Abstract
Microbe-associated molecular patterns are recognized by Toll-like receptors of the innate immune system. This recognition enables a rapid response to potential pathogens but does not clearly provide a way for the innate immune system to discriminate between virulent and avirulent microbes. We find that pulmonary infection of mice with type 3 translocation-competent Pseudomonas aeruginosa triggers a rapid inflammatory response, while infection with isogenic translocation-deficient mutants does not. Discrimination between translocon-positive and -negative bacteria requires caspase-1 activity in bone marrow-derived cells and interleukin-1 receptor signaling. Thus, the activation of caspase-1 by bacteria expressing type 3 secretion systems allows for rapid recognition of bacteria expressing conserved functions associated with virulence.
The innate immune system provides a means for vertebrate and invertebrate animals to respond rapidly to potential bacterial, viral, or protozoan pathogens. Unlike the B- and T-cell receptors of the adaptive immune system, the receptors that trigger innate immune responses are germ line encoded and recognize molecular patterns common to microbes but absent from the host (20). Although these pattern recognition receptors, as exemplified by Toll-like receptors (TLRs), can discriminate between host and microbe, the microbial-associated molecular patterns that they recognize are expressed by both pathogenic microorganisms and minimally virulent commensal or environmental microbes. Much effort has been devoted, therefore, to understanding how hosts limit inflammatory responses to most microorganisms while retaining the ability to respond appropriately to a potential pathogen (45).
Macrophages mount a proinflammatory response to a variety of Gram-negative and Gram-positive bacteria via inflammasome activation (4). Although the bacterial triggers of inflammasome activation are incompletely characterized, monomeric flagellin present in the macrophage cytosol strongly activates the inflammasome via the adaptor IPAF/Nlrc4 (29). However, flagellin-independent signaling pathways for inflammasome activation via IPAF/Nlrc4 and other cytosolic adaptors also exist, as nonflagellated bacteria trigger robust inflammasome activation (43, 44). The molecular trigger for inflammasome activation in this situation remains unknown. However, Gram-negative organisms lacking a functional type 3 secretion apparatus do not activate the macrophage inflammasome (4), suggesting that some component of this apparatus (or a ligand that it translocates into the cell) may be sensed directly or indirectly. As these specialized protein secretion/translocation systems are frequently required for bacterial virulence, the inflammasome response is thus directed to a broadly conserved bacterial structure or function that is linked to pathogenesis.
The Gram-negative pathogen Pseudomonas aeruginosa is capable of both acutely infecting and persistently colonizing the human respiratory tract (28). Rapid neutrophil recruitment to the airways appears to be a critical determinant in controlling P. aeruginosa replication in the lungs following acute infection (22, 32). P. aeruginosa expresses a type 3 secretion system (T3SS) that has been associated with increased virulence in murine pneumonia models and with worse clinical outcomes in human patients with ventilator-associated pneumonia (15, 38). However, a substantial proportion of P. aeruginosa clinical strains recovered from patients (with or without cystic fibrosis) no longer express a functional T3SS (19, 34). We asked whether the presence or absence of a T3SS, in otherwise isogenic P. aeruginosa strains, affected the initial host response to bacterial infection.
MATERIALS AND METHODS
Mice.
All animal work was conducted according to relevant national and international guidelines. Protocols for all animal studies were approved by the Yale Institutional Animal Care and Use Committee. Caspase-1−/− mice backcrossed onto C57BL/6 (N10) mice were provided by Richard Flavell (23). Interleukin-1R1-deficient (IL-1R1−/−) mice on a C57BL/6 background were purchased from the Jackson Laboratory. Wild-type C57BL/6 and C57BL/6 Ly5.2 mice were purchased from the Jackson Laboratory or NCI (NIH). All mice were housed in a specific-pathogen-free facility in microisolator cages. Age- (8 to 10 weeks) and sex-matched littermates were used for all experiments. In subgroups of mice as indicated below, IL-1Ra (anakinra) (10 mg/kg of body weight) was administered intraperitoneally immediately prior to bacterial infection and on the 3 days prior to infection at 24-h intervals.
For bone marrow chimeras, 3- to 4-week-old recipient mice were gamma irradiated (a total of 1,050 rad delivered in 2 doses at a 4-h interval) prior to receiving 3 × 106 to 6 × 106 donor bone marrow cells (retro-orbital). Chimerism was assessed by fluorescence-activated cell sorting (FACS) on splenic leukocytes (Ly5.1/CD45.1 and Ly5.2/CD45.2; BD Biosciences). In all mice except one (excluded from further analysis), recipient genotype leukocytes made up less than 15% of the population.
Bacterial strains and growth conditions.
All bacterial strains were maintained as frozen stocks at −80°C and freshly streaked to Vogel-Bonner minimal (VBM) or Luria-Bertani (LB) agar prior to each experiment. P. aeruginosa strain PA103 is a nonflagellated clinical isolate that produces two T3SS effectors, ExoU and ExoT (27). The effector mutants PA103ΔU (exoU deletion) and PA103ΔUT (exoU and exoT deleted, effectorless but translocation competent), the type 3 translocon-negative mutant PA103pcrV::Tn5, the T3SS-negative mutant PA103pscJ::Tn5, and the deletion mutants PA103ΔpcrV, PA103ΔpopB, and PA103ΔpopD have been described previously (12, 16, 43). Although pcrGVH popBD are encoded in an operon, the in-frame unmarked deletions of PA103ΔpcrV, PA103ΔpopB, and PA103ΔpopD are not polar: in each, the other two secreted translocon proteins are detected by Western blotting and enzyme-linked immunosorbent assay (ELISA), and cell-based phenotypes that require effector translocation are complemented by a plasmid encoding the deleted protein but not by an empty vector (26; also data not shown).
P. aeruginosa strain PAK is a flagellated isolate that translocates three T3SS effectors, ExoT, ExoS, and ExoY (25). An effectorless mutant of PAK, PAKΔSTY, was kindly provided by Arne Riestch. An in-frame deletion of popD in PAKΔSTY was generated via homologous recombination as previously described (43). Candidate isolates were screened by PCR and confirmed by Western blotting (data not shown).
Animal infections.
Bacteria grown overnight in LB medium in a 37°C shaker were diluted 1:50 and grown for 2 to 4 h in LB at 37°C (250 rpm). The log-phase culture was again diluted, to an optical density at 600 nm (OD600) of 0.06, and grown for an additional 2 h (37°C, 250 rpm). Bacteria were washed, resuspended, and diluted in phosphate-buffered saline (PBS) to achieve an inoculum of ca. 1/100 of the 50% lethal dose (LD50) for PA103 or PAK, which corresponds to ca. 5 × 104 or 5 × 105 CFU, respectively (25, 41). Animals were lightly anesthetized with 30% (vol/vol) isoflurane in propylene glycol and infected intranasally. The inoculum dose was determined by plating serial dilutions of the inoculum to VBM agar. Mice were euthanized 4 h postinfection (h.p.i.) or 12 h.p.i. Bronchoalveolar lavage was performed 3 times with 1 ml PBS prior to dissection of lungs. Single-cell lung suspensions were prepared as described previously (24) and plated to VBM agar to determine the bacterial load in tissues.
