Abstract
The Gram-positive pathogen Enterococcus faecalis is a leading agent of nosocomial infections, including urinary tract infections, surgical site infections, and bacteremia. Among the infections caused by E. faecalis, endocarditis remains a serious clinical manifestation and unique in that it is commonly acquired in a community setting. Infective endocarditis is a complex disease, with many host and microbial components contributing to the formation of bacterial biofilm-like vegetations on the aortic valve and adjacent areas within the heart. In the current study, we compared the pathogenic potential of the vancomycin-resistant E. faecalis V583 and three isogenic protease mutants (ΔgelE, ΔsprE, and ΔgelE ΔsprE mutants) in a rabbit model of enterococcal endocarditis. The bacterial burdens displayed by GelE− mutants (ΔgelE and ΔgelE ΔsprE mutants) in the heart were significantly lower than those of V583 or the SprE− mutant. Vegetations on the aortic valve infected with GelE− mutants (ΔgelE and ΔgelE ΔsprE mutants) also showed a significant increase in deposition of fibrinous matrix layer and increased chemotaxis of inflammatory cells. In support of a role for proteolytic modulation of the immune response to E. faecalis, we also demonstrate that GelE can cleave the anaphylatoxin complement C5a and that this proteolysis leads to decreased neutrophil migration in vitro. In vivo, a decreased heterophil (neutrophil-like cell) migration was observed at tissue sites infected with GelE-producing strains but not at those infected with SprE-producing strains. Taken together, these observations suggest that of the two enterococcal proteases, gelatinase is the principal mediator of pathogenesis in endocarditis.
Enterococci are leading causes of hospital-acquired infections, including bacteremia, surgical site infections, and urinary tract infections (31). However, one of the most serious clinical manifestations of enterococcal infection is endocarditis, with mortality rates ranging from 15 to 20% (23). Enterococci, most commonly E. faecalis, are the third leading cause of infective endocarditis (21). Enteroccoci cause subacute-chronic endocarditis and are the causative agents of up to 20% of native valve endocarditis and 15% of prosthetic valve endocarditis (21, 23). Unlike other enterococcal infections, endocarditis is most often community acquired, although recent studies indicate that there is a significant risk of acquiring enterococcal endocarditis in a clinical environment (7, 8).
The pathological progression of infective endocarditis initially involves the development of vegetations on heart valves, followed by embolization and dissemination to other body sites (15). In experimental endocarditis in rabbits, mortality is often associated with embolization to secondary infectious sites, including blood vessels of the heart, brain, and kidneys (10). Occasionally the emboli occlude blood vessels in the secondary infection sites, leading to tissue damage. Previous studies indicated that the presence of extracellular proteases (GelE and SprE) significantly increased mortality in animal infection models, but the relative contribution of each protease in experimental endocarditis has not been examined to date (10, 35).
Multiple bacterial species produce extracellular proteases that contribute to pathogenesis through manipulation of the host immune response (28). These proteases target several components of the host innate immune system, including complement, antimicrobial peptides (AMPs), cytokines, and cytokine receptors (28). Complement C3a is an anaphylatoxin involved in activation and recruitment of eosinophils but is limited in its ability to activate and recruit neutrophils (3, 4, 6, 19). Compared to C3a, the complement C5a is at least 100-fold more potent in activation and recruitment of neutrophils (6). Determination of the effects of E. faecalis proteases on C5a is of particular importance because of the relevance of neutrophil recruitment for bacterial clearance. In addition, thrombin activation that is commonly observed as a consequence of microbial infection on the heart valve results in direct C5 cleavage generating functional C5a in the absence of C3 (16).
The E. faecalis proteases GelE and SprE are cotranscribed through regulation by the fsr regulatory system (29, 30). SprE has been shown to contribute to disease in animal models (5, 30, 34, 36), but mechanistically how it contributes is not known at this time. Gelatinase is a zinc-metalloprotease (18) that is related to aureolysin from Staphylococcus aureus and elastase from Pseudomonas aeruginosa (28). Gelatinase is known for its contribution to biofilm formation (12, 38) and is also thought to contribute to virulence through degradation of a broad range of host substrates, including collagen, fibrinogen, fibrin, endothelin-1, bradykinin, LL-37, and complement components C3 and C3a (18, 19, 26, 27, 33, 39). The broad substrate specificity of GelE probably contributes significantly to the complexity of endocarditis pathology, but specific mechanistic contributions to endocarditis have not been elucidated. We sought to elucidate the specific contributions of each protease to endocarditis as well as assess mechanisms that are likely associated with increased pathogenesis.
MATERIALS AND METHODS
Experimental endocarditis.
