Abstract
The regulation of the response of Candida albicans to hypoxic (low-oxygen) conditions is poorly understood. We used microarray and other transcriptional analyses to investigate the role of the Upc2 and Bcr1 transcription factors in controlling expression of genes involved in cell wall metabolism, ergosterol synthesis, and glycolysis during adaptation to hypoxia. Hypoxic induction of the ergosterol pathway is mimicked by treatment with sterol-lowering drugs (ketoconazole) and requires UPC2. Expression of three members of the family CFEM (common in several fungal extracellular membranes) of cell wall genes (RBT5, PGA7, and PGA10) is also induced by hypoxia and ketoconazole and requires both UPC2 and BCR1. Expression of glycolytic genes is induced by hypoxia but not by treatment with sterol-lowering drugs, whereas expression of respiratory pathway genes is repressed. However, Upc2 does not play a major role in regulating expression of genes required for central carbon metabolism. Our results indicate that regulation of gene expression in response to hypoxia in C. albicans is complex and is signaled both via lowered sterol levels and other unstudied mechanisms. We also show that induction of filamentation under hypoxic conditions requires the Ras1- and Cdc35-dependent pathway.
The human fungal pathogen Candida albicans grows on superficial and internal sites in the infected host, areas that differ significantly in the availability of oxygen (26). Under low-oxygen (hypoxic) conditions, C. albicans switches from yeast to hyphal growth, a phenotypic change that has been associated with invasion and virulence (24, 30, 54). Exposure to hypoxia results in increased expression of genes involved in ergosterol synthesis and glycolysis and reduced expression of oxidative phosphorylation (3, 69). There is also an increase in expression of some cell wall and hyphal-specific genes (69), which is reflected in changes in the cell wall proteome (72).
Although the response to hypoxia has been well studied in fungi such as Saccharomyces cerevisiae and Schizosaccharomyces pombe, much less is known about the response in C. albicans. In S. pombe, a decrease in oxygen is detected by the reduction in sterol synthesis, which requires 12 molecules of oxygen to convert one squalene to ergosterol (38). Sterol content is sensed by Sre1 and Scp1, which are homologs of the human sterol regulatory element binding protein (SREBP) and Scap (SREBP cleavage-activating protein) (38, 77). Sre1 and Scp1 are localized in the membrane, where they remain when sterol and oxygen levels are high. When sterol levels drop, the N terminus of Sre1 is cleaved off, and it enters the nucleus, where it acts as a transcriptional regulator. Sre1 is the major regulator of the hypoxic response in S. pombe and regulates expression of >100 genes (38). It also functions in a sterol-independent manner (39). SREBPs are also important for sensing oxygen levels in the pathogenic fungi Cryptococcus neoformans and Aspergillus fumigatus (7, 13, 85).
There are no obvious SREBP orthologs in S. cerevisiae or in any of the Candida clade species, although there are some similarities with other proteins, such as Cph2 and Tye7 in C. albicans (7). Tye7 is required for regulation of glycolytic gene expression (3), but as the similarity with SREBP does not extend beyond the basic helix-loop-helix domains, it is very unlikely that Tye7 is a member of the SREBP family. The role of Cph2 in hypoxic regulation has not been studied. However, C. albicans (and S. cerevisiae) lacks orthologs of Scap (7).
S. cerevisiae cells sense oxygen via the levels of heme and sterols (19, 20, 34). Biosynthesis of heme regulates the activity of the transcriptional regulators Hap1 and the CCAAT-binding complex Hap2/3/4/5 (90). Targets of Hap1 include the repressor Rox1. When the oxygen levels drop, heme is no longer synthesized and no longer binds to Hap1. As a result, ROX1 is not expressed and no longer represses expression of >100 genes involved in the hypoxic response (6, 49, 76).
In a parallel pathway, sterol depletion leads to activation of the regulators Upc2 and Ecm22, which control expression of a subset of hypoxic genes (19, 80). Upc2 and Ecm22 are paralogs resulting from the whole-genome duplication in yeast (86) and are especially similar at the amino terminus, where there is a Zn(2)-Cys(6) binuclear transcription factor domain, and at the carboxy terminus, where they contain four predicted transmembrane segments (53). Upc2 and Ecm22 bind a sequence motif known as the sterol regulatory element (SRE) in the promoters of their target genes. They regulate expression of ergosterol biosynthesis genes and the DAN/TIR family of cell wall proteins (1, 80). Despite the sequence similarity between Upc2 and Ecm22, there are some differences in function. Ecm22 is bound to the promoters of ergosterol synthesis genes in normoxia. Under hypoxic conditions, or when sterol levels drop, Ecm22 is lost from the promoters and is replaced by Upc2 (19, 20). Upc2 also binds to SREs in the promoters of the DAN/TIR genes. Ecm22 is required for the repression of hypoxic genes in normoxia, whereas Upc2 (whose expression is repressed in normoxia) is associated with hypoxic induction.
Davies and Rine (19) showed that blocking ergosterol production in normoxia by using lovastatin or ketoconazole results in induction of expression of UPC2 and its target genes. This is not alleviated by the addition of heme, demonstrating that in S. cerevisiae, hypoxic induction of UPC2 responds directly to reduced sterols. The promoter of UPC2 also contains Rox1 binding sites; expression is therefore also regulated indirectly by Hap1 (65).
The responses of C. albicans and S. cerevisiae to hypoxia are similar and are associated with increased expression of ergosterol synthesis, cell wall composition, and glycolytic genes and reduced expression of components of the respiratory chain, ATP synthesis, and the citric acid cycle (3, 49, 69, 90). However, the homolog of ROX1 in C. albicans (RFG1) plays no role in the regulation of expression in response to hypoxia and instead is a general regulator of filamentation (42, 45).
UPC2 in C. albicans is a homolog of both S. cerevisiae ECM22 (ScECM22) and S. cerevisiae UPC2 (ScUPC2), with 64% similarity to each (70). C. albicans UPC2 (CaUPC2) is required for expression of the ergosterol biosynthesis pathway in hypoxia and for the response to azole drugs (53, 61, 70). Deletion of UPC2 does not reduce expression of ergosterol synthesis genes in normoxia (25, 53, 70), whereas gain-of-function mutations result in higher expression (25, 33, 35). Like ScUpc2, CaUpc2 binds an SRE in the promoters of the majority of the ergosterol synthesis genes (25, 61, 70, 91). CaUpc2p also regulates its own expression (36, 37, 91). Other regulators of the hypoxic response in C. albicans include Efg1 (69) and the Ace2 transcription factor (54), which are required for the filamentous response to hypoxia.
Here, we investigate the role of Upc2 in regulating the global response of C. albicans during adaptation to hypoxia. We show that lowering sterol levels mimics the hypoxic induction of the ergosterol pathway and of the family CFEM (common in several fungal extracellular membranes) of glycosylphosphatidylinositol (GPI)-anchored cell wall proteins. We also investigate the role of Bcr1, which is a major regulator of CFEM expression (57, 59). Hypoxic induction of the ergosterol pathway is dependent on Upc2 only, whereas induction of CFEM genes requires both Upc2 and Bcr1. The lowering of sterol levels also induces both sets of genes. In contrast, expression of glycolytic genes does not result from lowered sterol levels.
MATERIALS AND METHODS
Strains and media.
The strains used are shown in Table 1 and were grown in YPD (1% yeast extract, 2% peptone, 2% glucose). Where specified, ketoconazole was added at a final concentration of 0.04 μg ml−1. At this concentration, expression of the ergosterol family genes is induced, but inhibition of growth is minimized. Experiments were carried out using an Invivo2 400 hypoxic chamber. The oxygen concentration was maintained at 1% O2 by varying the concentration of nitrogen. For analysis of colony morphology, overnight cultures of cells were diluted with sterile water to a concentration of 1,000 cells ml−1, and 100 μl was spread on YPD plates and incubated at 37°C and 1% O2 or 21% O2 for 5 days. Liquid YPD was preconditioned at 1% O2 for no less than 4 h before strains were subcultured.
Table 1.
Strains used in this study
| C. albicans strain | Genotype or description | Reference |
|---|---|---|
| SC5314 | Wild type | 29 |
| DAY286 | ura3::λimm434/ura3::λimm434 pARG4::URA3::arg4::hisG/arg4::hisG his1::hisG/his1::hisG | 21 |
| TW14920 | upc2::URA3/upc2::ARG4 | 70 |
| CJN702 | ura3::λimm434/ura3::λimm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG::pHIS1 bcr1::ARG4/bcr1::URA3 | 59 |
| CJN698 | ura3::λimm434/ura3::λimm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG::pHIS1-BCR1 bcr1::ARG4/bcr1::URA3 | 59 |
| JCO188 | ura3::λimm434/ura3::λimm434 aqy1::hisG-URA3-hisG/aqy1::hisG | 12 |
| CDH107 | ura3/ura3 ras1::hisG-URA3-hisG/ras1::hisG | 51 |
| CR216 | ura3::λimm434/ura3::λimm434 cdc35::hisG-URA3hisG/cdc35::hisG | 64 |
| RM1000 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG | 55 |
Fluorescence microscopy.