Cell counts and differentials.
Cells from bronchoalveolar lavage (BAL) fluid were collected by centrifugation. Red blood cells (RBC) were lysed (RBC lysing buffer; Sigma, St. Louis, MO) before total cell counts were determined using a hematocytometer. Cytospin samples were prepared using a Shandon cytocentrifuge (Thermo Scientific, MI) and dried prior to staining with a Diff-Quik stain kit (IMEB, Inc., San Marcos, CA). At least 200 cells were counted to determine macrophage and neutrophil percentages in BAL fluid.
BAL fluid cytokine detection assay.
Supernatants prepared from the first milliliter of BAL fluid were stored at −20°C and assayed using the Beadlyte 23-plex (Upstate) or Bio-Plex suspension array system (Bio-Rad, Hercules, CA) according to the manufacturer's protocol.
Alveolar macrophage isolation and infection.
Alveolar macrophages were harvested from bronchoalveolar lavage fluid as described previously (3) and confirmed to be >95% pure by Diff-Quik staining. Adherent cells were infected with bacteria (prepared as described above for animal infections) at a multiplicity of infection (MOI) of 10. Supernatant was sampled at hourly intervals postinfection and stored at −20°C. IL-1β was measured by ELISA (R&D Systems) according to the manufacturer's protocol.
MTEC isolation and infection.
Murine tracheal epithelial cells (MTEC) were isolated from C57BL/6 mouse tracheas following the protocol published by You et al. (46). MTEC were used for experiments after 14 to 21 days of culture at an air-liquid interface. Bacteria were cultured as described for animal infections, and 5 × 104 CFU were applied to the apical surface of MTEC monolayers in a 100-μl final volume. Supernatants were sampled at indicated times and stored at −20°C. KC levels were measured by ELISA (R&D Systems) according to the manufacturer's protocol.
Western blotting.
Bacterial strains were cultured overnight in T3SS-inducing MinS medium with 10 mM nitrilotriacetic acid as the calcium chelator (36). Cultures were centrifuged at 6,000 × g for 15 min to remove bacteria; secreted proteins were precipitated with 10% (vol/vol) trichloroacetic acid. The precipitated proteins were boiled in 4% SDS, separated by SDS-PAGE, and transferred to polyvinylidene difluoride (PVDF) membrane. The membrane was probed with anti-type 3 polyclonal, anti-PcrV monoclonal, or anti-PopD monoclonal antiserum as described previously (26).
Statistical analysis.
Statistical analysis was carried out using Prism 5 software (GraphPad). Pairwise comparisons of median values were carried out using the Mann-Whitney test. Comparisons made between three or more groups were carried out using the Kruskal-Wallis test, followed by Dunn's multiple-comparison posttest (alpha = 0.05).
RESULTS
The host immune response discriminates between bacteria that do and do not express a functional T3SS.
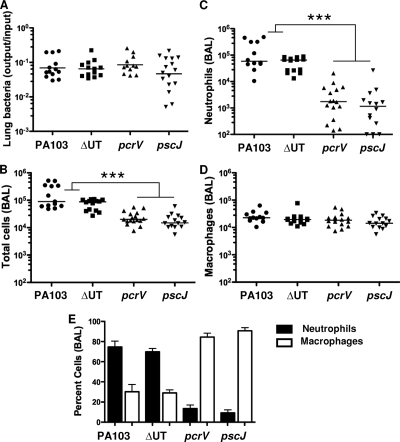
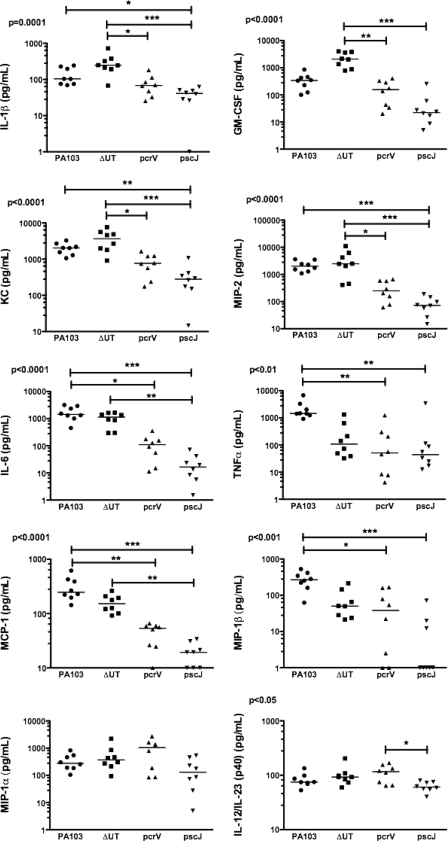
PA103 is a well-characterized serogroup O11 P. aeruginosa strain initially isolated from a human pneumonia patient (27). It secretes ExoU and ExoT, two of the four known Pseudomonas T3SS effectors. We infected C57BL/6 mice intranasally with 5 × 104 CFU (about 1/100 of the LD50) of PA103, the isogenic effectorless mutant (ΔUT), an isogenic T3SS mutant (pscJ::Tn5), or the isogenic translocation-deficient mutant (pcrV::Tn5). The pcrV::Tn5 mutant assembles a T3SS needle and can secrete effectors into culture medium but does not express the three proteins (PcrV, PopB, and PopD) required for the assembly of the translocon pore and subsequent translocation of effectors across a host cell membrane (16). Mice were sacrificed 4 h.p.i., and bronchoalveolar lavage (BAL) fluid and lung tissue were collected aseptically. Bacteria were recovered in comparable numbers from lung tissue (Fig. 1A) and BAL fluid (not shown) at this time point. Only wild-type and effectorless (ΔUT) bacteria, however, triggered neutrophil recruitment to the airways by 4 h.p.i. (Fig. 1C). Cytokine levels in BAL fluid were determined using Luminex multiplex cytokine assay kits. Several cytokines and chemokines, among them IL-1β, granulocyte-macrophage colony-stimulating factor (GM-CSF), KC, and macrophage inflammatory protein 2 (MIP-2), were present at significantly higher levels in BAL fluid from mice infected with bacteria possessing an intact T3SS than in BAL fluid from mice infected with mutants lacking either the T3SS apparatus or the translocon components (Fig. 2). Other cytokines, such as tumor necrosis factor alpha (TNF-α) and MIP-1β, were secreted in the largest amounts after infection by bacteria expressing the effectors ExoU and ExoT, as well as a T3SS apparatus (Fig. 2).