New Zealand White rabbits weighing approximately 2 kg were anesthetized by intramuscular injection with ketamine (25 mg/kg) and xylazine (20 mg/kg). The right carotid artery was exposed for catheterization by surgical incision, and a polyethylene catheter with an internal diameter of 0.86 mm (Becton Dickinson, MD) was introduced in the right carotid artery and advanced until it traversed the aortic valve into the left ventricle. Proper catheter placement was determined by feeling the resistance and noting the pulsation of the catheter line. Wound clips were used to close the incision, and all rabbits recovered without complications. Groups of six to eight catheterized rabbits were injected with 1 ml of a diluted culture (107 CFU) of E. faecalis strain V583, VT01 (ΔgelE), VT02 (ΔsprE), or VT03 (ΔgelE ΔsprE) (32, 38) via the marginal ear vein 24 h after catheter insertion. Two negative-control rabbits received sterile saline. To prepare the bacteria for injection, enterococci (V583, VT01, VT02, and VT03) were grown to stationary phase, washed twice, and diluted to a final cell density of 107 CFU/ml in sterile saline. The rabbits were euthanized 48 h after the bacterial challenge by intraperitoneal administration of sodium pentobarbital. Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to the principles stated in the Guide for the Care and Use of Laboratory Animals (24).
Determination of bacterial burden.
Animals with macroscopic valvular vegetations and proper catheter placement were analyzed for data in this study. Blood was drawn just prior to euthanasia to determine bacterial CFU in blood at the time of sacrifice. At the time of sacrifice, aortic valve vegetations were removed, weighed, homogenized in 1.0 ml of sterile phosphate-buffered saline (PBS), pH 7.4, and quantitatively cultured by plating serial dilutions on Todd-Hewitt broth (THB) agar plates. To determine the extent to which emboli formed from aortic valve vegetations, enterococci present in the remaining portions of the heart, as well as the spleen, liver, and kidneys, were also assessed by plate count. Harvested organs were introduced into 3 ml of sterile PBS, pH 7.4, and thoroughly homogenized with a tissue homogenizer. Tissue homogenates were serially diluted and plated on THB agar, and colonies were counted after overnight incubation at 37°C. Bacterial loads were expressed as log10 CFU per gram of tissue.
Histology.
The walls of the aorta and aortic valves exhibiting vegetations from representative rabbits infected with V583, VT01, VT02, and VT03 and mock-infected controls were fixed in 10% buffered formalin for histopathology. For general histology, aortic valves were embedded in paraffin and serial sections (5 μm thick) were stained either with H&E (hematoxylin and eosin) or by Gram staining.
Image analysis and statistical analysis.
Images were obtained at a final magnification of ×400 and analyzed using ImageJ software (NIH, Bethesda, MD). The average thickness of the matrix layer (generally thought to be composed of host fibrin, fibronectin, plasma proteins, and platelets [22]) and bacterial biomass was determined from regions of the vegetation that were adherent to the underlying endothelial layer. Measures of four randomly picked regions from the base of the endothelial layer to the tip of the bacterial biomass and from the tip of the biofilm biomass to the edge of the matrix layer were considered respective thicknesses of the biofilm bacterial biomass and matrix layer. Thicknesses were averaged and expressed as mean ± standard deviation (SD).
For quantitative analysis of heterophils within the matrix layer of the aortic vegetations, histological images were initially converted to an 8-bit format and a threshold was applied to contrast heterophils from the background. Heterophils were counted from images using dimensions obtained from a training data set. In cases where heterophils overlapped, the watershed algorithm was applied to delineate heterophil boundaries before counting particles. The total number of heterophils from each bacterial treatment was normalized to the area of the surrounding matrix layer and reported as the number of heterophils per 10 mm2 (10,000 μm2).
Statistical analysis of heterophil counts, matrix layer thickness, and bacterial tissue burdens was carried out with GraphPad software (San Diego, CA). One-way analysis of variance followed by a Newman-Keuls post hoc test was carried out to determine statistical significance. A P value of <0.05 was considered to be statistically significant.
GelE and SprE purification.
GelE was purified as previously described with some minor differences (12). Briefly, two liters of Todd-Hewitt broth (THB) was inoculated with 20 ml of an overnight culture of the GelE-overexpressing E. faecalis strain FA2-2 harboring the pML29 plasmid (12). The 2.0-liter culture was incubated at 37°C for 24 h. Bacteria were removed by centrifugation for 30 min at 15,000 × g. The recovered supernatants were filter sterilized and incubated at 37°C for 24 h with 10 μg/ml RNase A and 1.0 U/ml DNase. GelE was precipitated from the supernatant upon the addition of ammonium sulfate to 60% saturation followed by incubation overnight at 4°C. The mixture was centrifuged for 30 min at 27,500 × g, and the pellets were recovered by dissolving them in 150 ml of GelE buffer (50 mM Tris and 1 mM CaCl2, pH 7.8). The 150-ml sample was applied to a CL-4B column (2.5 × 17 cm) at a flow rate of 5.0 ml/min using a Bio-Rad BioLogic LP. The column was washed with six column volumes of GelE buffer. Five-milliliter fractions were collected as GelE was eluted from the column by washing with three column volumes of 50% ethylene glycol (vol/vol) in GelE buffer. To assay enzyme activity, 10 μl from each fraction was spotted on a THB agar plate containing 1.5% skim milk. Fractions showing proteolytic activity on the THB-1.5% skim milk plates were pooled and dialyzed extensively against 5.0 mM sodium phosphate (pH 7.0) using dialysis tubing (Mr cutoff of 12,000 to 14,000). After dialysis, the protease purity was checked by SDS-PAGE and the gel was silver stained. Purified GelE was aliquoted and stored at 20°C. Each aliquot was tested for activity on a THB-1.5% skim milk plate prior to use. SprE was purified as previously described (37). Both purified proteases were further analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) analysis, and molecular mass determinations were 32,866.3 Da for GelE and 25,717.1 Da for SprE.