Cells were picked from a plate, suspended in 100 μl calcofluor white solution (1 mg ml−1 in 10 mM NaOH), and incubated at room temperature for 5 min with gentle agitation. Images were obtained using a ColorView II (Soft Imaging Systems) camera mounted on an Olympus BX40 fluorescence microscope with analySIS software.
Transcriptional profiling.
Cells were grown overnight in 5 ml YPD at 30°C, 200 rpm, and 21% O2, subcultured to an A600 of 0.2 in 50 ml YPD medium, and grown for 3.5 h at 30°C under different conditions, such as variable oxygen concentrations (21% or 1%) or in the presence of ketoconazole (0.04 μg ml−1) for 3.5 h. RNA was extracted using the RiboPure yeast kit, obtained from Ambion. For the hypoxia experiments, eight biological replicates (C. albicans DAY286) were used; in six replicates, the normoxic sample was labeled with Cy3, and the hypoxic samples were labeled with Cy5, and in two, the dyes were swapped. Four biological replicates were used to determine the response to ketoconazole; in two, the untreated samples were labeled with Cy5, and the treated samples were labeled with Cy3. For the upc2Δ study, both wild-type (DAY286) and deletion strains were grown in YPD under hypoxic conditions. Nine biological replicates were used; in six, the wild-type sample was labeled with Cy3, and the upc2Δ deletion strain was labeled with Cy5. cDNA synthesis was carried out using 24 μg RNA, 0.2 pmol C. albicans-specific reverse transcriptase (RT) primers mix (Eurogentec), and RNase-free water in a volume of 18.5 μl. Labeling, hybridization, washing, and scanning were carried out as described in Rossignol et al. (67), except that hybridization was carried out in an Agilent SureHyb hybridization chamber. The microarrays used in this study were designed from assembly 21 of the C. albicans genome using eArray from Agilent Technologies (design ID 017942). A total of 6,101genes (including 12 mitochondrial genes) are represented by two sets of probes, both spotted in duplicate. Probes are randomly distributed. Four copies of each array were printed on a single slide (4 × 44,000) and hybridized individually.
Data analysis.
All microarray data were analyzed using the LIMMA package from the Bioconductor project (http://bioconductor.org) (71). Preprocessing of the data was carried out by applying Lowess normalization and no background correction, and duplicated probes were considered within each array as technical replicates (89). This assumption allows us to take full advantage of the platform design, analyzing the within-array replicate spots using a pooled correlation method. Probes with log fold changes of less than 1 or P values of greater than 0.05 were discarded.
Clustering analysis.
Each of the arrays in these experiments make a direct comparison with a common reference (wild type), allowing the application of clustering techniques to identity functional groups of genes among different experimental conditions. First, Lowess normalization was applied to each array separately. Second, quantile normalization was used to allow log ratios to be compared across arrays. Genes that were differentially expressed in at least one experiment were selected. The normalized data set was analyzed using hierarchical clustering (Euclidean distance and complete linkage) and k-means clustering (23). An optimal number of 9 clusters was adopted, as suggested by the evaluation of the Rand index, implemented in CRAN (http://cran.r-project.org/).
GO analysis.
Gene ontology (GO) analysis was carried out using the GO Term Finder at the CGD (Candida Genome Database) (http://www.candidagenome.org/cgi-bin/GO/goTermFinder). Upregulated and downregulated genes were analyzed separately.
Quantitative reverse transcriptase PCR (qRT-PCR).
A supplementary treatment with DNase I was used to ensure that there was no DNA contamination. DNase I-treated RNA (2 μg) was then incubated at 70°C for 10 min with 0.1 μg oligo(dT). This was added to a final mix of 20 μl containing 4 μl 5× RT reaction buffer (Promega), 1 μl 10 mM deoxynucleoside triphosphates (dNTPs), 1 μl RNasin (40 U μl−1; Promega), and 1 μl Moloney murine leukemia virus reverse transcriptase (MMLV) RT (200 U μl−1; Promega) and incubated at 37°C for 1 h and then at 95°C for 2 min. Primers (see Table S1 in the supplemental material) were designed using Primer3Plus online software (available at http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) using quantitative PCR (qPCR) settings. 18S RNA was used as an endogenous control. A 10-μl cocktail containing 0.5 μg of cDNA was used as a template for amplification, with 2× SYBR green PCR master mixes (Applied Biosystems) and 0.3 μM of each primer, on an ABI 7900HT sequence detection system. The program consisted of a denaturing step at 50°C for 2 min and an activation step at 95°C for 10 min. cDNA from four replicates was used, except for with the wild type in normoxia treated with ketoconazole, in which that from three replicates was used, and expression of ERG11 under hypoxic conditions was determined using two replicates.
Microarray data accession number.
The original data and a description of the arrays used have been deposited in the Gene Expression Omnibus database under the series accession number GSE24076.
RESULTS
Induction of gene expression by hypoxia.
We first tested the transcriptional response of C. albicans during adaptation to hypoxia by comparing the profile of cells grown for 3.5 h at 1% oxygen in an hypoxic chamber to that of cells grown in atmospheric oxygen. Our results confirmed those reported by Setiadi et al. (69) and Askew et al. (3), who used cultures bubbled with 99% nitrogen. We identified 409 genes with differential expression in hypoxia (see Table S2 in the supplemental material). Gene ontology (GO) analysis indicated that genes with increased expression are associated with glycolysis and steroid/ergosterol synthesis, whereas downregulated genes are associated with the tricarboxylic acid (TCA) cycle, oxidative phosphorylation, and iron transport (see Table S3 in the supplemental material). We also observed significant increases in expression of three related cell wall genes (RBT5, PGA7, and PGA10) (see Table S2 in the supplemental material). These each contain an 8-cysteine domain, belong to the family CFEM (common in several fungal extracellular membranes), and are associated with biofilm development and heme binding (62, 82).
To determine if the hypoxic response could be mimicked by lowering sterol levels, we treated cells with ketoconazole, which targets the activity of Erg11 (84). We used a low level of drug (0.04 μg ml−1) and a short treatment time (3.5 h), which allows us to investigate the initial response to drug treatment and compare it to the early response to hypoxia. We identified 200 genes with increased expression and 151 genes with decreased expression following exposure to ketoconazole (see Table S4 in the supplemental material). Upc2 is a major regulator of expression of ergosterol genes in C. albicans and other fungi, and its own expression is induced during hypoxia (37, 53, 61, 70). We therefore examined if Upc2 plays a general role in regulating the hypoxic response of C. albicans by comparing the transcriptional profile of a upc2 deletion isolate with that of a wild-type isolate, both grown under hypoxic conditions. We identified 528 genes that are differentially expressed in the upc2 deletion isolate; 259 are upregulated, and 269 are downregulated (see Table S5 in the supplemental material).
Expression analysis of all data sets.
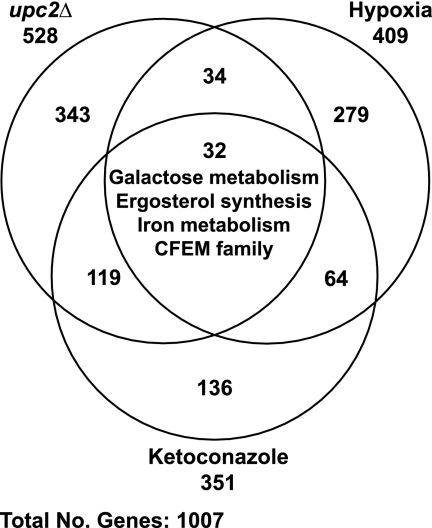
A summary of the array data from all three experiments is shown in Fig. 1. The degree of overlap is relatively small; only 32 genes are differentially expressed in hypoxia, following ketoconazole treatment, and in a upc2 deletion background (Table 2). However, this group includes many of the major classes of genes differentially expressed in hypoxia, as follows: members of the CFEM family, genes involved in ergosterol synthesis, and genes required for iron assimilation and galactose metabolism. Expression of CFEM, ergosterol, and iron metabolism genes is induced by hypoxia and ketoconazole and reduced in the upc2 deletion background, whereas expression of galactose genes is reduced in hypoxia and increased following treatment with ketoconazole or in the upc2 deletion background (Table 2).
Fig. 1.
Genes with differential expression in all three microarray data sets (hypoxia, ketoconazole, and upc2Δ). The overlap between each data set was determined using Microsoft Access. Genes with differential expression in all experiments are listed in Table 2.
Table 2.