FIG. 1.
Bacteria that do not express a translocation-competent T3SS fail to elicit rapid neutrophil recruitment to the lung. C57BL/6 mice were infected intranasally with 5 × 104 CFU of PA103, ΔUT, pcrV::Tn5, or pscJ::Tn5 bacteria and then sacrificed at 4 h.p.i. (A) Bacterial recovery from lung tissue expressed as the ratio of output (CFU per lung) over input (inoculum per mouse); bars indicate geometric means. Bacterial recovery did not differ significantly between groups (P = 0.4, Kruskal-Wallis test). (B to D) Total cells (B), neutrophils (C), and macrophages (D) recovered from BAL fluid; lines indicate median values. Each point represents the value for an individual animal. Significantly more total cells and neutrophils were recovered from BAL fluid from PA103 and ΔUT strain-infected animals than from pcrV or pscJ strain-infected animals (P < 0.001, Kruskal-Wallis test followed by Dunn's posttest comparison). (E) Percentages of neutrophils and macrophages in BAL fluid samples; error bars indicate means ± standard errors of the means. ***, P < 0.001 for Dunn's multiple-comparison posttest. Data shown are from 3 independent experiments with 4 to 6 mice per group.
FIG. 2.
Many BAL cytokines are secreted in response to translocon-positive P. aeruginosa. The Bio-Plex suspension array system (Bio-Rad) was used to measure a panel of chemokines and cytokines in BAL fluid collected at 4 h.p.i. from C57BL/6 mice infected with 5 × 104 CFU PA103 or the indicated mutant. Each point represents the value for an individual animal; lines indicate median values. Points graphed on the x axis indicate values below the detection limit of the assay. Median values differ significantly for all cytokines except MIP-1α; P values were calculated using the Kruskal-Wallis test. Dunn's multiple-comparison test was used to evaluate all pairwise comparisons. All significant differences are indicated on each graph (*, P < 0.05; **, P < 0.01; ***, P < 0.001). These results were replicated in an independent experiment in which cytokine and chemokine levels were determined using a Beadlyte 23-plex kit (Upstate) on mice infected with these same isogenic mutants (n = 5 to 6 animals per strain).
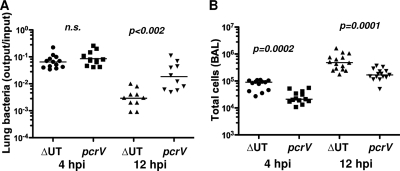
Groups of mice infected with translocon-positive (ΔUT) and translocon-negative (pcrV::Tn5) bacteria were also sacrificed at 12 h.p.i. At this time point, the number of bacteria recovered from ΔUT strain-infected lung tissue was substantially lower than the number recovered from pcrV::Tn5 strain-infected lungs, while the number of cells in the BAL fluid remained significantly higher in ΔUT strain-infected mice (Fig. 3). Thus, the proinflammatory response to translocon-positive bacteria was substantially greater than that elicited by isogenic translocon-negative organisms or T3SS mutants and resulted in more rapid clearance of translocon-positive bacteria from the lung.
FIG. 3.
Bacteria lacking a T3SS translocon are cleared less rapidly from the lung. C57BL/6 mice were infected intranasally with 5 × 104 CFU of ΔUT or pcrV:Tn5 bacteria and then sacrificed at 12 h.p.i.; 4-h.p.i. data are reproduced from Fig. 1 for comparison. (A) Bacterial recovery from lung tissue expressed as output (CFU per lung) over input (CFU per inoculum); lines indicate geometric means. (B) Total cells recovered in BAL fluid. Each point represents the value for an individual animal. P values for pairwise comparisons are indicated (Mann-Whitney test; n.s., not significant). The 12-h.p.i. data are from 2 independent experiments with 5 to 6 mice per group.
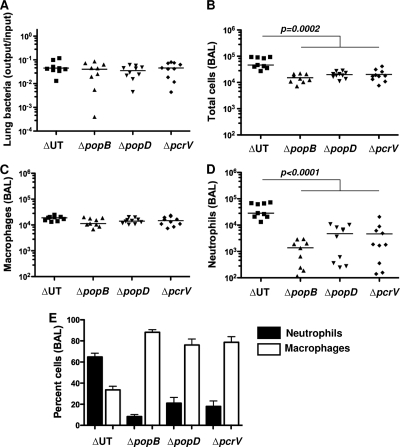
All three translocon proteins are required for bacteria to elicit rapid neutrophil recruitment to the airway.
The proteinaceous translocation pore is formed by oligomerization of PopD and PopB in the eukaryotic plasma membrane (39). PcrV is not detected within this oligomeric pore but is required to trigger assembly of the PopB/PopD translocation channel (8). In the absence of PcrV, P. aeruginosa cannot elicit any of the phenotypes associated either with membrane insertion of the translocon pore (e.g., RBC hemolysis) or with delivery of T3SS effectors to the host cell cytosol (13). We previously demonstrated that inflammasome activation in bone marrow-derived macrophages is observed following infection with translocation-competent ΔUT bacteria but that neither pcrV::Tn5 bacteria nor any of the single translocon protein mutants (ΔpopB, ΔpopD, or ΔpcrV) activated caspase-1 (43). We tested whether all three translocon proteins were likewise required for rapid neutrophil recruitment to the airways following P. aeruginosa infection. As seen by the results in Fig. 4, each of the three proteins was required, suggesting that this response directly or indirectly reflects the assembly of a translocation-competent pore.
FIG. 4.
Both translocon pore components and PcrV are required for rapid neutrophil recruitment after infection. C57BL/6 mice were infected intranasally with 5 × 104 CFU of the indicated bacterial mutants and then sacrificed at 4 h.p.i. (A) Bacterial recovery from lung tissue is expressed as output (CFU per lung) over input (CFU per inoculum) and did not differ significantly between groups; lines show geometric means. (B to D) Total cells (B), macrophages (C), and neutrophils (D) recovered in BAL fluid. Each point represents the value for an individual animal; lines indicate median values. The Kruskal-Wallis test was used to compare numbers of cells present in BAL fluid; the total numbers of cells (P = 0.0002) and neutrophils (P < 0.0001) differ significantly between ΔUT bacteria and the other three mutants. (E) Percentages of cells in BAL fluid; bars indicate means ± standard errors of the means. Data shown are from mice infected in 4 independent experiments with 4 to 5 mice per group.
The translocon influences the innate immune response to flagellated Pseudomonas.
