (strain SEY6210 plus pADH-yEGFP), or with C. albicans hyphal cells (strain SKY38). Sc, Ca, and Cg, S. cerevisiae, C. albicans, and C. glabrata, respectively. The percent internalization ratio was calculated by scoring yeast cells as red or green and as CW positive (outside) or CW negative (inside), as described in Materials and Methods. Representative single-focal-plane images of these assays are shown in panel B, with arrows highlighting single macrophages that have taken up both red and green yeast cells at various ratios.
J774 murine macrophages prefer yeast cells over hyphal cells.
We applied these competition assays to ask how fungal dimorphism influences the uptake of yeast cells by J774 macrophages. There are conflicting reports in the literature regarding the importance of the yeast form versus the hyphal form during phagocytosis (15, 32). We tested the uptake efficiency of C. albicans in competition with S. cerevisiae or C. glabrata, which do not form hyphal cells, or with C. albicans hyphal cells. We chose to compare S. cerevisiae, C. glabrata, and C. albicans because they represent the most commonly studied and important pathogenic and nonpathogenic yeasts. Equal numbers of yEmRFP-expressing S. cerevisiae (or C. glabrata) and yEGFP-expressing C. albicans yeast cells were mixed and cocultured with J774 macrophages at an MOI of 5. To rule out contributions of complement, competition assays were performed in medium supplemented with heat-inactivated serum. After ∼40 min, CW was added and the percent internalization ratio by macrophages was calculated as described above. The results demonstrated a marked bias by J774 macrophages for uptake of S. cerevisiae versus C. albicans (∼5:1 ratio) (Fig. 4A and B). At higher MOIs, this bias was even more pronounced, with single J774 cells commonly containing 20 or more S. cerevisiae yeast cells (data not shown). A similar preference was observed for C. glabrata, a haploid pathogenic yeast that, despite its genus name, is more closely related to S. cerevisiae than to C. albicans. In competition assays, we observed an ∼10-fold preference by J774 macrophages for uptake of C. glabrata when C. glabrata was coincubated with C. albicans (Fig. 4A and B). The increased competitive uptake efficiency of S. cerevisiae and C. glabrata relative to that of C. albicans was independent of fluorescent proteins, since reversing the fluorescent color of the competitor strains had no effect on these biases (data not shown). The increased number of C. glabrata or S. cerevisiae yeast cells observed within macrophages was not due to yeast cell division within the phagosome, since the total number of C. glabrata or S. cerevisiae cells per well remained constant and did not increase during the course of this 40-min experiment (data not shown). Together, these results demonstrated that S. cerevisiae and C. glabrata are recognized and taken up by J774 macrophages better than C. albicans.
As a first step toward investigating the determinant that influences phagocytosis by J774 macrophages, we performed time-lapse imaging to measure the kinetics of phagocytosis. Unlike GFP, yEmRFP red fluorescence is not rapidly bleached by repeated fluorescence excitation exposure. This photostability allowed us to perform high-resolution dual-channel time-lapse imaging of yeast cells and macrophages simultaneously during phagocytosis. Fluorescence (3.5 s per exposure) and light images (∼0.1 s per exposure) were simultaneously captured every minute for 120 to 180 min after addition of yeast cells to macrophages (see Materials and Methods and Fig. S2 in the supplemental material). These experiments demonstrated that the initial rate of C. albicans uptake by macrophages is >4 times slower than that for S. cerevisiae and C. glabrata (compare Fig. S2a and b in the supplemental material). Under identical conditions, J774 macrophages began to ingest C. albicans after ∼30 min of coincubation (Fig. S2a). In contrast, uptake of C. glabrata (Fig. S2b) and S. cerevisiae (data not shown) began after 5 to 7 min of coincubation. Thus, the decreased internalization efficiency of C. albicans may be explained in part by a kinetic difference with which these yeast cells are recognized and ingested by J774 macrophages.
We also examined whether J774 macrophages have a bias toward C. albicans yeast cells versus hyphal cells. Red fluorescent C. albicans cells were induced to form hyphae by preincubation at 37°C in the presence of 10% serum for variable lengths of time. These red fluorescent hyphal cells were mixed with an equal number of green yEGFP-expressing C. albicans yeast cells, cocultured with macrophages for 40 min, and scored as inside or outside macrophages after staining of the cells with CW, as described above. The results of this experiment demonstrated a marked preference by J774 macrophages for C. albicans yeast cells over hyphal cells (∼10:1 ratio) (Fig. 4A). This preference was observed even when the competitor yeast cells were induced to form hyphae at 37°C for as little as 10 min before they were mixed with yeast cells before coculturing with macrophages (data not shown). Thus, when given a choice, J774 macrophages prefer to take up C. albicans yeast cells rather than hyphal cells.