C5a degradation.
His-tagged and nontagged versions of recombinant human complement C5a were commercially obtained from BioVision (Mountain View, CA). Human complement protein C5a (His tag) (molecular mass, 12 kDa) was incubated with purified GelE or SprE to determine the ability of the proteases to hydrolyze C5a. C5a (0.5 μg = 41.6 pmol) was incubated at 2-fold molar excess with either GelE or SprE in a total volume of 25.0 μl in GelE buffer (50 mM Tris, pH 7.8, 1 mM CaCl2) or SprE buffer (50 mM Tris, pH 7.4, 5 mM CaCl2) for 20 min at 37°C. A 15.0-μl aliquot from each sample was analyzed on a Tris-Tricine 10 to 20% gradient gel (Invitrogen) by silver staining as previously described (25). The remaining 10 μl was desalted using a ZipTip (Millipore, Bedford, MA) following the manufacturer's instructions. Samples were eluted from the ZipTip in a solution of 50% acetonitrile containing 0.1% trifluoroacetic acid, mixed with 2,5-dihydroxy benzoic acid (Sigma, Saint Louis, MO), and spotted on a Bruker aluminum plate for MALDI-TOF analysis. Samples were analyzed using a Bluker Ultraflex II mass spectrometer.
HL-60 growth and differentiation.
The human promyelocytic leukemia HL-60 cells (ATCC CCL-240) were grown in Iscove's modified Dulbecco's medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin, and 50 μg/ml streptomycin at 37°C with 5% CO2.
It is known that HL-60 cells can be differentiated into neutrophil-like cells upon the addition of dimethyl sulfoxide (DMSO) (13) and that differentiated HL-60 (dHL-60) cells are a reliable substitute for isolated neutrophils in chemotaxis and migration studies (14, 40). HL-60 cells for use in downstream applications were differentiated as previously described (14). Briefly, HL-60 cells were incubated for 5 days in Iscove's modified Dulbecco's medium supplemented with 1.2% DMSO at a concentration of 5 × 105 cells/ml. Cell differentiation was evaluated by analyzing CD11b expression on the surface of HL-60 and dHL-60 cells by flow cytometry. Briefly, HL-60 and dHL-60 cells were harvested and resuspended in culture medium to a concentration of 106 cells/ml. The cells were washed three times in 200 μl of stain medium containing PBS (pH 7.0), 10% fetal bovine serum, and 0.2% sodium azide. The Fc receptors were blocked with FcR block (BD Biosciences, San Jose, CA), followed by incubation on ice for 15 min with anti-CD11b allophycocyanin (APC)-conjugated antibodies (BioLegend, San Diego, CA) or anti-F4/80 fluorescein isothiocyanate (FITC)-conjugated antibodies (eBiosciences, San Diego, CA) as a negative control. Cells were washed three times in stain medium, resuspended to a final volume of 500 μl, and analyzed by flow cytometry using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA) at a flow rate of ∼200 cells per second. Data were analyzed using the WinList software program (VerityHouse, Topsham, ME).
dHL-60 transmigration assay.
Differentiated HL-60 (dHL-60) cells were labeled with carboxyfluorescein diacetate, succinimidyl ester (CFDA-SE), prior to the migration assay. Briefly, dHL-60 cells were pelleted and resuspended in three milliliters of PBS containing 0.1% bovine serum albumin (BSA) at a concentration of 106 cells/ml followed by the addition of an equal volume of CFDA-SE in PBS at a concentration of 20 μM. The cells were incubated with CFDA-SE for 10 min at 37°C and subsequently washed three times with Dulbecco modified Eagle medium (DMEM) supplemented with human serum albumin (HSA) (5.0 mg/ml) and HEPES (15 mM). Washed cells were resuspended in DMEM-HSA-HEPES at a concentration of 106 cells/ml, and 100 μl of cells was aliquoted into the upper chamber (3.0-μm polyester membrane) of a 24-well Transwell (Corning) plate. A volume of 600 μl of DMEM-HSA-HEPES containing either C5a (10−9 M) alone or C5a incubated with GelE for 20 min was added to the lower wells prior to the addition of upper chambers. Culture medium containing GelE alone was used in the lower wells as a negative control. The dHL-60 cells were allowed to migrate toward the bottom chamber for 70 min at 37°C. Cells that had migrated to the bottom well were collected, washed three times in PBS, and lysed with 0.2 M NaOH. The amount of CFDA-SE present from the cell lysates was measured spectrofluorometrically with excitation at 492 nm and emission at 571 nm on a Perkin Elmer Victor 3 fluorescent plate reader. Fluorescence values for the negative control were subtracted from the samples, and data were analyzed as percent fluorescence with C5a alone set to 100%. Statistical analysis was performed using GraphPad Prism software.
RESULTS
Tissue bacterial burdens.