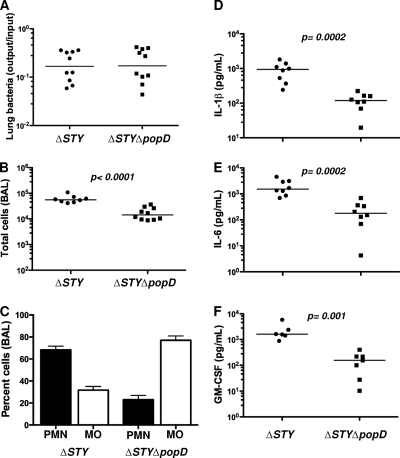
PA103 does not express flagellin, a potent ligand for both extracellular TLR5 and intracellular Nod-like receptors (NLRs) (17, 29). However, many clinical isolates express flagellin (Fla positive [Fla+]) (34), and we hypothesized that the T3SS-dependent responses observed in PA103-infected mice might be overshadowed by responses directed to flagellin. We therefore constructed a translocon-negative mutant (ΔSTY ΔpopD) of the Fla+ strain PAKΔSTY and compared it to the isogenic effectorless parent (ΔSTY). C57BL/6 mice were sacrificed 4 h after intranasal inoculation of 5 × 105 CFU of the ΔSTY or ΔSTY ΔpopD strain. Both bacterial strains were recovered from lung tissue in equal numbers at this time point (Fig. 5A). Again, the number of inflammatory cells in the airways was greater for the translocon-positive strain (Fig. 5B), and this difference was accounted for by neutrophil recruitment (Fig. 5C). IL-1β, IL-6, GM-CSF, KC, MIP-1β, and monocyte chemoattractant protein 1 (MCP-1) levels in the BAL fluid were significantly higher in ΔSTY strain-infected mice than in translocon-deficient strain-infected animals, while TNF-α, MIP-1α, and IL-12/IL-23(p40) levels were not significantly different between the two groups (Fig. 5D to F and data not shown). Thus, the presence of an intact T3SS influences neutrophil recruitment to the lung in a similar fashion following infection with either Fla+ or Fla-negative (Fla−) P. aeruginosa.
FIG. 5.
Translocon-dependent recognition dominates the early response to Fla+ P. aeruginosa. C57BL/6 mice were infected intranasally with 5 × 105 CFU of PAKΔSTY or ΔSTY ΔpopD bacteria and then sacrificed at 4 h.p.i. Each point represents the value for an individual animal. (A) Bacterial recovery from lung tissue expressed as output (CFU per lung) over input (CFU per inoculum). The geometric means do not differ. (B) Total cells recovered in BAL fluid; bars indicate median values. (C) Percentages of macrophages and neutrophils in BAL fluid (means ± standard errors of the means). PMN, polymorphonuclear leukocytes; MO, macrophages. (D to F) Cytokines in BAL fluid measured by Luminex. Median values are indicated and differ significantly (P < 0.001). P values for pairwise comparisons were calculated using the Mann-Whitney test. Data shown are from 2 independent experiments with 5 mice per group.
Isolated airway epithelial cells do not discriminate between T3SS-positive and -negative bacteria, but alveolar macrophages do.
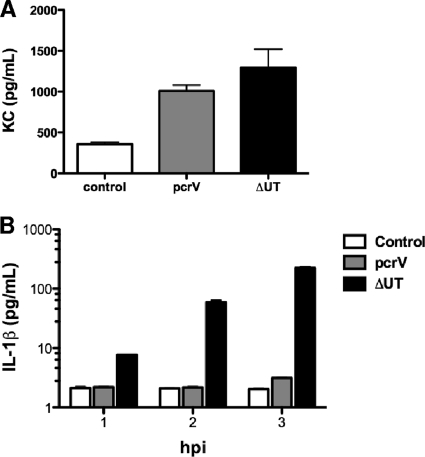
Both bone marrow-derived and non-bone marrow-derived cell types respond to P. aeruginosa in vitro (37). We isolated and cultured murine tracheal epithelial cells (MTEC) from C57BL/6 mice at an air-liquid interface and then infected them with wild-type and mutant P. aeruginosa strains. MTEC secreted similar amounts of the neutrophil chemoattractant KC in response to infection with either T3SS-positive or -negative bacteria (Fig. 6A). Isolated alveolar macrophages, however, produced IL-1β in a T3SS-dependent manner (Fig. 6B). This response mirrored what we had observed previously for PA103-infected bone marrow-derived macrophages (43) and was consistent with the caspase-1-dependent response of alveolar macrophages infected with other P. aeruginosa strains (11). Of note, we were unable to detect IL-1β secretion by MTEC infected with either T3SS-positive or -negative bacteria under these conditions (level of detection, 10 pg/ml; data not shown).
FIG. 6.
Alveolar macrophages but not MTEC respond differently to infection with a translocon-positive versus a translocon-negative P. aeruginosa strain. (A) MTEC were differentiated at an air-liquid interface on Transwell inserts and then infected with 5 × 104 CFU of ΔUT or pcrV bacteria. Bars indicate KC measured in supernatant by ELISA at 4 h.p.i. (means ± standard errors of the means) for 5 to 6 wells; the results are representative of 3 independent experiments and were comparable for MOIs between 0.1 and 10. Each well is plated with MTEC harvested from ca. one mouse trachea. (B) Alveolar macrophages were infected with ΔUT or pcrV bacteria (MOI 10), and secreted IL-1β was measured by ELISA at the indicated times postinfection. Bars indicate means ± standard deviations of the results for triplicate wells; the results are representative of two independent experiments.
Caspase-1 activity is required for discrimination between translocon-positive and -negative bacteria.
Flagellin is dispensable for caspase-1 activation by P. aeruginosa (43); however, an intact T3SS is absolutely required for this process (10, 11, 30, 43). The presence of elevated levels of IL-1β in BAL fluid from mice infected with translocon-positive but not translocon-negative bacteria suggested that caspase-1 activation and subsequent IL-1β secretion might underlie the lung's ability to discriminate between the presence and the absence of an intact T3SS. If this were so, we reasoned that mice lacking caspase-1 would respond similarly to infection with translocon-positive and translocon-negative bacteria. Age and sex-matched caspase-1−/− mice were infected intranasally with ΔUT or pcrV::Tn5 bacteria and then sacrificed at 4 h.p.i. The number of bacteria recovered from lung tissue (Fig. 7A) and BAL fluid (not shown) was indistinguishable between the two strains. Unlike wild-type mice, however, caspase-1−/− mice infected with either translocon-positive or -negative bacteria had identical numbers of total cells, macrophages, and neutrophils present in BAL fluid at this time point (Fig. 7B to D). The IL-1β levels in BAL fluid were also identical and only slightly elevated over the levels present in BAL fluid from uninfected mice (Fig. 7E). Thus, though other proteases can cleave and activate pro-IL-1β, our observations suggest that the majority of IL-1β is produced by caspase-1-mediated cleavage early during P. aeruginosa ΔUT pulmonary infection (7).