Defects in cell wall mannan but not in glucan or chitin decrease the competitive uptake efficiency of yeast cells by macrophages.
Why are S. cerevisiae and C. glabrata taken up faster and more efficiently than C. albicans? One possibility is that a critical cell wall recognition ligand is more abundant in S. cerevisiae and C. glabrata than in C. albicans. Alternatively, C. albicans may contain surface molecules that attenuate recognition and uptake by macrophages. To distinguish between these possibilities and analyze the relative contribution of cell wall components to uptake by macrophages, we constructed a collection of green and red fluorescently tagged S. cerevisiae and C. albicans mutants with altered levels of cell wall chitin, glucan, and mannan (Table 1) to analyze by competition assays. The role of mannan was examined using S. cerevisiae or C. albicans strains with defects in the mannosyltransferases required for outer chain elongation of N-linked (mnn10Δ, och1Δ, hoc1Δ) or O-linked (pmt1Δ, pmt2Δ, or pmt4Δ) protein mannosylation (3, 13, 35, 38). The role of glucan was examined using mutants defective in β1,6-glucan synthesis (kre1Δ or kre6Δ) or in β1,3-glucan synthesis (fks1Δ) (4, 16, 33). C. albicans CAI4 has three copies of the FKS1/GSC1 gene, but disruption of two of these three alleles in an fksΔ/Δ/+ strain decreases cell wall β1,3-glucan levels by greater than 50% (33), while in S. cerevisiae fks1Δ mutants, the levels are reduced by greater than 80% (17). The role of chitin was examined using mutants lacking the Chs3 chitin synthase, which is responsible for most (∼90%) of the chitin synthesis in S. cerevisiae and C. albicans (7, 48).
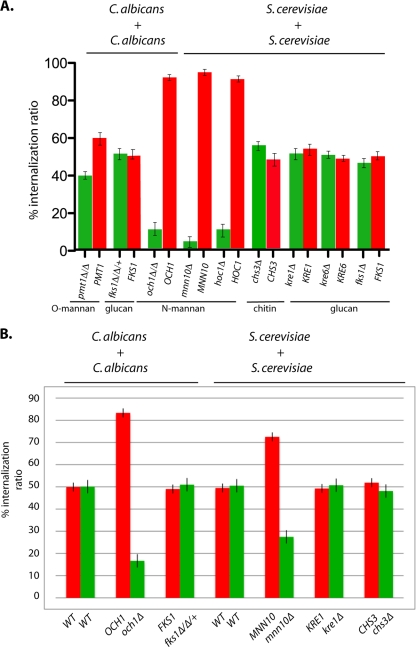
Competition assays between isogenic red fluorescent yeast cells and green fluorescent cell wall mutants were performed to measure the effect of each mutation on competitive uptake by J774 macrophages, as described above. Interestingly, reduced levels of mannan, either N or O linked, led to a significant reduction in the competitive uptake of yeast cells by macrophages, while lowered levels of chitin or glucan did not (Fig. 5). This was true of both S. cerevisiae and C. albicans, in which loss of OCH1 or MNN10, which affects N-linked mannan, reduced competitive fungal uptake by almost 10-fold. Complete loss of protein O-mannosylation leads to inviability (21, 38), but even the partial loss of PMT protein O-linked mannosyltransferase function led to reproducibly reduced rates of competitive fungal uptake. In contrast, reduction of glucan or chitin levels had little, if any, effect. The reduced competitive uptake fitness of mannan mutants was not correlated with cell wall-related growth or clumping phenotypes, since some mannan mutants, for instance, pmt1, pmt4, hoc1, and mnn10, have little, if any, growth phenotypes (13, 35, 38), while some glucan mutants, such as kre6Δ, grow very slowly and clump significantly yet are taken up by macrophages as well as their wild-type competitor is. It should be emphasized that decreased uptake of these mannan mutants was not readily observed when these strains were incubated alone with macrophages; rather, these differences in uptake reflected an increased preference by macrophages for yeast cells containing mannan in their cell wall that is detected during competition. These results demonstrate that defects in mannan biosynthesis that affect the outer layer of the cell wall decreased the competitive efficiency of yeast recognition for uptake by J774 macrophages, while decreased levels of the inner wall components of glucan or chitin did not.