Genes with differential expression in hypoxia, following ketoconazole treatment, and in a upc2Δ background
| Locus tag | Gene | Description | Log fold change |
||
|---|---|---|---|---|---|
| Hypoxia | Ketoconazole treatment | upc2Δ background | |||
| Ergosterol synthesis pathway | |||||
| orf19.767 | ERG3 | C-5 sterol desaturase | 2.47 | 1.59 | −2.34 |
| orf19.922 | ERG11 | Lanosterol 14-alpha-demethylase | 1.49 | 1.14 | −1.89 |
| CFEM family | |||||
| orf19.5636 | RBT5 | GPI-anchored cell wall protein involved in hemoglobin utilization | 2.77 | 1.40 | −2.17 |
| orf19.5635 | PGA7 | Hyphal surface antigen | 1.90 | 1.16 | −1.48 |
| Iron metabolism | |||||
| orf19.5634 | FRP1 | Predicted ferric reductase | 3.39 | 2.53 | −1.97 |
| orf19.4215 | FET34 | Multicopper ferroxidase | 2.63 | 0.75 | −1.05 |
| Galactose metabolism | |||||
| orf19.3675 | GAL7 | Galactose-1-phosphate uridyl transferase | −1.57 | 1.13 | 1.16 |
| orf19.3672 | GAL10 | UDP-glucose 4-epimerase | −1.47 | 1.59 | 1.83 |
| orf19.3670 | GAL1 | Galactokinase | −1.30 | 1.95 | 1.30 |
| Miscellaneous | |||||
| orf19.1691 | Uncharacterized | 6.33 | 2.45 | −1.71 | |
| orf19.3548.1 | WH11 | Cytoplasmic protein | 5.64 | 2.13 | 1.36 |
| orf19.2959.1 | Uncharacterized | 2.65 | 2.15 | 1.92 | |
| orf19.251 | ThiJ/PfpI protein family | 2.22 | 1.01 | 1.02 | |
| orf19.3932 | Uncharacterized | 2.09 | 1.80 | 2.40 | |
| orf19.675 | Similar to cell wall proteins | 2.03 | 1.30 | 1.25 | |
| orf19.260 | SLD1 | Sphingolipid Δ8 desaturase | 1.94 | 1.33 | −1.10 |
| orf19.7218 | RBE1 | Putative cell wall protein | 1.73 | −0.89 | −2.01 |
| orf19.5103 | Uncharacterized | 1.73 | −0.93 | −1.00 | |
| orf19.6117 | Uncharacterized | 1.68 | 0.93 | 1.10 | |
| orf19.4477 | CSH1 | Member of aldo-keto reductase family | 1.57 | 1.87 | 2.34 |
| orf19.4255 | ECM331 | GPI-anchored protein | 1.28 | 1.75 | 1.99 |
| orf19.6637 | Uncharacterized | 1.27 | 0.91 | 1.67 | |
| orf19.6877 | Uncharacterized | 1.19 | 1.35 | 1.34 | |
| orf19.2030 | Uncharacterized | 1.10 | 1.38 | 1.81 | |
| orf19.6554 | Uncharacterized | 1.01 | 1.43 | 1.95 | |
| orf19.4082 | DDR48 | Immunogenic stress-associated protein | 0.82 | 4.70 | 4.18 |
| orf19.999 | Uncharacterized | 0.79 | 1.08 | 1.08 | |
| orf19.4476 | Uncharacterized | 0.78 | 1.92 | 1.76 | |
| orf19.5125 | Uncharacterized | 0.67 | 1.38 | 1.59 | |
| orf19.4506 | LYS22 | Homocitrate synthase | −0.99 | 1.48 | 1.39 |
| orf19.4716 | GDH3 | NADP-glutamate dehydrogenase | −1.39 | 1.00 | 2.36 |
| orf19.670.2 | Uncharacterized | −3.12 | 1.32 | 1.30 | |
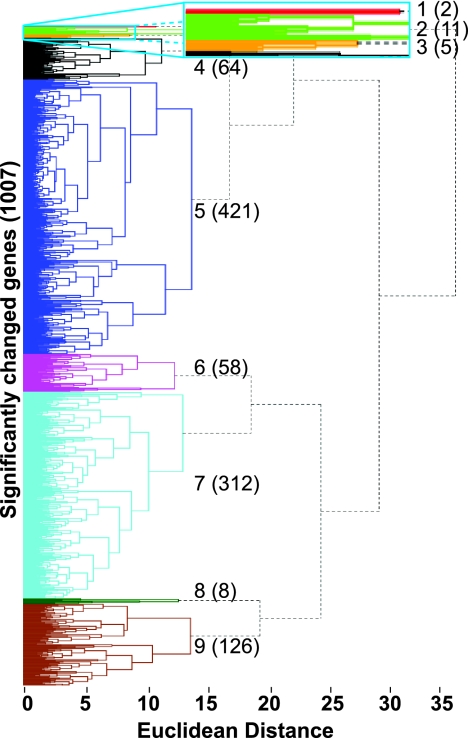
We also used both k-means and hierarchical cluster analysis to characterize genes with similar patterns of expression in the three data sets (1,007 nuclear genes). Genes were grouped based on their parameters of expression. A comparison of both methods using the Rand index (88) suggested that the optimal number of clusters ranged from 7 to 9. Based on manual examination, nine major gene clusters were selected (Fig. 2). The major GO classes represented in each cluster are shown in Table 3, and the genes in each cluster are listed in Table S6 in the supplemental material.
Fig. 2.
Hierarchical cluster analysis of all three microarray data sets (hypoxia, ketoconazole, and upc2Δ). Analysis of 1,007 genes in total. Each cluster is represented by a different color. The numbers to the right represent the clusters, with the number of genes in each cluster shown in parentheses. The blue rectangular box at the top is a close-up view of tightly bunched clusters 1, 2, and 3. The major GO category represented by each cluster is shown in Table 3.
Table 3.
Major GO categories in clusters shown in Fig. 3
| Cluster | No. of genes | GO processes |
|---|---|---|
| 1 | 2 | No significant function |
| 2 | 11 | Ergosterol metabolism and sterol/steroid metabolism |
| 3 | 5 | Iron ion transport, copper ion transport, and metal ion transport |
| 4 | 64 | Steroid/sterol metabolism and ergosterol biosynthesis |
| 5 | 421 | rRNA maturation, ribosomal subunit, and ribosome assembly |
| 6 | 58 | Glycolysis, glucose metabolism, and alcohol metabolism |
| 7 | 312 | Fatty acid catabolism, lipid catabolism, and organic acid catabolism |
| 8 | 8 | No significant function |
| 9 | 126 | Galactose metabolism and amino acid metabolism |
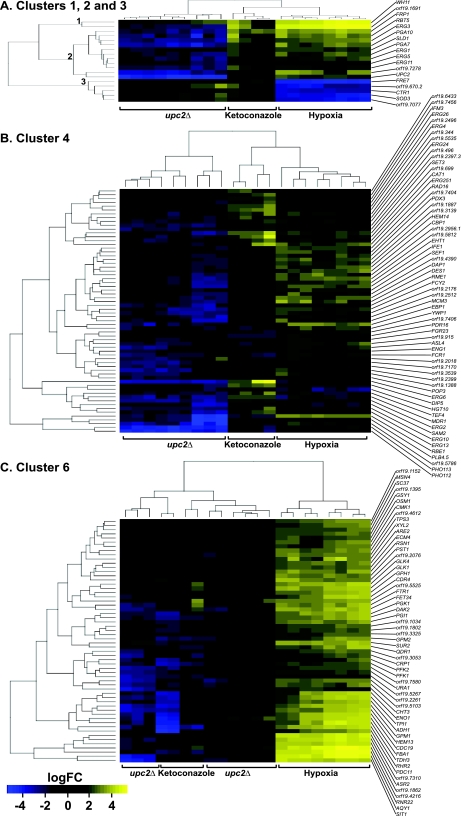
Cluster 1 contains only two genes, WH11 and orf19.1691 (Fig. 3A), which group together separately from the rest of the data (Fig. 3B). WH11 encodes a cytoplasmic protein that is expressed in white-phase yeast cells (73); little is known about the function of orf19.1691, apart from the observation that its expression is induced in filaments or following exposure to fluconazole (17, 41). Expression of both genes is dramatically increased in hypoxia and following treatment with ketoconazole. However, induction of orf19.1691 is dependent on Upc2, whereas expression of WH11 is not. It is likely that the separation of orf19.1691 from genes in cluster 2 is an artifact related to the very high expression levels observed.
Fig. 3.
Gene expression in selected clusters. (A) Genes in clusters 1, 2, and 3. Cluster 1 contains WH11 and orf19.1691 of unknown function. Cluster 2 consists of some ergosterol synthesis genes and the CFEM family. Cluster 3 contains iron metabolism genes. (B) Genes in cluster 4. Cluster 4 contains mainly ergosterol synthesis genes, with some transporters (major facilitator transporter superfamily), cell wall genes, hypha-specific genes, and genes with unknown function. (C) Genes in cluster 6. Cluster 6 consists mainly of genes involved in glycolysis and iron metabolism. Genes in yellow have increased expression, genes in blue have reduced expression, and genes in black are unchanged. logFC, log fold change.