FIG. 7.
Caspase-1−/− mice fail to discriminate between translocon-positive and -negative strains of P. aeruginosa early after infection. Age- and sex-matched caspase-1−/− mice were infected intranasally with 5 × 104 CFU of ΔUT or ΔpcrV bacteria and then sacrificed at 4 h.p.i. Each point represents the value for an individual animal. (A) Bacterial recovery from lung tissue is expressed as output (CFU per lung) over input (CFU per inoculum). Geometric means are indicated. (B to D) Total cells (B), macrophages (C), and neutrophils (D) recovered in BAL fluid are shown; bars indicate median values. There were no significant differences between groups (Mann-Whitney test). (E to H) Cytokine levels in BAL fluid were measured by Luminex. Cytokines levels in BAL fluid samples from 2 uninfected mice (UI) are shown for comparison. Median values do not differ significantly for ΔUT versus pcrV strain-infected animals (Mann-Whitney test). N.D., not detected. Data shown are from mice infected in two independent experiments with 5 to 6 per group.
Caspase-1 expression in bone marrow-derived cells is required for rapid neutrophil recruitment.
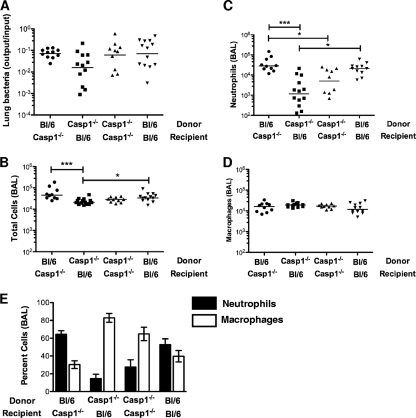
Epithelial cells are not considered to be a significant source of IL-1β (7). A recent paper, however, suggested that caspase-1 activation in gut stromal cells is required for IL-1β production and mucosal inflammation in response to Salmonella enterica serovar Typhimurium infection (33). We addressed the possibility that lung epithelial cells play an analogous role during P. aeruginosa pulmonary infection by constructing bone marrow chimeras between C57BL/6 and caspase-1−/− mice and then infecting the animals with the translocon-positive ΔUT strain. Animals were euthanized 4 h postinfection, and BAL fluid was examined for evidence of neutrophil recruitment to the airways. Irradiated C57BL/6 mice transplanted with caspase-1−/− bone marrow had significantly fewer total cells and neutrophils recruited to airways at 4 h.p.i. than either irradiated caspase-1 knockout mice reconstituted with C57BL/6 mouse bone marrow or irradiated C57BL/6 controls reconstituted with C57BL/6 mouse bone marrow (Fig. 8B to D). Thus, the genotype of the donor cells determined whether infection with a translocon-positive P. aeruginosa strain elicited rapid neutrophil recruitment to the airways and resulted in a shift to a neutrophil-predominant BAL fluid differential (Fig. 8E).
FIG. 8.
Rapid neutrophil recruitment requires caspase-1 expression in a radiosensitive cell compartment. Bone marrow chimeras were constructed using irradiated 3- to 4-week-old caspase-1−/− (Ly5.1) and C57BL/6 (Ly5.2) mice as donors or recipients. Chimeric animals were infected with 5 × 104 CFU of ΔUT bacteria intranasally and then sacrificed at 4 h.p.i. Chimerism was confirmed by FACS analysis of splenic lymphocytes. Each point represents the value for an individual animal. (A) Bacterial recovery from lung tissue is expressed as the ratio of output CFU to input inoculum; lines indicate geometric means (P = 0.09, Kruskal-Wallis test). (B to D) Total cells (B), neutrophils (C), and macrophages (D) present in BAL fluid are shown; lines indicate median values, which differ significantly by Kruskal-Wallis test for total cells (P = 0.0006) and neutrophils (P = 0.0002). P values for all pairwise comparisons were calculated with Dunn's multiple-comparison test and are indicated whenever the P value is <0.05 (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (E) Percentages of neutrophils and macrophages in BAL fluid (means ± standard errors of the means). Data for each chimeric group were generated in two independent experiments with 4 to 9 mice per group.
IL-1R-dependent pathways are required for discrimination between translocon-positive and -negative bacteria.
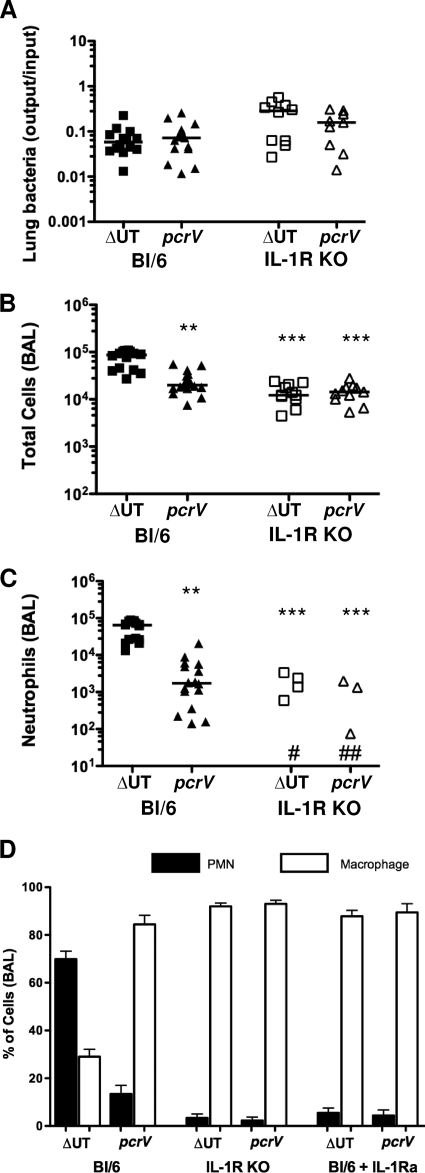
Inflammasome activation leads to macrophage production of IL-1β and IL-18. If this is a key signal in the recognition of the translocon-positive bacteria, we reasoned that it should require intact IL-1R or IL-18R signaling pathways. IL-18 appears to be dispensable for pulmonary innate immune responses to P. aeruginosa (35, 40). We therefore focused on IL-1R by infecting IL-1R−/− animals with ΔUT or pcrV::Tn5 bacteria and then sacrificing the animals 4 h postinfection. Although we recovered substantial numbers of bacteria from lung tissue and BAL fluid (Fig. 9A and data not shown), total cell numbers in the BAL fluid of IL-1R−/− mice infected with the different strains were indistinguishable (Fig. 9B), and no neutrophils were observed in the majority of BAL fluid samples (level of detection, ca. 102 cells) from either group of mice (Fig. 9C). Elevated levels of IL-1β were detected only in BAL fluid from mice infected with translocon-positive bacteria (Fig. 10B). C57BL/6 mice treated with the IL-1R antagonist IL-1Ra (anakinra) prior to infection with translocon-positive or -negative bacteria phenocopied IL-1R−/− mice (Fig. 9D). Thus, IL-1R signaling is required for neutrophil recruitment following ΔUT strain infection. As IL-1α levels in BAL fluid from ΔUT or pcrV strain-infected mice are minimally elevated over background (uninfected) values (data not shown), the requirement for IL-1R likely reflects a requirement for IL-1β signals.