Fig. 5.
Defects in cell wall mannan but not glucan or chitin decrease the competitive uptake of yeast cells by both J774 and primary macrophages. Green fluorescent yeast strains containing mutations that affect protein mannosylation or glucan or chitin synthesis were mixed with an equal number of red fluorescent isogenic wild-type (WT) strains and coincubated with macrophages. After 40 min, CW was added and fluorescent yeast cells were visually scored as being in or out of macrophages to calculate the percent internalization ratio, as described in Materials and Methods. Data sets represent an average of three different experiments (n > 300 cells per experiment). The genotypes of the yeasts affected in N-linked mannan (och1, mnn10, hoc1), O-linked mannan (pmt1, pmt2, pmt4), β1,3-glucan (fks1), β1,6-glucan (kre1, kre6), or chitin (chs3) are listed in Table 2. Competition assays were performed using J744 cells (A) or primary bone marrow-derived macrophages (B) infected with yeast cells at an MOI of 5, as described in Materials and Methods.
To determine if mannan-dependent fungal recognition is a general property of murine macrophages, we tested competitive fungal uptake by primary murine macrophages isolated from bone marrow that were used without passaging. There were several notable differences between the phagocytic behavior of J774 cells and that of bone marrow-derived macrophages. The primary cells appeared to be saturated for yeast uptake at lower MOIs; unlike a population of J774 cells which contained multiple macrophages that were observed to take up five or more yeast cells per macrophage, primary macrophages usually ingested one and seldom more than two yeast cells per macrophage, even at high MOIs. In addition, we noticed that primary macrophages ingested yeast more quickly (within 20 to 25 min) and induced yeast hyphal elongation within the phagosome more rapidly (data not shown). Despite these differences, primary macrophages displayed the same biases toward yeast cell wall mutants during competition assays. S. cerevisiae and C. albicans mutants defective in N-linked mannosylation were reduced in their efficiency of competitive fungal uptake by up to 10-fold, while those with defects in glucan or chitin synthesis were taken up as well as their wild-type competitors were (Fig. 5B). These results suggest that mannan-dependent recognition during fungal uptake is a general property of murine macrophages.
DISCUSSION
Interactions between the human fungal pathogen Candida albicans and its host alter the infective environment in ways that favor survival of the host or of the pathogen. Understanding this extraordinarily complex process requires methods for studying host-pathogen interactions in living cells. In this report, we demonstrate the feasibility of real-time analyses of fungal infection in live cells and animals, using a codon-optimized photostable RFP. With the availability of green and red fluorescent proteins that are compatible with the noncanonical C. albicans genetic code, we used competition assays to measure the relative contributions of different dimorphic states and of cell wall components to fungal recognition and uptake by murine macrophages. These assays demonstrated the marked preferences by J774 and primary macrophages for yeast uptake and that mannan, but not glucan or chitin, plays a key role in influencing this bias during the initial steps of fungal recognition.
An important technical advance we describe is the proof of concept for the use of yEmRFP as a phenotypically neutral fungal biomarker. Despite high levels of production, yEmRFP does not appear to adversely affect any aspect of yeast growth or virulence, and fungal fluorescence is not attenuated during the course of infection (25) (Table 3). Thus, this yEmRFP biomarker may prove generally useful in proteomic or transcriptional profiling of fungal infections that require separation of microbial from host cells in infected organs. As highlighted by our study of yeast phagocytosis, this yEmRFP biomarker offers significant advantages for monitoring macrophage-fungal interactions, not the least of which is its exceptional photostability. We anticipate that the technology and assays described in this report offer a substantially time- and cost-reduced route for sensitive fluorescence-based assays of fungal and host factors that influence pathogenesis in the context of live cells in all systems, mammalian as well as invertebrate, that can be applied to high-throughput formats.