Cluster 2 contains 11 genes, whose expression is induced in hypoxia and by ketoconazole and reduced in a upc2 deletion background (Fig. 3A). This cluster contains four ergosterol synthesis genes (ERG1, ERG3, ERG5, and ERG11). UPC2 is also represented in this cluster, reflecting the fact that it is deleted in the experimental strain. The four ergosterol synthesis genes are among those required for the oxygen-dependent steps of the pathway (see Fig. 7). Cluster 2 also includes three members of the CFEM family (RBT5, PGA7, and PGA10).
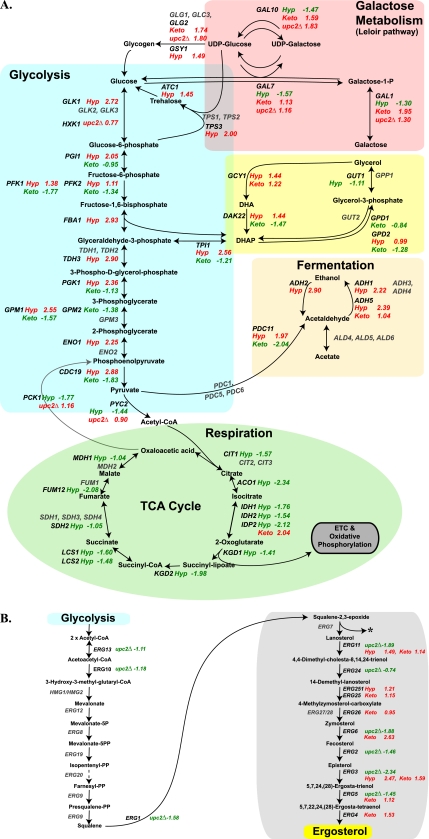
Fig. 7.
Changes in gene expression in central carbon metabolism (A) and ergosterol synthesis (B). The oxygen-dependent part of the ergosterol pathway in highlighted with a gray box. The log fold change in expression is shown; genes with increased expression are shown in red, those with decreased expression are shown in green, and genes that were not detected or are not differentially expressed are shown in gray. Hyp, hypoxia; Keto, ketoconazole; CoA, coenzyme A.
Cluster 3 contains 5 genes, whose expression is reduced in hypoxia but is increased by ketoconazole and in a upc2 deletion background (Fig. 3A). Four are involved in copper and iron ion transport (FRE7, CTR1, SOD3, and orf19.7707). orf19.670.2 has no known function, but its expression pattern suggests that it may also play a role in metal transport.
Cluster 4 contains 64 genes, whose expression is induced in hypoxia. Some have increased expression following exposure to ketoconazole, and most have reduced expression in the upc2 deletion background (Fig. 3B). Many of the genes are involved in sterol biosynthesis and sterol metabolic processes. This cluster also contains SEF1, a putative regulator of iron uptake (50), and FCR1, PDR16, and MDR1, which are associated with drug resistance (22, 31, 75).
Cluster 6 contains 58 genes, whose expression is increased in hypoxia but not in ketoconazole (Fig. 3C). Deletion of UPC2 has little effect on expression, reflected in the fact that the data from the upc2 deletion arrays are intermingled with the ketoconazole data. GO analysis identified a significant overrepresentation of genes involved in glycolysis (Table 3). The cluster also contains some genes associated with iron transport (FTR1 and FET34) and heme biosynthesis (HEM13) and an iron-dependent ribonucleoside diphosphate reductase gene (RNR22).
The remaining clusters are either very large (clusters 5, 7, and 9) or very heterogeneous (cluster 8) and are not discussed. We do, however, note that the galactose metabolism genes cluster together (cluster 9) (Table 3).
Confirmation of array data using qRT-PCR.
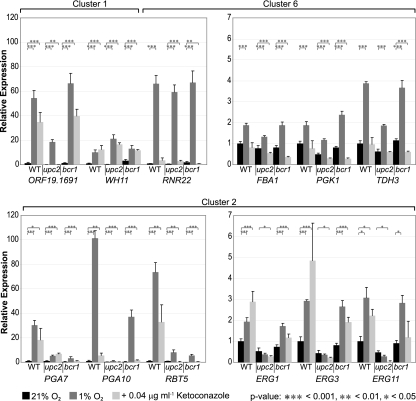
To confirm our array data, we selected the two outlying genes, orf19.1691 and WH11, from cluster 1, the ergosterol genes ERG1, ERG3, and ERG11 and the CFEM genes PGA7, PGA10, and RBT5 from cluster 2, and the iron-dependent ribonucleoside diphosphate reductase gene RNR22 and the glycolytic genes FBA1, PGK1, and TDH3 from cluster 6. Expression was determined by qRT-PCR following exposure to hypoxia, to ketoconazole, and in a upc2 deletion background. Because expression of the CFEM genes is known to be regulated by the transcription factor Bcr1 (2, 57, 59), we also measured expression in a bcr1 deletion background (Fig. 4).
Fig. 4.
Quantitative RT-PCR analysis of gene expression. Genes from clusters 1, 2, and 6 are indicated. Four independent biological replicates were used, except for with treatment of the wild type (WT) with ketoconazole (3 replicates) and measurement of ERG11 expression (2 replicates). Expression was normalized against 18S rRNA and is shown relative to expression of the wild type, with associated standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
qRT-PCR confirmed that expression of genes in the oxygen-dependent steps of the ergosterol pathway (cluster 2) is induced by hypoxia and ketoconazole (Fig. 4). Induction is completely lost when upc2 is deleted, but deletion of bcr1 has very little effect.
Expression of the CFEM genes is greatly induced in hypoxia and somewhat less induced by ketoconazole (Fig. 4). The smallest change observed was for ketoconazole induction of PGA10 expression, and even here, expression is induced by >5-fold. Induction of these genes requires both Upc2 and Bcr1. In the absence of BCR1, in particular, basal levels of expression of all three genes are reduced to effectively zero.
Hypoxia results in a relatively small increase in expression of glycolytic genes (from 1.5- to 3.8-fold) in cluster 6, and ketoconazole has no effect (Fig. 4). Hypoxic induction is slightly reduced in a upc2Δ background but not in a bcr1Δ background. Expression of RNR22 is greatly induced in hypoxia and not by ketoconazole, and again, deletion of upc2 or bcr1 has no effect.
Expression of orf19.1691 and WH11 (outliers in cluster 1) is markedly induced in hypoxia and also by ketoconazole (Fig. 4). These are among the largest changes in expression observed in any experiment. Hypoxic induction of WH11 is independent of Upc2 and Bcr1, and expression of orf19.1691 is reduced in the upc2 deletion background, as suggested by the array experiments.
All of the transcriptional profiling experiments described here were performed in C. albicans strains that carry URA3 deletions and, therefore, also carry deletions of the adjacent gene IRO1 (15). IRO1 is related to AFT1 from S. cerevisiae, which is required for iron utilization (28, 87). However, IRO1 is not an ortholog of AFT1, and its role in iron regulation in C. albicans is unclear (50). Because many genes with increased expression under low-iron conditions are also induced by hypoxia (50), we used quantitative RT-PCR to determine whether the gene expression changes we observed also occur when wild-type C. albicans cells are exposed to low oxygen levels (see Fig. S1 in the supplemental material). We showed that during growth under low-oxygen conditions, expression of orf19.1691, TDH3 (a representative glycolytic gene), ERG1, and ERG11 is increased, similar to the response of the ura3/iro1 deletion strain (see Fig. S1). Expression of PGA7 and PGA10 is also increased, but the magnitude of induction is much less (3- to 7-fold in the wild-type strain compared to 30- to 100-fold in the ura3/iro1 deletion strain). The global response to hypoxia is therefore not affected in the iro1 deletion background, but it is likely that IRO1 plays a role in regulating expression of the CFEM family.
Identification of genes directly regulated by Upc2.
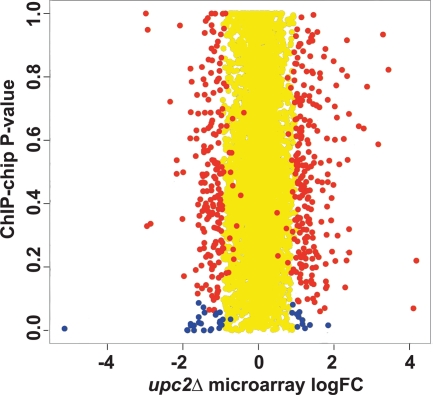
Znaidi et al. (91) identified gene promoters that are bound directly by Upc2 by using chromatin immunoprecipitation (ChIP) analysis of an three-hemagglutinin (HA3)-tagged protein, which acts as a gain-of-function mutation. We compared the fold changes from our upc2Δ gene expression microarrays with the binding score P values from the ChIP-microarray chip (ChIP-chip) data (Fig. 5). We used stringent measurements to identify 38 genes that are shared between the data sets (Fig. 5, blue spots); 25 genes have decreased expression, and 13 have increased expression in the upc2Δ strain grown under hypoxic conditions (Table 4). The scores for Upc2 binding to the promoters are highly significant, and the genes lie at the extremes (low and high) of the expression data (Fig. 5). It is therefore likely that Upc2p directly regulates these genes. The outlying gene with the greatest reduction in expression (Fig. 5, bottom left corner) is UPC2 itself.