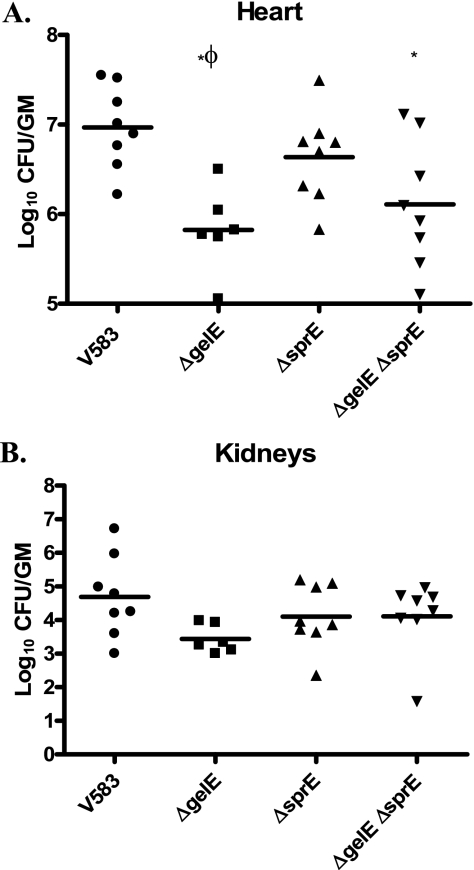
Bacterial burdens were determined from the aortic vegetations, heart tissues, kidneys, blood, liver, and spleen of rabbits (containing endocardial catheters) infected with V583 (parental strain) and ΔgelE (VT01), ΔsprE (VT02), and ΔgelE ΔsprE (VT03) isogenic extracellular protease mutants. Figure 1 shows the log10 CFU per gram of tissue from the heart after resection of the aortic valve and associated vegetations (Fig. 1A) and the kidneys (Fig. 1B). Compared to the value for V583, the mean numbers of CFU for VT01 and VT03 per of gram of heart tissue were significantly decreased by 14-fold and 7.2-fold, respectively (P < 0.05). Conversely, the mean bacterial burdens in the hearts of rabbits infected with VT02 (GelE+ SprE−) were significantly increased by 6.5-fold compared to those for rabbits infected with VT01 (GelE− SprE+) (P < 0.05). No significant differences could be observed in the mean bacterial burdens of heart tissues infected with either V583 and VT02 (GelE+ SprE−) or VT01 (GelE− SprE+) and VT03 (GelE− SprE−). Other tissues harvested from the rabbits, including aortic valve vegetations, the kidneys, spleen, and liver as well as blood, did not display significant differences in bacterial burden for any of the E. faecalis strains (Fig. 1B and data not shown).
FIG. 1.
Enterococcal burdens in the rabbit heart and kidneys. Vital organs of rabbits infected with E. faecalis (parental and isogenic protease mutants) were harvested following catheter-induced enterococcal endocarditis as described in Materials and Methods. (A) Mean bacterial burdens for V583 (parental), VT01 (ΔgelE), VT02 (ΔsprE), and VT03 (ΔgelE ΔsprE) are represented as log10 CFU/g of homogenized heart tissue. (B) Mean bacterial burdens in the pooled kidneys from each rabbit (n = 6 to 8). *, significant P values of less than 0.05 relative to V583; Φ, significant P values of less than 0.05 relative to the ΔsprE mutant (VT02).
Histology of aortic valve vegetations.
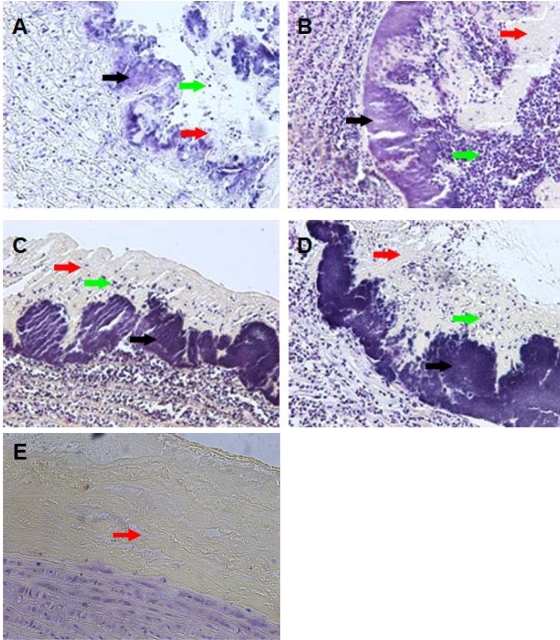
Based on the historic findings of Gutschik et al. (10), we hypothesized that the architectures of the vegetations might be different among the wild-type and isogenic protease mutants. As illustrated in Fig. 2A to D, vegetations were observed on the aortic valve for all strains tested. Histological examination of infected lesions showed bacterial colonization on the endothelial lining of the ascending aorta, which mostly appeared as a smooth layer (approximately 25 μm to 50 μm in thickness) closely interspersed with towerlike projections that rose up to ∼150 μm. Additionally, aortic vegetations showed a variable deposition of a matrix layer across the bacterial biomass depending on the proteolytic nature of the strain. The matrix layer also showed variable infiltration of heterophils depending on the protease phenotype of the E. faecalis strain. The mock-infected control animal showed the deposition of a matrix layer in response to catheterization but did not exhibit signs of heterophil recruitment to this site (Fig. 2E).
FIG. 2.