Although there has been a great deal of progress in identifying mammalian receptors that mediate different signaling cascades during phagocytosis, the identities of the molecules that mediate the initial recognition event between the phagocyte and fungus remain undefined because of technical limitations. Use of competition assays that specifically monitor the efficiency of uptake in the context of the infective environment allowed us to identify factors that influence these earliest steps of fungal recognition by macrophages. Our results pointed to a clear hierarchical preference by J774 macrophages, in which S. cerevisiae and C. glabrata yeast cells are preferred over C. albicans yeast cells and C. albicans yeast cells are preferred over hyphal cells. C. albicans is the most pathogenic of these three organisms, yet it is recognized and taken up the most poorly by these macrophages. This observation raises the question of whether inefficient fungal uptake by phagocytes may be an evasion tactic used by C. albicans to improve its virulence capability. There is precedent for the use of this strategy by other fungi, for instance, the coating of Aspergillus fumigatus with hydrophobins (1). Inefficient uptake of C. albicans by J774 cells could be mediated by the presence of molecules that mask C. albicans surface recognition ligands or, alternatively, by a regulated reduction in the ligands themselves.
Of the major cell wall components, only defects in mannan biosynthesis severely affected S. cerevisiae as well as C. albicans competitive uptake by both J774 cells and primary macrophages (Fig. 5), thus implicating mannose or some mannosylated protein as a critical recognition ligand. The sole dependency on mannan for increased competitive uptake efficiency was unexpected because cell wall β1,3-glucan recognition by dectin-1 has been proposed to be a key event in mediating recognition of yeast by macrophages and triggering phagocytosis (5, 6, 20, 49). Numerous studies demonstrate a dectin-1-dependent activation of host immune responses, but our data are consistent with the idea that this activation must occur after the initial recognition of mannan or a mannosylated protein in murine macrophages. Since the complete loss of any one cell wall component is lethal, our analyses relied on partial-loss-of-function mutants. Thus, the possibility that the residual amount of carbohydrates remaining in the wall of glucan or chitin mutants may be sufficient for recognition by a macrophage receptor cannot be ruled out. However, the observation that the competitive uptake fitness of yeast by macrophages is affected by only a reduced amount of mannan implies that it is the only component whose levels can be altered to influence this stage of uptake, in a manner that is analogous to the limiting component of a chemical reaction. It is notable that C. albicans contains less cell wall mannan than S. cerevisiae or C. glabrata; the mannose-to-glucose ratio of the C. albicans cell wall is almost half that of the C. glabrata cell wall (14). The C. albicans cell wall is also distinguished from the S. cerevisiae and C. glabrata cell walls by the presence of terminal β1,2-linked mannose, whose level affects fungal antigenic properties (44, 47), and by the presence of dityrosine (45), which is known to confer resistance to a variety of harsh conditions. It will be of interest to investigate if the absence of these surface molecules biases the recognition preference of macrophages toward C. albicans.
Supplementary Material
ACKNOWLEDGMENTS
Support was provided by a grant from the National Institutes of Health (NIH R01 A1 047837) to J.B.K.
We thank James Bliska and Galina Romanov for providing primary bone marrow-derived macrophages. We also thank Joachim Ernst for providing the set of C. albicans pmtΔ strains, Toshiuko Mio for the C. albicans gsc1Δ strains, and Hector Mora-Montes and Neil Gow for the C. albicans och1Δ/och1Δ strain.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 10 September 2010.
REFERENCES
- 1.Aimanianda V., Bayry J., Bozza S., Kniemeyer O., Perruccio K., Elluru S. R., Clavaud C., Paris S., Brakhage A. A., Kaveri S. V., Romani L., Latge J. P. 2009. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117–1121 [DOI] [PubMed] [Google Scholar]
- 2.Averbeck N., Gao X. D., Nishimura S., Dean N. 2008. Alg13p, the catalytic subunit of the endoplasmic reticulum UDP-GlcNAc glycosyltransferase, is a target for proteasomal degradation. Mol. Biol. Cell 19:2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates S., Hughes H. B., Munro C. A., Thomas W. P., MacCallum D. M., Bertram G., Atrih A., Ferguson M. A., Brown A. J., Odds F. C., Gow N. A. 2006. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 281:90–98 [DOI] [PubMed] [Google Scholar]