Fig. 5.
Comparison of genes that are differentially expressed in a upc2Δ deletion background in microarrays to those bound by Upc2. The log fold changes in expression of the genes in the upc2Δ arrays (x axis) were plotted against the P values from the ChIP-chip array data from Znaidi et al. (91), which identified promoters bound by Upc2 (y axis). Yellow spots represent genes with a log fold change of expression of less than 1. Red spots represent genes with significant changes in expression (P < 0.05) in the upc2Δ array data but with a P value greater than 0.1 (nonsignificant) in the ChIP-chip data. Blue spots represent 38 genes with significant changes in expression in the upc2Δ arrays and with significant binding in the ChIP-chip experiments. The genes are listed in Table 4.
Table 4.
Genes with differential expression in a upc2Δ strain whose promoters are bound by Upc2
| Locus tag | Gene | Description | Mean log fold changea | Mean BRb |
|---|---|---|---|---|
| Downregulated genes | ||||
| Ergosterol biosynthesis | ||||
| orf19.391 | UPC2 | Transcriptional regulator of ergosterol biosynthetic genes | −5.14 | 1.90 |
| orf19.922 | ERG11 | Lanosterol 14-alpha-demethylase | −1.89 | 7.80 |
| orf19.1631 | ERG6 | Δ24-sterol C-methyltransferase | −1.88 | 2.90 |
| orf19.406 | ERG1 | Squalene epoxidase | −1.58 | 1.50 |
| orf19.6026 | ERG2 | C-8 sterol isomerase | −1.46 | 1.80 |
| orf19.5178 | ERG5 | Putative C-22 sterol desaturase | −1.45 | 3.95 |
| orf19.1591 | ERG10 | Acetyl coenzyme A acetyltransferase | −1.11 | 1.15 |
| orf19.1598 | ERG24 | C-14 sterol reductase | −0.74 | 1.65 |
| Multidrug transport | ||||
| orf19.5604 | MDR1 | Multidrug efflux pump of plasma membrane | −1.27 | 3.95 |
| Miscellaneous | ||||
| orf19.2251 | AAH1 | Similar to serine/threonine dehydratases | −1.80 | 1.60 |
| orf19.1996 | CHA1 | Uncharacterized | −1.79 | 1.70 |
| orf19.6090 | NSR1 | Uncharacterized | −1.71 | 1.70 |
| orf19.1691 | Uncharacterized | −1.71 | 3.95 | |
| orf19.1030 | FPR3 | Uncharacterized | −1.58 | 1.60 |
| orf19.5010 | DIM1 | Similar to S. cerevisiae Dim1p | −1.58 | 2.75 |
| orf19.3291 | HMT1 | Major type I protein arginine methyltransferases (PRMT) | −1.52 | 1.65 |
| orf19.18 | IMH3 | IMP dehydrogenase | −1.43 | 1.90 |
| orf19.2575 | Uncharacterized | −1.25 | 1.95 | |
| orf19.1901 | MCM3 | Uncharacterized | −1.13 | 1.60 |
| orf19.657 | SAM2 | S-adenosylmethionine synthetase | −1.04 | 2.25 |
| orf19.5066 | Uncharacterized | −1.03 | 1.85 | |
| orf19.496 | MSH1 | Uncharacterized | −1.02 | 1.50 |
| orf19.6253 | RPS23A | Ribosomal protein | −1.02 | 1.60 |
| orf19.2309.2 | RPL2B | Uncharacterized | −0.97 | 1.70 |
| orf19.4889 | HOL2 | Predicted membrane transporter | −0.94 | 10.75 |
| Upregulated genes | ||||
| orf19.2593 | BIO2 | Biotin synthase | 0.89 | 1.50 |
| orf19.6311 | Uncharacterized | 0.91 | 1.60 | |
| orf19.251 | ThiJ/PfpI protein family | 1.02 | 1.50 | |
| orf19.2699 | ABP1 | Protein similar to S. cerevisiae Abp1p | 1.03 | 1.70 |
| orf19.1789.1 | LYS1 | Mitochondrial homoisocitrate dehydrogenase | 1.06 | 1.60 |
| orf19.1307 | Uncharacterized | 1.07 | 1.50 | |
| orf19.4833 | MLS1 | Malate synthase | 1.09 | 1.60 |
| orf19.1350 | Uncharacterized | 1.16 | 18.50 | |
| orf19.1267 | CAJ1 | Uncharacterized | 1.17 | 1.65 |
| orf19.7204 | Uncharacterized | 1.23 | 2.20 | |
| orf19.4711 | Uncharacterized | 1.36 | 1.65 | |
| orf19.3282 | BMT3 | β-Mannosyltransferase | 1.49 | 1.90 |
| orf19.6840 | Uncharacterized | 1.85 | 1.70 |
Change in expression in a upc2Δ strain in hypoxia.
BR, binding ratio (from reference 91).
Genes with decreased expression in the upc2 deletion background that are also bound by Upc2 include ergosterol synthesis genes (ERG1, ERG2, ERG5, ERG6, ERG11, and UPC2), the multidrug efflux pump MDR1, a predicted membrane transporter, HOL2, the unknown orf19.1691, and ribosomal genes RPS23A and RPL2.
Genes with increased expression in the upc2 deletion background include orf19.251, whose expression is induced both in hypoxia and by ketoconazole (see Tables S2 and S4 in the supplemental material). Expression of orf19.6311, MLS1, BMT3, and orf19.6840 is also increased in the absence of UPC2 and in the presence of ketoconazole; however, no changes in expression were observed following exposure to hypoxia (see Tables S2 and S4).
Regulation of filamentation in response to hypoxia.
C. albicans strains grow as filaments under several environmental conditions, including under low-oxygen conditions (8, 24, 54). The signaling pathways have been well studied, and most require Ras1, a GTP-bound protein that regulates the cyclic AMP (cAMP) signal transduction cascade and the mitogen-activated protein kinase (MAPK) cascade (51). Many different signals that induce hyphal growth have been identified, from nutritional signals such as N-acetylglucosamine and methionine, to quorum sensing molecules such as farnesol, to environmental changes in CO2, pH, and osmotic stress. Most sensors activate downstream pathways via cAMP and protein kinase A or MAP kinases (18). We therefore tested the effect of deleting RAS1, a GTP-bound protein that regulates the cAMP signal transduction cascade and the MAPK cascade (51), and CDC35, which encodes adenylate cyclase, which synthesizes cAMP (64). The phenotypic effect was compared with that of deletions of upc2 and bcr1.
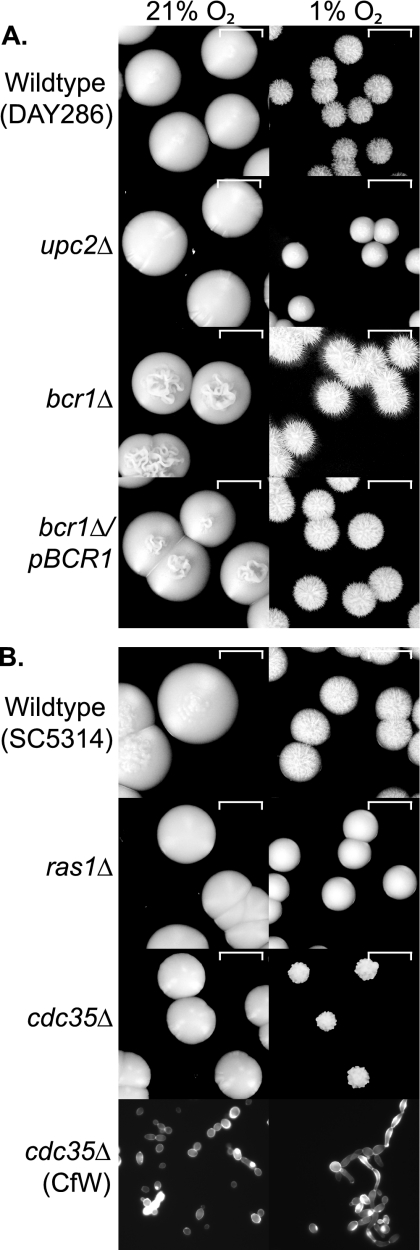
Figure 6 A shows that the upc2 deletion strain grows slowly in reduced oxygen and does not generate hyphae. The bcr1 deletion, however, is hyperfilamentous in hypoxia, and filamentation levels return to normal when BCR1 is reintroduced. Filamentation is absent in ras1 and cdc35 deletion strains in hypoxia (Fig. 6B). Both grow more slowly when oxygen is reduced, and colonies from the cdc35 deletion strain have an unusual “popcorn” appearance that contains pseudohyphae but no hyphae (detected by staining chitin with calcofluor white). These results suggest that Upc2 is required for filamentation under hypoxic conditions and that oxygen sensing induces hyphal development via the downstream cAMP-dependent pathway.