FIG. 9.
IL-1R signaling is required for rapid neutrophil recruitment following infection with translocon-positive P. aeruginosa. IL-1R−/− mice were infected intranasally with 5 × 104 CFU of ΔUT or ΔpcrV bacteria and then sacrificed at 4 h.p.i. Each point represents the value for an individual animal. Data from C57BL/6 animals infected identically are shown for comparison. (A) Bacterial recovery from lung tissue is expressed as output (CFU per lung) over input (CFU per inoculum); lines indicate geometric means. (B, C) Total cells (B) and neutrophils (C) recovered in BAL fluid are shown; median values differ significantly (P < 0.0001, Kruskal-Wallis test), and significant pairwise comparisons are indicated (**, P < 0.01; ***, P < 0.001 [Dunn's multiple-comparison test]). No neutrophils were detected in BAL fluid from seven ΔUT (#) and eight pcrV (##) strain-infected mice. (D) Differential cell counts in BALs from C57BL/6, IL-1R knockout, and C57BL/6 mice pretreated with anakinra (IL-1Ra) were determined at 4 h.p.i. with the indicated bacterial strains. Data shown are from three independent experiments with 4 to 6 mice per group. IL-1R KO, IL-1R−/− (knockout) mice.
FIG. 10.
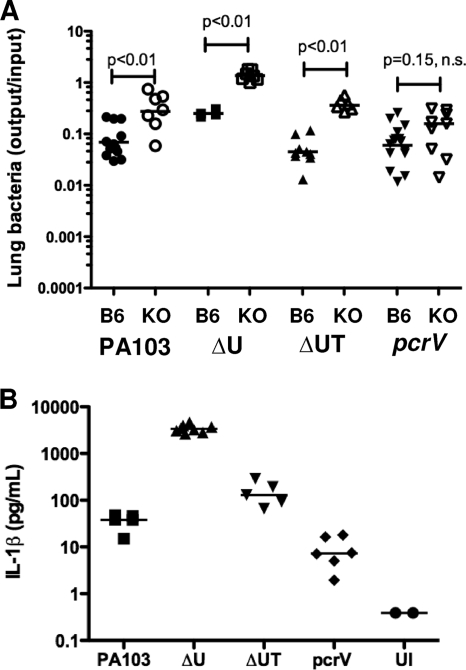
Bacterial strains that induce IL-1β are cleared less efficiently by IL-1R−/− mice. (A) Bacterial recovery from lung tissue (output/input) for C57BL/6 (B6) or IL-1R−/− (KO) animals infected with 5 × 104 CFU of the indicated isogenic strains at 4 h.p.i. Lines indicate geometric means; pair-wise comparisons were carried out with the Mann-Whitney test. (B) IL-1β levels in BAL fluid collected from IL-1R−/− mice infected with the indicated isogenic strains at 4 h.p.i. Median values differ significantly (P < 0.001, Kruskal-Wallis test). n.s., not significant; UI, uninfected. Data shown are from 2 to 3 independent experiments with 4 to 5 mice per group.
Increased numbers of translocon-positive bacteria are recovered from the IL-1R−/− mouse lung early after infection.
Bacterial recovery from C57BL/6 or IL-1R−/− mouse lungs was compared at 4 h.p.i. for a number of isogenic PA103 mutants. In the absence of IL-1R signaling, significantly greater numbers of PA103 (4.0-fold increase in the geometric mean), ΔU (5.4-fold), and ΔUT (8.3-fold) bacteria were recovered from lung tissue (Fig. 10A). In contrast, the recovery of the translocon-negative pcrV strain was not significantly increased from IL-1R−/− mouse lungs (1.7-fold increase in the geometric mean). We also measured IL-1β in BAL fluid collected from infected IL-1R−/− mice (Fig. 10B). Bacterial expression of the effector ExoU was associated with a significant reduction in IL-1β production, as seen by comparing the levels measured in BAL fluid from PA103-infected mice (median, 38 pg/ml) versus the levels in BAL fluid from PA103ΔU-infected animals (median, 3,373 pg/ml) or PA103ΔUT-infected animals (median, 129 pg/ml). This is consistent with our prior observation that ExoU appears to inhibit caspase-1 activation and IL-1β secretion by infected macrophages in vitro (43). Conversely, ExoT expression was associated with higher levels of IL-1β in BAL fluid. We had previously observed more IL-1β secretion by bone marrow-derived macrophages infected with ΔU than with ΔUT P. aeruginosa (43), while Ichikawa et al. had reported that lung A549 cells infected with ExoT+ P. aeruginosa had higher levels of IL-1β transcript than cells infected with isogenic ExoT− bacteria in a microarray analysis (18). Lastly, although other cell types, including neutrophils, can process and secrete IL-1β via caspase-1-independent mechanisms (14), the majority of cells recovered from the airways of infected IL-1R−/− mice were macrophages (Fig. 9 and data not shown), suggesting that they were the likely source of the IL-1β measured.
DISCUSSION
In this paper, we describe a rapid and dominant response of the lung to P. aeruginosa infection that is directed specifically at bacteria expressing a translocation-competent T3SS. This response manifests as the recruitment of neutrophils to the airways and results in more rapid bacterial clearance. The recognition of T3SS-positive bacteria requires host caspase-1 expression in bone marrow-derived cells and is accompanied by increased IL-1β levels in bronchoalveolar lavage fluid. IL-1 signaling is necessary for rapid neutrophil recruitment, as IL-1R−/− mice or IL-1Ra-pretreated animals have few to no neutrophils in the airways even after infection with T3SS-positive bacteria. These translocon-dependent responses dominate the initial host response even to P. aeruginosa strains expressing potent TLR ligands, such as flagellin. The recognition of a widely conserved behavior of Gram-negative organisms that is also correlated with virulence may allow the immune system to rapidly and preferentially respond to relatively small inocula of likely pathogens while minimizing activation by commensal flora and other organisms of more limited pathogenic potential.