- 4.Boone C., Sommer S. S., Hensel A., Bussey H. 1990. Yeast KRE genes provide evidence for a pathway of cell wall beta-glucan assembly. J. Cell Biol. 110:1833–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown G. D., Gordon S. 2001. Immune recognition. A new receptor for beta-glucans. Nature 413:36–37 [DOI] [PubMed] [Google Scholar]
- 6.Brown G. D., Herre J., Williams D. L., Willment J. A., Marshall A. S., Gordon S. 2003. Dectin-1 mediates the biological effects of beta-glucans. J. Exp. Med. 197:1119–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulawa C. E., Miller D. W., Henry L. K., Becker J. M. 1995. Attenuated virulence of chitin-deficient mutants of Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 92:10570–10574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celada A., Gray P. W., Rinderknecht E., Schreiber R. D. 1984. Evidence for a gamma-interferon receptor that regulates macrophage tumoricidal activity. J. Exp. Med. 160:55–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaffin W. L. 2008. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 72:495–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi J. H., Roos J., Dean N. 1996. The OST4 gene of Saccharomyces cerevisiae encodes an unusually small protein required for normal levels of oligosaccharyltransferase activity. J. Biol. Chem. 271:3132–3140 [DOI] [PubMed] [Google Scholar]
- 11.Cormack B. P., Bertram G., Egerton M., Gow N. A., Falkow S., Brown A. J. 1997. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology 143:303–311 [DOI] [PubMed] [Google Scholar]
- 12.Cormack B. P., Falkow S. 1999. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 151:979–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean N., Poster J. 1996. Molecular and phenotypic analysis of the S. cerevisiae MNN10 gene identifies a family of related glycosyltransferases. Glycobiology 6:73–81 [DOI] [PubMed] [Google Scholar]
- 14.de Groot P. W., Kraneveld E. A., Yin Q. Y., Dekker H. L., Gross U., Crielaard W., de Koster C. G., Bader O., Klis F. M., Weig M. 2008. The cell wall of the human pathogen Candida glabrata: differential incorporation of novel adhesin-like wall proteins. Eukaryot. Cell 7:1951–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.d'Ostiani C. F., Del Sero G., Bacci A., Montagnoli C., Spreca A., Mencacci A., Ricciardi-Castagnoli P., Romani L. 2000. Dendritic cells discriminate between yeasts and hyphae of the fungus Candida albicans. Implications for initiation of T helper cell immunity in vitro and in vivo. J. Exp. Med. 191:1661–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douglas C. M., D'Ippolito J. A., Shei G. J., Meinz M., Onishi J., Marrinan J. A., Li W., Abruzzo G. K., Flattery A., Bartizal K., Mitchell A., Kurtz M. B. 1997. Identification of the FKS1 gene of Candida albicans as the essential target of 1,3-beta-d-glucan synthase inhibitors. Antimicrob. Agents Chemother. 41:2471–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douglas C. M., Foor F., Marrinan J. A., Morin N., Nielsen J. B., Dahl A. M., Mazur P., Baginsky W., Li W., el-Sherbeini M., et al. 1994. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-d-glucan synthase. Proc. Natl. Acad. Sci. U. S. A. 91:12907–12911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filler S. G. 2006. Candida-host cell receptor-ligand interactions. Curr. Opin. Microbiol. 9:333–339 [DOI] [PubMed] [Google Scholar]
- 19.Fonzi W. A., Irwin M. Y. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gantner B. N., Simmons R. M., Underhill D. M. 2005. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24:1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentzsch M., Tanner W. 1996. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 15:5752–5759 [PMC free article] [PubMed] [Google Scholar]
- 22.Goodridge H. S., Wolf A. J., Underhill D. M. 2009. Beta-glucan recognition by the innate immune system. Immunol. Rev. 230:38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guthrie C., Fink G. R. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:3–20 [PubMed] [Google Scholar]
- 24.Kapteyn J. C., Ram A. F., Groos E. M., Kollar R., Montijn R. C., Van Den Ende H., Llobell A., Cabib E., Klis F. M. 1997. Altered extent of cross-linking of beta1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall beta1,3-glucan content. J. Bacteriol. 179:6279–6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keppler-Ross S., Noffz C., Dean N. 2008. A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genetics 179:705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleinegger C. L., Lockhart S. R., Vargas K., Soll D. R. 1996. Frequency, intensity, species, and strains of oral Candida vary as a function of host age. J. Clin. Microbiol. 34:2246–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klis F. M., Boorsma A., De Groot P. W. 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast 23:185–202 [DOI] [PubMed] [Google Scholar]