Fig. 6.
Disruption of hypoxia-induced filamentation. (A) Hypoxia-induced filamentation is abolished in a upc2Δ background and increased in a bcr1Δ background. Overnight cultures of the C. albicans wild-type (DAY286), upc2 deletion (TW14920), and bcr1 deletion (CJN702) strains and a reconstituted BCR1 strain (CJN698) were diluted to 20 cells μl−1. Approximately 100 cells were plated onto YPD agar and incubated at 37°C for 5 days in normoxia (21% O2) or in a hypoxic chamber at 1% O2. (B) Filamentation is abolished by deleting ras1 or cdc35. Overnight cultures of the C. albicans wild-type (SC5314), ras1 deletion (CDH107), and cdc35 deletion (CR216) strains were treated as described in the legend to panel A. The bottom panel shows cells picked from the edge of the cdc35 deletion strain colonies in 21% O2 and 1% O2 and stained with calcofluor white (CfW).
DISCUSSION
In an attempt to better understand how C. albicans responds to hypoxia, we carried out transcriptional analysis using whole-genome microarrays. The transcriptional responses of C. albicans to hypoxia and to azole drugs have been reported previously (3, 52, 69), and Dunkel et al. (25) identified some targets of Upc2 using a gain-of-function mutation under normoxic conditions. However, our analysis allowed us to compare the responses and to ascertain the global role of Upc2 during adaptation to hypoxia. We identified 9 gene clusters that are differentially expressed following exposure to hypoxic conditions, including genes involved in glycolysis and ergosterol metabolism and members of the CFEM family (see Tables S2 and S3 in the supplemental material) (3, 69).
Upc2 plays a role in regulating expression of genes in clusters 1, 2, and 4 (except for WH11), which are induced both in hypoxia and in ketoconazole (Fig. 3). The clusters are predominantly discriminated by the magnitude of the response, with genes in clusters 1 and 2 showing greater increases in expression in response to hypoxia. Hypoxic induction of genes in these clusters is likely to be regulated in response to lowered sterol levels, because treatment with the azole drug mimics the hypoxic response. Motifs similar to that of the SRE (binding site for Upc2 [25, 61]) are overrepresented in promoters of genes in the three clusters (data not shown).
Cluster 2 contains four genes from the oxygen-dependent part of the ergosterol pathway (plus UPC2); most of the remaining ergosterol synthesis genes (together with two that are not O2 dependent) are found in cluster 4 (Fig. 3C and 7 B). Expression is reduced in a upc2 deletion background, and Upc2 binds to the promoters of many genes (Table 4). Our results support previous observations that Upc2 directly regulates expression of these ergosterol synthesis genes and of the multidrug transporter MDR1 (25, 33, 53, 61, 70). It is also likely that Upc2 directly regulates expression of orf19.1691, which is essentially uncharacterized (Table 4; Fig. 3A).
Cluster 2 also contains three CFEM family members, RBT5, PGA7, and PGA10, and at least two of which are necessary for endocytosis of hemoglobin (83). There are five members of the CFEM family in C. albicans, PGA7, PGA10, RBT5, WAP1/CSA1, and CSA2, which are part of a larger fungal family of glycosylphosphatidylinositol (GPI) proteins (46). In some species (such as the plant pathogen Magnaporthe grisea), CFEM members are involved in pathogenesis (47). In C. albicans, the CFEM family is associated with adhesion and biofilm formation (62) and in acquisition of iron from heme (82, 83). Levels of Pga10 and Rbt5 in the cell wall are increased when oxygen is limited (72). Expression of the CFEM family genes is reduced in a upc2 deletion background. However, it is unlikely that Upc2 directly activates expression of the CFEM family, because it does not bind directly to the promoters (91) and the promoters do not contain SREs (25; data not shown). Indirect regulation may occur through Upc2-dependent control of an additional as-yet-unidentified transcription factor. One candidate is IRO1, because hypoxic induction of CFEM genes is less marked in strains carrying an intact IRO1 gene (see Fig. S1 in the supplemental material).
Cluster 3 contains genes associated with iron and copper transport (Table 3; Fig. 3A). Expression is greatly reduced during adaptation to hypoxia in wild-type cells and is increased in hypoxia in cells with upc2 deletion. Expression of other iron metabolism genes is increased in hypoxia, and expression of some is reduced in the upc2 deletion background (e.g., FRP1 and FET34) (Table 2). Upc2 may therefore indirectly regulate expression of some iron response genes. For genes in this cluster, changes in sterol levels do not signal the hypoxic response, because treatment with ketoconazole does not result in reduced expression. However, control of iron-responsive genes is complex and also requires the transcription factors Sfu1 (50), Efg1 (74), Rim101, and Cbf (4).
The major role of Upc2 during the hypoxic response appears to be as an activator of expression. However, expression of some genes is increased in a upc2 deletion background in hypoxia (see Table S5 in the supplemental material). It is therefore likely that for some genes, Upc2 acts as a repressor.
Cluster 6 contains several genes associated with glycolysis (Fig. 3C and 7). Expression is greatly induced in hypoxia but is relatively unaffected by treatment with ketoconazole or by deletion of upc2. For these genes, hypoxic induction is therefore not signaled via the lowering of sterol levels and is not dependent on UPC2 (Fig. 3 and 4). C. albicans must therefore have another mechanism for sensing low oxygen levels. In mammalian cells, the shift to glycolytic metabolism is regulated by hypoxic activation of the HIF1 transcription factor (14, 79); however, there are no fungal orthologs of HIF1, at least not in the Saccharomyces/Candida clades. In S. cerevisiae, expression of glycolytic genes is regulated by Gcr1, Gcr2, and Tye7 (5, 56, 78). C. albicans has no orthologs of GCR1 and GCR2, and Askew et al. (3) showed that expression of glycolytic genes in this species is controlled by the transcription factors Gal4 and Tye7. Both of these factors are required for induction under hypoxic conditions, and expression of each is rapidly induced upon exposure to hypoxia. However, the mechanisms by which Gal4 and Tye7 respond to changing oxygen levels are not known. The transcription factors Ace2 and Efg1 also upregulate expression of glycolytic genes in C. albicans in normoxia (54, 69), and Efg1 transiently upregulates expression in hypoxia (74). Again, the mechanisms used to correlate expression with oxygen levels are not known.
Expression of genes associated with the TCA pathway, oxidative phosphorylation, and ATP synthesis are reduced in hypoxia (Fig. 7). The regulation of respiratory gene expression in C. albicans has not been well studied (40). However, it is possible (and indeed likely) that it is similar to that in S. cerevisiae, and that regulation requires transcription factors such as those in the HAP family (40, 44, 48, 63).
The changes in central carbon metabolism genes during adaptation to hypoxia in C. albicans (Fig. 7) resemble those reported in other fungi such as S. cerevisiae (48, 49, 90), S. pombe (38), Candida parapsilosis (66), and the aquatic ascomycete Blastocladiella emersonii (11) and in hypoxic mammalian cells (27). Hypoxic induction of glycolytic gene expression is an ancient conserved response and probably represents a shift to fermentative metabolism in the absence of oxygen (81). Interestingly, in the obligatory aerobe C. neoformans, exposure to hypoxic conditions has very little effect on expression of glycolytic genes (16), whereas in Trichoderma reesei (also an obligate aerobe), expression of glycolysis is inhibited in anoxia, and the organism is unable to survive (9). Changes in regulation of the hypoxic response may therefore be associated with the evolution of obligate aerobic metabolism.
Our results show that the transcription factor Bcr1 is required for expression of the CFEM family under both hypoxic and normoxic conditions. It is unlikely, however, to be a major hypoxic regulator, because deletion of BCR1 has little effect on hypoxic induction of the ergosterol pathway or of glycolytic genes. The main role of Bcr1 is regulating expression of genes important for biofilm development, including members of the CFEM family, the ALS adhesins, and the cell wall gene HWP1 (57–60). It is not known if Bcr1 plays a role in hypoxic adaptation in biofilms, where many of its target genes are expressed (74).