Histology of aortic vegetations. Panels A, B, C, D, and E are representative images of Gram-stained cross-sections (5 μm) of vegetations formed on the ascending aorta of rabbits infected with V583, VT01 (ΔgelE), VT02 (ΔsprE), and VT03 (ΔgelE ΔsprE) and an uninfected control, respectively (magnification, ×200). Black arrows point to E. faecalis biomass on the surface of the endothelium. Red arrows point to deposited matrix layer composed mostly of platelets and fibrin. Green arrows point to influx of heterophils and other immune cell infiltrates.
Matrix layer deposition.
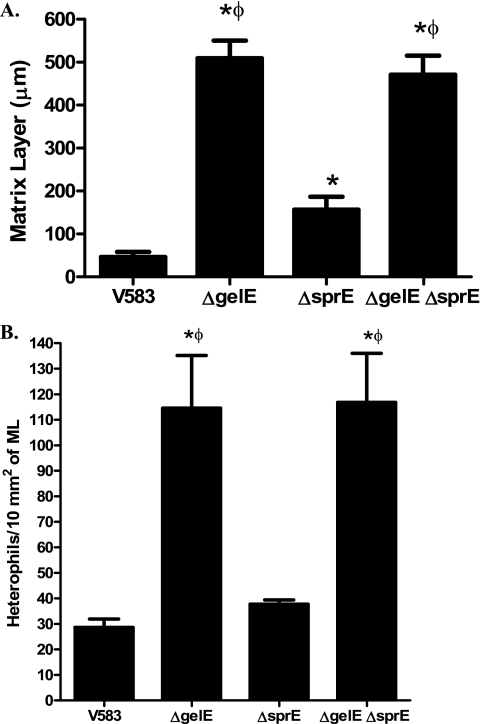
The relative thickness of the matrix layer was variable among different extracellular protease mutants (Fig. 3 A). VT01 (GelE− SprE+) and VT03 (GelE− SprE−) displayed an ∼10-fold thicker matrix layer than V583 (P < 0.05), suggesting a critical role for GelE in regulating matrix layer turnover. This observation is consistent with earlier reports of GelE's ability to hydrolyze fibrin (39), a major constituent of the matrix layer. Interestingly, compared to the parental strain (V583), VT02 (GelE+ SprE−) exhibited an ∼3.4-fold increase in the thickness of the matrix layer (P < 0.05), suggesting a possible role for SprE in degradation of the matrix layer. However, as the relative thicknesses of the matrix layers of VT01 (GelE− SprE+) and VT03 (GelE− SprE−) were not significantly different, the observed role of SprE in matrix turnover in the absence of GelE may arguably be minor.
FIG. 3.
(A) Matrix layer (ML) of animals infected with wild-type and extracellular protease mutants. Differences in the ML thickness were determined from histological images of vegetations (magnification, ×400). The lengths between the E. faecalis biomass layer and the upper edges of ML from eight random regions of vegetations from each strain were measured and reported as mean thickness (μm, mean ± standard error of the mean [SEM]). (B) Quantification of heterophil chemotaxis in the hearts of rabbits infected with E. faecalis. Differences in the numbers of heterophils that have migrated to the bacterial vegetations were determined from histological images (magnification, ×400) and were normalized to the area of ML surrounding them. Heterophils were counted using ImageJ software from four random images of vegetations from each strain and reported as the total number of heterophils trapped per 10 mm2 of ML (mean ± SEM). *, significant P values of less than 0.05 relative to V583; Φ, significant P values of less than 0.05 relative to the ΔsprE mutant (VT02).
Heterophil recruitment.
Based on histological differences (Fig. 2), there appeared to be differences in how rabbit heterophils (neutrophil-like) migrate to infected tissues. We sought to quantify the differences in heterophil recruitment between rabbits infected with the various proteolysis-proficient and -deficient strains. The numbers of heterophils/10 mm2 of matrix layer were determined from four aortic valve sections containing vegetations for each strain. Figure 3B shows that rabbits infected with strains lacking GelE (VT01 and VT03) had 3- to 4-fold-higher numbers of heterophils/10 mm2 in the matrix layer than rabbits infected with strains producing GelE (V583 and VT02) (P < 0.05). There was no significant difference in the amount of heterophils/10 mm2 of matrix layer between rabbits infected with VT01 (GelE−) or VT03 (GelE− SprE−) or between rabbits infected with V583 or VT02 (SprE−), suggesting that GelE plays the primary role in limiting heterophil recruitment at infected sites.
GelE and SprE degradation of C5a.