- 28.Klis F. M., de Groot P., Hellingwerf K. 2001. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 39(Suppl. 1):1–8 [PubMed] [Google Scholar]
- 29.Klis F. M., Sosinska G. J., de Groot P. W., Brul S. 2009. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res. 9:1013–1028 [DOI] [PubMed] [Google Scholar]
- 30.Kollar R., Reinhold B. B., Petrakova E., Yeh H. J., Ashwell G., Drgonova J., Kapteyn J. C., Klis F. M., Cabib E. 1997. Architecture of the yeast cell wall. Beta(1 → 6)-glucan interconnects mannoprotein, beta(1 → 3)-glucan, and chitin. J. Biol. Chem. 272:17762–17775 [DOI] [PubMed] [Google Scholar]
- 31.Levitz S. M. 2010. Innate recognition of fungal cell walls. PLoS Pathog. 6:e1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcil A., Gadoury C., Ash J., Zhang J., Nantel A., Whiteway M. 2008. Analysis of PRA1 and its relationship to Candida albicans-macrophage interactions. Infect. Immun. 76:4345–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mio T., Adachi-Shimizu M., Tachibana Y., Tabuchi H., Inoue S. B., Yabe T., Yamada-Okabe T., Arisawa M., Watanabe T., Yamada-Okabe H. 1997. Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in beta-1,3-glucan synthesis. J. Bacteriol. 179:4096–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murad A., Lee P. I. D., Broadbent I., Barelle C., Brown A. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325–327 [DOI] [PubMed] [Google Scholar]
- 35.Neiman A. M., Mhaiskar V., Manus V., Galibert F., Dean N. 1997. Saccharomyces cerevisiae HOC1, a suppressor of pkc1, encodes a putative glycosyltransferase. Genetics 145:637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Netea M. G., Brown G. D., Kullberg B. J., Gow N. A. 2008. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 6:67–78 [DOI] [PubMed] [Google Scholar]
- 37.Pappas P. G., Kauffman C. A., Andes D., Benjamin D. K., Jr., Calandra T. F., Edwards J. E., Filler S. G., Jr., Fisher J. F., Kullberg B. J., Ostrosky-Zeichner L., Reboli A. C., Rex J. H., Walsh T. J., Sobel J. D. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 48:503–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prill S. K., Klinkert B., Timpel C., Gale C. A., Schroppel K., Ernst J. F. 2005. PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 55:546–560 [DOI] [PubMed] [Google Scholar]
- 39.Pujol C., Bliska J. B. 2003. The ability to replicate in macrophages is conserved between Yersinia pestis and Yersinia pseudotuberculosis. Infect. Immun. 71:5892–5899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rida P. C., Nishikawa A., Won G. Y., Dean N. 2006. Yeast-to-hyphal transition triggers formin-dependent Golgi localization to the growing tip in Candida albicans. Mol. Biol. Cell 17:4364–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8:4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reference deleted.
- 43.Sandrock C., Siddiqui J. 2009. The value of a risk model for early-onset candidemia. Crit. Care 13:1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shibata N., Ikuta K., Imai T., Satoh Y., Satoh R., Suzuki A., Kojima C., Kobayashi H., Hisamichi K., Suzuki S. 1995. Existence of branched side chains in the cell wall mannan of pathogenic yeast, Candida albicans. Structure-antigenicity relationship between the cell wall mannans of Candida albicans and Candida parapsilosis. J. Biol. Chem. 270:1113–1122 [DOI] [PubMed] [Google Scholar]
- 45.Smail E. H., Briza P., Panagos A., Berenfeld L. 1995. Candida albicans cell walls contain the fluorescent cross-linking amino acid dityrosine. Infect. Immun. 63:4078–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timpel C., Strahl-Bolsinger S., Ziegelbauer K., Ernst J. F. 1998. Multiple functions of Pmt1p-mediated protein O-mannosylation in the fungal pathogen Candida albicans. J. Biol. Chem. 273:20837–20846 [DOI] [PubMed] [Google Scholar]
- 47.Trinel P. A., Faille C., Jacquinot P. M., Cailliez J. C., Poulain D. 1992. Mapping of Candida albicans oligomannosidic epitopes by using monoclonal antibodies. Infect. Immun. 60:3845–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valdivieso M. H., Mol P. C., Shaw J. A., Cabib E., Duran A. 1991. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae. J. Cell Biol. 114:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willment J. A., Brown G. D. 2008. C-type lectin receptors in antifungal immunity. Trends Microbiol. 16:27–32 [DOI] [PubMed] [Google Scholar]
- 50.Wilson R. B., Davis D., Mitchell A. P. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.