Culturing under hypoxic conditions induces filamentation in C. albicans (10, 24, 30, 54), which requires the transcription factors Efg1, Ace2, Czf1, and Upc2 (24, 30, 54). However, before this study, the signal transduction pathway regulating filamentation in hypoxia had not been investigated. We show that hypoxia-induced filamentation is dependent on both Ras1 and Cdc35. It is not clear, however, whether oxygen directly stimulates the adenylyl cyclase to generate cAMP (similar to CO2) or if it acts via Ras1, similar to peptidoglycan and serum (32). Aquaporin (Aqy1) was considered a possible membrane sensor of oxygen because its expression is greatly induced in hypoxia (Fig. 3C; see also Table S2 in the supplemental material); however, gene deletions have no effect on filamentation (not shown).
The response of pathogenic fungi under hypoxic conditions is important for several virulence attributes, from infection to biofilm formation (26, 66, 68, 69, 74). We have shown that lowered sterol levels result in increased expression of a subset of hypoxic genes through the activity of Upc2. However, other regulators, including Ace2 (43), Efg1 (74), Tye7 (3), and some not yet identified, also play major roles.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Science Foundation Ireland 08/IN.1/B1865 and the Irish Research Council for Science, Engineering and Technology.
We are grateful to Ted White, Fritz Muehlschlegel, Aaron Mitchell, and Patrick van Dijck for providing strains and to the anonymous reviewer who suggested that we investigate the role of IRO1.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 24 September 2010.
REFERENCES
- 1.Abramova N. E., Cohen B. D., Sertil O., Kapoor R., Davies K. J., Lowry C. V. 2001. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics 157:1169–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argimon S., Wishart J. A., Leng R., Macaskill S., Mavor A., Alexandris T., Nicholls S., Knight A. W., Enjalbert B., Walmsley R., Odds F. C., Gow N. A., Brown A. J. 2007. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryot. Cell 6:682–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Askew C., Sellam A., Epp E., Hogues H., Mullick A., Nantel A., Whiteway M. 2009. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog. 5:e1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baek Y. U., Li M., Davis D. A. 2008. Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot. Cell 7:1168–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker H. V. 1991. GCR1 of Saccharomyces cerevisiae encodes a DNA binding protein whose binding is abolished by mutations in the CTTCC sequence motif. Proc. Natl. Acad. Sci. U. S. A. 88:9443–9447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becerra M., Lombardia-Ferreira L. J., Hauser N. C., Hoheisel J. D., Tizon B., Cerdan M. E. 2002. The yeast transcriptome in aerobic and hypoxic conditions: effects of hap1, rox1, rox3 and srb10 deletions. Mol. Microbiol. 43:545–555 [DOI] [PubMed] [Google Scholar]
- 7.Bien C. M., Espenshade P. J. 2010. Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot. Cell 9:352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas S., Van Dijck P., Datta A. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71:348–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonaccorsi E. D., Ferreira A. J., Chambergo F. S., Ramos A. S., Mantovani M. C., Farah J. P., Sorio C. S., Gombert A. K., Tonso A., El-Dorry H. 2006. Transcriptional response of the obligatory aerobe Trichoderma reesei to hypoxia and transient anoxia: implications for energy production and survival in the absence of oxygen. Biochemistry 45:3912–3924 [DOI] [PubMed] [Google Scholar]
- 10.Brown D. H., Jr., Giusani A. D., Chen X., Kumamoto C. A. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651–662 [DOI] [PubMed] [Google Scholar]
- 11.Camilo C. M., Gomes S. L. 2010. Transcriptional response to hypoxia in the aquatic fungus Blastocladiella emersonii. Eukaryot. Cell 9:915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carbrey J. M., Cormack B. P., Agre P. 2001. Aquaporin in Candida: characterization of a functional water channel protein. Yeast 18:1391–1396 [DOI] [PubMed] [Google Scholar]
- 13.Chang Y. C., Bien C. M., Lee H., Espenshade P. J., Kwon-Chung K. J. 2007. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol. Microbiol. 64:614–629 [DOI] [PubMed] [Google Scholar]
- 14.Chi J. T., Wang Z., Nuyten D. S., Rodriguez E. H., Schaner M. E., Salim A., Wang Y., Kristensen G. B., Helland A., Borresen-Dale A. L., Giaccia A., Longaker M. T., Hastie T., Yang G. P., van de Vijver M. J., Brown P. O. 2006. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 3:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chibana H., Uno J., Cho T., Mikami Y. 2005. Mutation in IRO1 tightly linked with URA3 gene reduces virulence of Candida albicans. Microbiol. Immunol. 49:937–939 [DOI] [PubMed] [Google Scholar]
- 16.Chun C. D., Liu O. W., Madhani H. D. 2007. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 3:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Copping V. M., Barelle C. J., Hube B., Gow N. A., Brown A. J., Odds F. C. 2005. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J. Antimicrob. Chemother. 55:645–654 [DOI] [PubMed] [Google Scholar]
- 18.Cottier F., Muhlschlegel F. A. 2009. Sensing the environment: response of Candida albicans to the X factor. FEMS Microbiol. Lett. 295:1–9 [DOI] [PubMed] [Google Scholar]
- 19.Davies B. S., Rine J. 2006. A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae. Genetics 174:191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies B. S., Wang H. S., Rine J. 2005. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Mol. Cell. Biol. 25:7375–7385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis D. A., Bruno V. M., Loza L., Filler S. G., Mitchell A. P. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Deken X., Raymond M. 2004. Constitutive activation of the PDR16 promoter in a Candida albicans azole-resistant clinical isolate overexpressing CDR1 and CDR2. Antimicrob. Agents Chemother. 48:2700–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Do J. H., Choi D. K. 2008. Clustering approaches to identifying gene expression patterns from DNA microarray data. Mol. Cells 25:279–288 [PubMed] [Google Scholar]
- 24.Doedt T., Krishnamurthy S., Bockmuhl D. P., Tebarth B., Stempel C., Russell C. L., Brown A. J., Ernst J. F. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15:3167–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunkel N., Liu T. T., Barker K. S., Homayouni R., Morschhauser J., Rogers P. D. 2008. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell 7:1180–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst J. F., Tielker D. 2009. Responses to hypoxia in fungal pathogens. Cell. Microbiol. 11:183–190 [DOI] [PubMed] [Google Scholar]
- 27.Fedele A. O., Whitelaw M. L., Peet D. J. 2002. Regulation of gene expression by the hypoxia-inducible factors. Mol. Interv. 2:229–243 [DOI] [PubMed] [Google Scholar]
- 28.Garcia M. G., O'Connor J. E., Garcia L. L., Martinez S. I., Herrero E., del Castillo Agudo L. 2001. Isolation of a Candida albicans gene, tightly linked to URA3, coding for a putative transcription factor that suppresses a Saccharomyces cerevisiae aft1 mutation. Yeast 18:301–311 [DOI] [PubMed] [Google Scholar]
- 29.Gillum A. M., Tsay E. Y., Kirsch D. R. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 30.Giusani A. D., Vinces M., Kumamoto C. A. 2002. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160:1749–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goffeau A., Park J., Paulsen I. T., Jonniaux J. L., Dinh T., Mordant P., Saier M. H., Jr 1997. Multidrug-resistant transport proteins in yeast: complete inventory and phylogenetic characterization of yeast open reading frames with the major facilitator superfamily. Yeast 13:43–54 [DOI] [PubMed] [Google Scholar]
- 32.Hall R. A., Cottier F., Muhlschlegel F. A. 2009. Molecular networks in the fungal pathogen Candida albicans. Adv. Appl. Microbiol. 67:191–212 [DOI] [PubMed] [Google Scholar]
- 33.Heilmann C. J., Schneider S., Barker K. S., Rogers P. D., Morschhauser J. 2010. An A643T mutation in the transcription factor Upc2p causes constitutive ERG11 upregulation and increased fluconazole resistance in Candida albicans. Antimicrob. Agents Chemother. 54:353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hickman M. J., Winston F. 2007. Heme levels switch the function of HapI of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol. Cell. Biol. 27:7414–7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoehamer C. F., Cummings E. D., Hilliard G. M., Morschhauser J., David Rogers P. 2009. Upc2p-associated differential protein expression in Candida albicans. Proteomics 9:4726–4730 [DOI] [PubMed] [Google Scholar]
- 36.Hoot S. J., Brown R. P., Oliver B. G., White T. C. 2010. The UPC2 promoter in Candida albicans contains two cis-acting elements that bind directly to Upc2p, resulting in transcriptional autoregulation. Eukaryot. Cell 9:1354–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoot S. J., Oliver B. G., White T. C. 2008. Candida albicans UPC2 is transcriptionally induced in response to antifungal drugs and anaerobicity through Upc2p-dependent and -independent mechanisms. Microbiology 154:2748–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes A. L., Todd B. L., Espenshade P. J. 2005. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120:831–842 [DOI] [PubMed] [Google Scholar]