The mechanisms by which virulence-associated bacterial features, such as type 3 or type 4 secretion systems, are sensed remain an area of active investigation. Recent papers have described cytokine transcriptional programs that are triggered when macrophages are infected with T3SS- or T4SS-positive bacteria (2, 42). Caspase-1 inflammasome activation in macrophages by T3SS- or T4SS-expressing bacteria and the subsequent cleavage, activation, and secretion of proinflammatory IL-1β, IL-18, and other protein targets (21) represent a distinct, posttranscriptional immune response to this virulence-associated bacterial signal (4). In vitro studies have suggested a number of potential mechanisms for triggering the NLRC4/IPAF inflammasome, which appears to play the dominant role in the response to T3SS-positive, flagellated and nonflagellated P. aeruginosa (43). These include “translocation” or “leakage” of bacterial molecules that are not bona fide T3SS substrates into the host cell cytoplasm (2, 29, 31), generation of danger signals associated with translocon insertion into the host cell membrane (45), or even translocon-associated potassium efflux (1). Our data underscore that a translocation-competent T3SS is required for the response we observe but do not rule out any of these proposed inflammasome triggers. Indeed, multiple T3SS-associated signals may be recognized during in vivo infection by the NLRC4/IPAF inflammasome. There is also evidence that more than one type of inflammasome may participate in the recognition of a T3SS- or T4SS-expressing bacterium, as illustrated by recent studies describing cell-based and whole-animal responses to secretion system-positive Yersinia and Legionella pneumophila (5, 6). Further studies are necessary to understand which of these proposed T3SS recognition mechanisms operate in vivo during P. aeruginosa infection and how they are integrated to generate the cellular and cytokine “signatures” that we have described.
Many of the effectors that are translocated through T3SS or T4SS structures function primarily to attenuate or subvert the host immune response to bacterial infection. A recent paper describes a Yersinia effector, YopK, which prevents inflammasome activation in response to T3SS-positive bacteria (5). Intriguingly, translocated YopK appears to interact not with components of the inflammasome but, rather, with the T3SS itself, suggesting that it might mask a component or function of the T3SS that is recognized by the inflammasome. P. aeruginosa encodes no protein with primary sequence homology to YopK. Unlike YopK, ExoU causes rapid cell death via a caspase-1-independent pathway. Thus, the ca. 10 to 15% of clinical and environmental isolates that secrete ExoU (9, 26) are unlikely to evade host recognition as pathogens and, indeed, elicit strong, proinflammatory responses in the lung (Fig. 1). The majority of P. aeruginosa isolates, however, are ExoU-negative, and a substantial proportion of human clinical isolates are phenotypically T3SS-negative (34). Further studies are necessary to address whether avoidance of inflammasome activation contributes to chronic colonization or persistence by T3SS-negative P. aeruginosa strains in human patients.
Acknowledgments
We thank Carla Weibel for excellent technical assistance. We thank Arne Rietsch for strain PAKΔSTY, and Richard Flavell for caspase-1−/− mice.
This work was supported by the National Institutes of Health (grant no. F31 AI-061882 to L.A.M.), the Patrick and Catherine Weldon Donaghue Medical Research Foundation (B.I.K.), and the Burroughs Wellcome Fund (B.I.K.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or NIH.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 7 September 2010.
REFERENCES
- 1.Arlehamn, C. S. L., V. Petrilli, O. Gross, J. Tschopp, and T. J. Evans. 2010. The role of potassium in inflammasome activation by bacteria. J. Biol. Chem. 285:10508-10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auerbuch, V., D. T. Golenbock, and R. R. Isberg. 2009. Innate immune recognition of Yersinia pseudotuberculosis type III secretion. PLoS Pathog. 5:e1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailie, M. B., T. J. Standiford, L. L. Laichalk, M. J. Coffey, R. Strieter, and M. Peters-Golden. 1996. Leukotriene-deficient mice manifest enhanced lethality from Klebsiella pneumonia in association with decreased alveolar macrophage phagocytic and bactericidal activities. J. Immunol. 157:5221-5224. [PubMed] [Google Scholar]
- 4.Brodsky, I. E., and D. Monack. 2009. NLR-mediated control of inflammasome assembly in the host response to bacterial pathogens. Semin. Immunol. 21:199-207. [DOI] [PubMed] [Google Scholar]
- 5.Brodsky, I. E., N. W. Palm, S. Sadanand, M. B. Ryndak, F. S. Sutterwala, R. A. Flavell, J. B. Bliska, and R. Medzhitov. 2010. A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe 7:376-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Case, C. L., S. Shin, and C. R. Roy. 2009. Asc and Ipaf inflammasomes direct distinct pathways for caspase-1 activation in responses to Legionella pneumophila. Infect. Immun. 77:1981-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinarello, C. A. 2009. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 27:519-550. [DOI] [PubMed] [Google Scholar]
- 8.Faudry, E., G. Vernier, E. Neumann, V. Forge, and I. Attree. 2006. Synergistic pore formation by type III toxin translocators of Pseudomonas aeruginosa. Biochemistry 45:8117-8123. [DOI] [PubMed] [Google Scholar]
- 9.Feltman, H., G. S. Schulert, S. Khan, M. Jain, L. Peterson, and A. Hauser. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659-2669. [DOI] [PubMed] [Google Scholar]
- 10.Franchi, L., J. Stoolman, T. Kanneganti, A. Verma, R. Ramphal, and G. Nunez. 2007. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur. J. Immunol. 37:3030-3039. [DOI] [PubMed] [Google Scholar]
- 11.Galle, M., P. Schotte, M. Haegman, A. Wullaert, H. J. Yang, S. Jin, and R. Beyaert. 2008. The Pseudomonas aeruginosa type III secretion system plays a dual role in the regulation of caspase-1 mediated IL-1β maturation. J. Cell Mol. Med. 12:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrity-Ryan, L., B. Kazmierczak, R. Kowal, J. Comolli, A. Hauser, and J. Engel. 2000. The arginine finger domain of ExoT is required for actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect. Immun. 68:7100-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goure, J., A. Pastor, E. Faudry, J. Chabert, A. Dessen, and I. Attree. 2004. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect. Immun. 72:4741-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greten, F. R., M. C. Arkan, J. Bollrath, L.-C. Hsu, J. Goode, C. Miething, S. I. Goktuna, M. Neuenhahn, J. Fierer, S. Paxian, N. van Rooijen, Y. Xu, T. O'Cain, B. B. Jaffee, D. H. Busch, J. Duyster, R. M. Schmid, L. Eckmann, and M. Karin. 2007. NF-κB is a negative regulator of IL-1β secretion as revealed by genetic and pharmacological inhibition of IKKβ. Cell 130:918-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauser, A., E. Cobb, M. Bodi, D. Mariscal, J. Valles, J. Engel, and J. Rello. 2002. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30:521-528. [DOI] [PubMed] [Google Scholar]