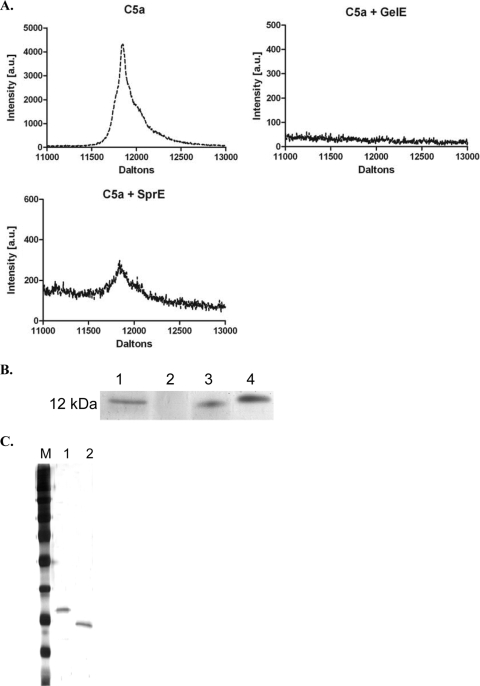
Because of its important role as a chemoattractant for neutrophils, we incubated purified GelE or SprE (Fig. 4 C) with human C5a to determine if either protease possessed proteolytic activity targeting C5a. We used Tris-Tricine gel analysis and MALDI-TOF analysis to determine activity of the enterococcal proteases toward C5a. Our results show that GelE completely degrades C5a during the course of the 20-min incubation with the C5a substrate at a 2-fold molar excess (Fig. 4A and B). These results are similar to the reported GelE activity toward C3a (26). Based on the limited role that SprE played in vivo at limiting heterophil recruitment, we also observed limited hydrolysis in vitro and only at a molar ratio of 2:1 (C5a relative to SprE), suggesting that C5a may not be an effective substrate for SprE. In contrast, GelE retains significant proteolytic activity toward C5a at higher molar ratios (10:1 and 100:1) of C5a relative to GelE (data not shown).
FIG. 4.
GelE degrades C5a. (A) MALDI-TOF spectra of C5a (∼12 kDa) alone, C5a incubated with GelE, and C5a incubated with SprE. Incubation of C5a with GelE results in complete hydrolysis of C5a in 20 min, whereas incubation of C5a with SprE results in ∼ 90% hydrolysis under similar conditions. (B) Silver-stained Tris-Tricine gel showing the molecular mass marker of ∼12 kDa (lane 1), C5a incubated with GelE (lane 2), C5a incubated with SprE (lane 3), and C5a alone (lane 4). (C) Silver-stained gel of the purified proteases: M designates the molecular weight ladder. Lane 1, GelE; lane 2, SprE. The purified proteases were subjected to MALDI-TOF, and the molecular masses were determined to be 32,866.3 Da for GelE and 25,717.13 Da for SprE (data not shown).
In vitro neutrophil chemotaxis in response to C5a incubated with GelE.
Based on the ability of GelE to cleave C5a and the fact that C5a is a powerful neutrophil chemoattractant, we determined if incubation of C5a with GelE decreased neutrophil chemotaxis in vitro. We used dHL-60 cells (differentiated neutrophil-like cell) in conjunction with Transwell migration assays to determine the effect of dHL-60 movement across a membrane in response to C5a or C5a incubated with GelE. Flow cytometry in conjunction with CD11b-specific antibodies was used to ensure that HL-60 cells incubated with DMSO had differentiated into neutrophil-like cells (data not shown). As previously described (14, 40), HL-60 cells displayed increased levels of CD11b on their surface following 5 days of incubation with DMSO, indicating differentiation into neutrophil-like cells.
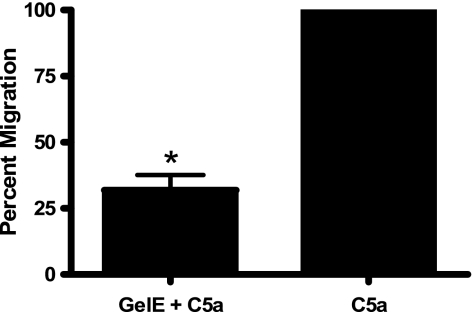
The dHL-60 cells (labeled with CFDA-SE) were allowed to migrate toward C5a or C5a previously incubated with GelE for 70 min. As expected from the results shown in Fig. 4, incubation of C5a with GelE resulted in a 60 to 70% reduction in neutrophil movement across the Transwell membrane compared to that of C5a alone (Fig. 5; *, P < 0.05).
FIG. 5.
Transwell migration assays. Incubation of C5a with GelE inhibits dHL-60 migration through Transwell membranes. Neutrophil-like dHL-60 cells were labeled with fluorogenic CFDA-SE and allowed to migrate through a 3.0-μm membrane in response to C5a or C5a previously incubated with GelE. Incubation of C5a with GelE significantly (*, P < 0.05) reduces dHL-60 chemotaxis compared to that of C5a alone.
DISCUSSION
Extracellular proteases from pathogenic bacteria assume many roles in manipulation and subversion of host innate immune responses (28). The E. faecalis extracellular proteases GelE and SprE are known to contribute to pathogenesis through contributions to biofilm production as well as degradation of important immune peptides (12, 26, 33, 38). Previous studies exploring the contribution of the secreted enterococcal proteases GelE and SprE in infective endocarditis caused by E. faecalis were unable to distinguish the relative contribution of either protease to disease pathology (10, 35). Gutschik et al. (10) compared proteolytic and nonproteolytic strains of E. faecalis in a rabbit endocarditis model, but these studies were not performed in an isogenic background and were unable to decouple the activity of GelE from that of SprE. More recently, Singh et al. (35) examined the contribution of the enterococcal proteases in a rat model of endocarditis. This study demonstrated an important role for the proteases by comparing wild-type OG1RF and an isogenic gelE insertion mutant, and it found that an insertion in gelE significantly increased the infectious dose required to induce endocarditis, compared to the wild type. However, as gelE and sprE are cotranscribed, the insertion mutation in gelE is known to abrogate the expression of sprE due to polar effects on downstream transcription (30), leaving a functional role for either protease in disease pathogenesis unclear.