- 39.Hughes B. T., Espenshade P. J. 2008. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 27:1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson D. C., Cano K. E., Kroger E. C., McNabb D. S. 2005. Novel regulatory function for the CCAAT-binding factor in Candida albicans. Eukaryot. Cell 4:1662–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kadosh D., Johnson A. D. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kadosh D., Johnson A. D. 2001. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21:2496–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelly M. T., MacCallum D. M., Clancy S. D., Odds F. C., Brown A. J., Butler G. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol. Microbiol. 53:969–983 [DOI] [PubMed] [Google Scholar]
- 44.Keng T. 1992. HAP1 and ROX1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2616–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khalaf R. A., Zitomer R. S. 2001. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics 157:1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kulkarni R. D., Kelkar H. S., Dean R. A. 2003. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28:118–121 [DOI] [PubMed] [Google Scholar]
- 47.Kulkarni R. D., Thon M. R., Pan H., Dean R. A. 2005. Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biol. 6:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwast K. E., Burke P. V., Poyton R. O. 1998. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol. 201:1177–1195 [DOI] [PubMed] [Google Scholar]
- 49.Kwast K. E., Lai L. C., Menda N., James D. T., III, Aref S., Burke P. V. 2002. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 184:250–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lan C. Y., Rodarte G., Murillo L. A., Jones T., Davis R. W., Dungan J., Newport G., Agabian N. 2004. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 53:1451–1469 [DOI] [PubMed] [Google Scholar]
- 51.Leberer E., Harcus D., Dignard D., Johnson L., Ushinsky S., Thomas D. Y., Schroppel K. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673–687 [DOI] [PubMed] [Google Scholar]
- 52.Liu T. T., Lee R. E., Barker K. S., Wei L., Homayouni R., Rogers P. D. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, andpyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49:2226–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.MacPherson S., Akache B., Weber S., De Deken X., Raymond M., Turcotte B. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 49:1745–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulhern S. M., Logue M. E., Butler G. 2006. The Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot. Cell 5:2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Negredo A., Monteoliva L., Gil C., Pla J., Nombela C. 1997. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 143:297–302 [DOI] [PubMed] [Google Scholar]
- 56.Nishi K., Park C. S., Pepper A. E., Eichinger G., Innis M. A., Holland M. J. 1995. The GCR1 requirement for yeast glycolytic gene expression is suppressed by dominant mutations in the SGC1 gene, which encodes a novel basic-helix-loop-helix protein. Mol. Cell. Biol. 15:2646–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nobile C. J., Andes D. R., Nett J. E., Smith F. J., Yue F., Phan Q. T., Edwards J. E., Filler S. G., Mitchell A. P. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nobile C. J., Mitchell A. P. 2006. Genetics and genomics of Candida albicans biofilm formation. Cell. Microbiol. 8:1382–1391 [DOI] [PubMed] [Google Scholar]
- 59.Nobile C. J., Mitchell A. P. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15:1150–1155 [DOI] [PubMed] [Google Scholar]
- 60.Nobile C. J., Nett J. E., Andes D. R., Mitchell A. P. 2006. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 5:1604–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliver B. G., Song J. L., Choiniere J. H., White T. C. 2007. cis-acting elements within the Candida albicans ERG11 promoter mediate the azole response through transcription factor Upc2p. Eukaryot. Cell 6:2231–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perez A., Pedros B., Murgui A., Casanova M., Lopez-Ribot J. L., Martinez J. P. 2006. Biofilm formation by Candida albicans mutants for genes coding fungal proteins exhibiting the eight-cysteine-containing CFEM domain. FEMS Yeast Res. 6:1074–1084 [DOI] [PubMed] [Google Scholar]
- 63.Pfeifer K., Kim K. S., Kogan S., Guarente L. 1989. Functional dissection and sequence of yeast HAP1 activator. Cell 56:291–301 [DOI] [PubMed] [Google Scholar]
- 64.Rocha C. R., Schroppel K., Harcus D., Marcil A., Dignard D., Taylor B. N., Thomas D. Y., Whiteway M., Leberer E. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12:3631–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosenfeld E., Beauvoit B. 2003. Role of the non-respiratory pathways in the utilization of molecular oxygen by Saccharomyces cerevisiae. Yeast 20:1115–1144 [DOI] [PubMed] [Google Scholar]
- 66.Rossignol T., Ding C., Guida A., d'Enfert C., Higgins D. G., Butler G. 2009. Correlation between biofilm formation and the hypoxic response in Candida parapsilosis. Eukaryot. Cell 8:550–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rossignol T., Logue M. E., Reynolds K., Grenon M., Lowndes N. F., Butler G. 2007. Analysis of the transcriptional response of Candida parapsilosis following exposure to farnesol. Antimicrob. Agents Chemother. 51:2304–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sellam A., Al-Niemi T., McInnerney K., Brumfield S., Nantel A., Suci P. A. 2009. A Candida albicans early stage biofilm detachment event in rich medium. BMC Microbiol. 9:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Setiadi E. R., Doedt T., Cottier F., Noffz C., Ernst J. F. 2006. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen sensing and Efg1p-regulatory networks. J. Mol. Biol. 361:399–411 [DOI] [PubMed] [Google Scholar]
- 70.Silver P. M., Oliver B. G., White T. C. 2004. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 3:1391–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smyth G. K., Speed T. 2003. Normalization of cDNA microarray data. Methods 31:265–273 [DOI] [PubMed] [Google Scholar]
- 72.Sosinska G. J., de Groot P. W., Teixeira de Mattos M. J., Dekker H. L., de Koster C. G., Hellingwerf K. J., Klis F. M. 2008. Hypoxic conditions and iron restriction affect the cell-wall proteome of Candida albicans grown under vagina-simulative conditions. Microbiology 154:510–520 [DOI] [PubMed] [Google Scholar]
- 73.Srikantha T., Soll D. R. 1993. A white-specific gene in the white-opaque switching system of Candida albicans. Gene 131:53–60 [DOI] [PubMed] [Google Scholar]
- 74.Stichternoth C., Ernst J. F. 2009. Hypoxic adaptation by Efg1 regulates biofilm formation by Candida albicans. Appl. Environ. Microbiol. 75:3663–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Talibi D., Raymond M. 1999. Isolation of a putative Candida albicans transcriptional regulator involved in pleiotropic drug resistance by functional complementation of a pdr1 pdr3 mutation in Saccharomyces cerevisiae. J. Bacteriol. 181:231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ter Linde J. J., Steensma H. Y. 2002. A microarray-assisted screen for potential HapI and Rox1 target genes in Saccharomyces cerevisiae. Yeast 19:825–840 [DOI] [PubMed] [Google Scholar]
- 77.Todd B. L., Stewart E. V., Burg J. S., Hughes A. L., Espenshade P. J. 2006. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol. Cell. Biol. 26:2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Uemura H., Fraenkel D. G. 1990. gcr2, a new mutation affecting glycolytic gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:6389–6396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vengellur A., Woods B. G., Ryan H. E., Johnson R. S., LaPres J. J. 2003. Gene expression profiling of the hypoxia signaling pathway in hypoxia-inducible factor 1alpha null mouse embryonic fibroblasts. Gene Expr. 11:181–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vik A., Rine J. 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6395–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Webster K. A. 2003. Evolution of the coordinate regulation of glycolytic enzyme genes by hypoxia. J. Exp. Biol. 206:2911–2922 [DOI] [PubMed] [Google Scholar]
- 82.Weissman Z., Kornitzer D. 2004. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol. 53:1209–1220 [DOI] [PubMed] [Google Scholar]
- 83.Weissman Z., Shemer R., Conibear E., Kornitzer D. 2008. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol. Microbiol. 69:201–217 [DOI] [PubMed] [Google Scholar]
- 84.White T. C., Holleman S., Dy F., Mirels L. F., Stevens D. A. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 46:1704–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Willger S. D., Puttikamonkul S., Kim K. H., Burritt J. B., Grahl N., Metzler L. J., Barbuch R., Bard M., Lawrence C. B., Cramer R. A., Jr 2008. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 4:e1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolfe K. H., Shields D. C. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708–713 [DOI] [PubMed] [Google Scholar]
- 87.Yamaguchi-Iwai Y., Dancis A., Klausner R. D. 1995. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 14:1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yeung K. Y., Ruzzo W. L. 2001. Principal component analysis for clustering gene expression data. Bioinformatics 17:763–774 [DOI] [PubMed] [Google Scholar]
- 89.Zahurak M., Parmigiani G., Yu W., Scharpf R. B., Berman D., Schaeffer E., Shabbeer S., Cope L. 2007. Pre-processing Agilent microarray data. BMC Bioinformatics 8:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zitomer R. S., Lowry C. V. 1992. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 56:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Znaidi S., Weber S., Al-Abdin O. Z., Bomme P., Saidane S., Drouin S., Lemieux S., De Deken X., Robert F., Raymond M. 2008. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot. Cell 7:836–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.