- 16.Hauser, A. R., P. J. Kang, S. J. M. Fleiszig, K. Mostov, and J. Engel. 1998. Defects in type III secretion correlate with internalization of Pseudomonas aeruginosa by epithelial cells. Infect. Immun. 66:1413-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi, F., K. Smith, A. Ozinsky, T. Hawn, E. Yi, D. Goodlett, J. Enge, S. AKira, D. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 18.Ichikawa, J. K., S. B. English, M. C. Wolfgang, R. Jackson, A. J. Butte, and S. Lory. 2005. Genome-wide analysis of host responses to the Pseudomonas aeruginosa type III secretion system yields synergistic effects. Cell. Microbiol. 7:1635-1646. [DOI] [PubMed] [Google Scholar]
- 19.Jain, M., D. Ramirez, R. Seshadri, J. F. Cullina, C. A. Powers, G. S. Schulert, M. Bar-Meir, C. L. Sullivan, S. A. McColley, and A. R. Hauser. 2004. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J. Clin. Microbiol. 42:5229-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janeway, C. A., and R. M. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197-216. [DOI] [PubMed] [Google Scholar]
- 21.Keller, M., A. Ruegg, S. Werner, and H.-D. Beer. 2008. Active caspase-1 is a regulator of unconventional protein secretion. Cell 132:818-831. [DOI] [PubMed] [Google Scholar]
- 22.Koh, A. Y., G. R. Priebe, C. Ray, N. van Rooijen, and G. B. Pier. 2009. Inescapable need for neutrophils as mediators of cellular innate immunity to acute Pseudomonas aeruginosa pneumonia. Infect. Immun. 77:5300-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuida, K., J. A. Lippke, G. Ku, M. W. Harding, D. J. Livingston, M. S. Su, and R. A. Flavell. 1995. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science 267:2000-2003. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski, M. A., and B. I. Kazmierczak. 2006. Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infect. Immun. 74:4462-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, V. T., R. S. Smith, B. Tummler, and S. Lory. 2005. Activities of Pseudomonas aeruginosa effectors secreted by the type III secretion system in vitro and during infection. Infect. Immun. 73:1695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, L., M. Ledizet, K. Kar, R. A. Koski, and B. I. Kazmierczak. 2005. An indirect enzyme-linked immunosorbent assay for rapid and quantitative detection of type III virulence phenotypes of Pseudomonas aeruginosa isolates. Ann. Clin. Microbiol. Antimicrobiol. 4:22-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, P. V. 1966. The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis: identity of the lethal toxins produced in vitro and in vivo. J. Infect. Dis. 116:481-489. [DOI] [PubMed] [Google Scholar]
- 28.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051-1060. [DOI] [PubMed] [Google Scholar]
- 29.Miao, E. A., C. M. Alpuche-Aranda, M. Dors, A. E. Clark, M. W. Bader, S. I. Miller, and A. Aderem. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7:569-575. [DOI] [PubMed] [Google Scholar]
- 30.Miao, E. A., R. K. Ernst, M. Dors, D. P. Mao, and A. Aderem. 2008. Pseudomonas aeruginosa activates caspase 1 through IPAF. Proc. Natl. Acad. Sci. U. S. A. 105:2562-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao, E. A., D. P. Mao, N. Yudkovsky, R. Bonneau, C. G. Lorang, S. E. Warren, I. A. Leaf, and A. Aderem. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. U. S. A. 107:3076-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morissette, C., C. Francoeur, C. Darmond-Zwaig, and F. Gervais. 1996. Lung phagocyte bactericidal function in strains of mice resistant and susceptible to Pseudomonas aeruginosa. Infect. Immun. 64:4984-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller, A. J., C. Hoffman, M. Galle, A. van den Broeke, M. Heikenwalder, L. Falter, B. Misselwitz, M. Kremer, R. Bayaert, and W.-D. Hardt. 2009. The S. Typhimurium effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe 6:125-136. [DOI] [PubMed] [Google Scholar]
- 34.Murray, T. S., M. Ledizet, and B. I. Kazmierczak. 2010. Swarming motility, secretion of type 3 effectors and biofilm formation phenotypes exhibited within a large cohort of Pseudomonas aeruginosa clinical isolates. J. Med. Microbiol. 59:511-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakasone, C., K. Kawakami, T. Hoshino, Y. Kawase, K. Yokota, K. Yoshino, K. Takeda, S. Akira, and A. Saito. 2004. Limited role for interleukin-18 in the host protection response to pulmonary infection with Pseudomonas aeruginosa in mice. Infect. Immun. 72:6176-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicas, T. I., and B. H. Iglewski. 1984. Isolation and characterization of transposon-induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect. Immun. 45:470-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raoust, E., V. Balloy, I. Garcia-Verdugo, L. Touqui, R. Ramphal, and M. Chignard. 2009. Pseudomonas aeruginosa LPS or flagellin are sufficient to activate TLR-dependent signaling in murine alveolar macrophages and airway epithelial cells. PLoS One 4:e7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 39.Schoehn, G., A. M. Di Guilmi, D. Lemaire, I. Attree, W. Weissenhorn, and A. Dessen. 2003. Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. EMBO J. 22:4957-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz, M. J., S. Knapp, S. Florquin, J. Pater, K. Takeda, S. Akira, and T. van der Poll. 2003. Interleukin-18 impairs the pulmonary host response to Pseudomonas aeruginosa. Infect. Immun. 71:1630-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultz, M. J., A. W. Rijneveld, S. Florquin, C. K. Edwards, C. A. Dinarello, and T. van der Poll. 2002. Role of interleukin-1 in the pulmonary immune response during Pseudomonas aeruginosa pneumonia. Am. J. Physiol. Lung Cell. Mol. Physiol. 282:L285-L290. [DOI] [PubMed] [Google Scholar]
- 42.Shin, S., C. L. Case, K. A. Archer, C. V. Nogueira, K. S. Kobayashi, R. A. Flavell, C. R. Roy, and D. S. Zamboni. 2008. Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 4:e1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutterwala, F. S., L. A. Mijares, L. Li, Y. Ogura, B. I. Kazmierczak, and R. A. Flavell. 2007. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204:3235-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki, T., L. Franchi, C. Toma, H. Ashida, M. Ogawa, Y. Yoshikawa, H. Mimuro, N. Inohara, C. Sasakawa, and G. Nunez. 2007. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 3:e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vance, R. E., R. R. Isberg, and D. A. Portnoy. 2009. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 6:10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You, Y., E. J. Richer, T. Huang, and S. L. Brody. 2002. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am. J. Physiol. Lung Cell. Mol. Physiol. 283:L1315-L1321. [DOI] [PubMed] [Google Scholar]