Here we show in a rabbit model of endocarditis using mutants with precise in-frame deletions of gelE, sprE, or both protease genes that the principal protease mediating increased bacterial burden at disseminated sites of infection is gelatinase. Surprisingly, at the primary site of colonization (the damaged aortic valve), we did not observe significant differences in the numbers of bacteria colonizing the aortic valve among the strains tested. This is, however, consistent with the findings reported by Gutschik et al. (10), as these authors reported no significant difference between the numbers of CFU colonizing the primary vegetation from proteolytic and nonproteolytic strains. We did, however, observe altered vegetation architecture consistent with the ability of gelatinase to hydrolyze fibrin. The matrix layer surrounding the bacteria was significantly diminished in a GelE+ background, which would allow the walled-off vegetation to more readily embolize and spread to adjacent or distal sites in the body. We found a significant correlation between the presence of GelE and bacterial burden in the remaining heart tissue, suggesting that the presence of GelE allows for dissemination from the primary site of colonization. Waters et al. (39) demonstrated a role for GelE in degrading fibrin. Fibrin is thought to be a principal component of the host-derived matrix layer enclosing the bacterial vegetations growing on damaged valves (20). Based on our histology findings, the fibrinolytic nature of gelatinase appears to contribute to dissemination from the primary vegetation site.
In addition to alterations to the matrix layer thickness, we observed that the presence of GelE contributed to altered heterophil recruitment at the primary site of infection (aortic valve). While there was no statistical correlation between strains for bacterial burden in the kidney and other organs, the data trended toward increased bacterial numbers in animals infected with GelE-expressing strains. We hypothesize that the absence of statistical correlation at distal sites is simply due to a timing and/or dosage effect. In the short course of the infection, bacteria must circulate to the damaged valve, colonize the valve, establish sufficient numbers to trigger the Fsr quorum response, and express the proteases. For ethical reasons, we did not use the 50% lethal dose (LD50) or the 50% toxic dose (TD50) as an outcome measure. However, in trying to establish an infectious dose that would not result in acute mortality over the short course of the experiment, we noted that 50% (two of four) of the rabbits infected with a dose of ∼108 CFU of GelE-producing strains (V583 or VT02) died due to acute embolization. In contrast, none of the animals (four of four) infected with a similar dose of GelE− strains (VT01 or VT03) succumbed to the infection, giving support to the notion that dosage and timing are important in this model. Our observation that heterophil recruitment was altered in the presence of GelE is consistent with the ability of this protease to alter the innate immune response. Makinen et al. (18, 19) demonstrated a broad substrate specificity for gelatinase, which included the ability to degrade insulin β-chain and bradykinin, displaying a tendency to favor cleavage sites containing a Leu, Phe, or Ile at the P′1 position and most basic and hydrophobic amino acids at the P2 and P1 positions. Schmidtchen et al. (33) showed that GelE was capable of cleaving the antimicrobial peptide LL-37. More recently, Park et al. (27) showed that GelE acts as a soluble C3 convertase leading to the turnover of human complement C3 in solution. This anti-C3 activity by GelE prevents the proper assemblage of the membrane attack complex on the surface of the offending pathogen, with subsequent release of the potent leukocyte chemoattractant C5a. Furthermore, any C3 bound and converted to iC3b on the surface of the pathogen is inactivated by GelE, thus preventing interaction of iC3b with its cognate neutrophil receptor, CR3.
As thrombin activation is also known to generate C5a independent of C3 activity (16), assessing the direct interaction of C5a with GelE and SprE is relevant. Several microbial proteases are known to specifically target C5a to prevent neutrophil migration to infected sites. ScpA of Streptococcus pyogenes is a cell-wall-anchored 130-kDa serine endopeptidase that specifically cleaves the complement factor C5a (2). By cleaving the chemotactic complement factor C5a, ScpA inhibits recruitment and activation of phagocytic cells to the infectious site (17). ScpB in group B streptococci has also been shown to contribute to cellular invasion and possesses sequence similarity to ScpA (1).
Our present in vitro data extend the role of GelE in modulating complement activity to C5a as well. The complement protein C5a is a potent inflammatory peptide with a broad spectrum of functions, including the modulation of cytokine production and induction of oxidative bursts, and also serves as powerful chemoattractant for neutrophils and monocytes (9, 11). While both proteases were capable of hydrolyzing C5a at near-equimolar ratios (2:1), only GelE continued to display activity at lower concentrations relative to C5a. The enzymatic activity toward C5a displayed by GelE correlated with altering the chemotactic migration of dHL-60 cells in an in vitro Transwell assay. It would appear that the ability of GelE to target the complement cascade at multiple levels (C3, iC3b, C3a, and now C5a) provides a likely corollary as to why this protease contributes to the pathogenesis of infection caused by E. faecalis. It is noteworthy that while some microbial pathogens, such as S. pyogenes and Streptococcus agalactiae, specifically target C5a, the role of GelE is a more broadly acting protease that E. faecalis uses to circumvent the complement cascade at multiple levels. The fact that SprE plays such a relatively inconsequential role in this infection model would suggest that it does not efficiently target the complement system and is of minor consequence in the rabbit endocarditis infection model. There appeared to be little effect on heterophil recruitment or bacterial burden when strains with (VT01) and without (VT03) SprE expression in the absence of GelE were compared.
Infective endocarditis is a complex disease with many bacterial and host factors contributing to diverse pathologies. Most virulence factors studied in relation to enterococcal endocarditis have focused on adherence (20). The extracellular proteases GelE and SprE are two known virulence factors that contribute to E. faecalis pathogenesis in other disease models. Elevated bacterial burden in the adjacent heart tissue of rabbits infected with the GelE-producing strains (V583 and VT02) is consistent with a crucial role for GelE in pathogenesis. Additionally, reduced heterophil recruitment to infection sites in animals infected with GelE-producing strains is consistent with the observation of C5a degradation. The role of SprE is more ambiguous than that of GelE. The presence of SprE does not significantly increase bacterial burden in the heart, as does GelE, nor does SprE inhibit heterophil recruitment in the matrix layer. Despite the indistinct role for SprE, it remains clear that GelE is a key contributor to the pathogenesis of E. faecalis in this infection model, thus adding to the ever-growing list of GelE contributions to pathogenesis and highlighting GelE as a promising target for therapeutic intervention against multidrug-resistant and virulent E. faecalis strains.
Acknowledgments
We are very grateful to John Tomich and Yasuaki Hiromasa for assistance with the MALDI-TOF experiments. We also extend our sincere thanks to Nathan Shankar and Arto Baghdayan (University of Oklahoma Health Sciences Center) for training on the rabbit endocarditis model.
This study was supported by Beginning Grants-in-Aid from the Heartland (0660072Z) and Midwest (0860084Z) affiliates of the American Heart Association (L.E.H.), NIH grant AI077782 (L.E.H.), NIH grant RR-P20 RR017686 from the IDeA Program of the National Center for Research Resources (L.E.H. and S.D.F.), NIH grant AI061691 (S.D.F.), and a grant-in-aid from the Terry C. Johnson Cancer Center at Kansas State University (V.C.T.).
Editor: J. L. Flynn
Footnotes
Published ahead of print on 16 August 2010.
REFERENCES
- 1.Cheng, Q., D. Stafslien, S. S. Purushothaman, and P. Cleary. 2002. The group B streptococcal C5a peptidase is both a specific protease and an invasin. Infect. Immun. 70:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cleary, P. P., U. Prahbu, J. B. Dale, D. E. Wexler, and J. Handley. 1992. Streptococcal C5a peptidase is a highly specific endopeptidase. Infect. Immun. 60:5219-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daffern, P. J., P. H. Pfeifer, J. A. Ember, and T. E. Hugli. 1995. C3a is a chemotaxin for human eosinophils but not for neutrophils. I. C3a stimulation of neutrophils is secondary to eosinophil activation. J. Exp. Med. 181:2119-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiScipio, R. G., P. J. Daffern, M. A. Jagels, D. H. Broide, and P. Sriramarao. 1999. A comparison of C3a and C5a-mediated stable adhesion of rolling eosinophils in postcapillary venules and transendothelial migration in vitro and in vivo. J. Immunol. 162:1127-1136. [PubMed] [Google Scholar]
- 5.Engelbert, M., E. Mylonakis, F. M. Ausubel, S. B. Calderwood, and M. S. Gilmore. 2004. Contribution of gelatinase, serine protease, and fsr to the pathogenesis of Enterococcus faecalis endophthalmitis. Infect. Immun. 72:3628-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez, H. N., P. M. Henson, A. Otani, and T. E. Hugli. 1978. Chemotactic response to human C3a and C5a anaphylatoxins. I. Evaluation of C3a and C5a leukotaxis in vitro and under simulated in vivo conditions. J. Immunol. 120:109-115. [PubMed] [Google Scholar]
- 7.Fernandez-Guerrero, M. L., L. Herrero, M. Bellver, I. Gadea, R. F. Roblas, and M. de Gorgolas. 2002. Nosocomial enterococcal endocarditis: a serious hazard for hospitalized patients with enterococcal bacteraemia. J. Intern. Med. 252:510-515. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez Guerrero, M. L., A. Goyenechea, C. Verdejo, R. F. Roblas, and M. de Gorgolas. 2007. Enterococcal endocarditis on native and prosthetic valves: a review of clinical and prognostic factors with emphasis on hospital-acquired infections as a major determinant of outcome. Medicine (Baltimore) 86:363-377. [DOI] [PubMed] [Google Scholar]
- 9.Guo, R. F., and P. A. Ward. 2005. Role of C5a in inflammatory responses. Annu. Rev. Immunol. 23:821-852. [DOI] [PubMed] [Google Scholar]
- 10.Gutschik, E., S. Moller, and N. Christensen. 1979. Experimental endocarditis in rabbits. 3. Significance of the proteolytic capacity of the infecting strains of Streptococcus faecalis. Acta Pathol. Microbiol. Scand. B 87:353-362. [PubMed] [Google Scholar]
- 11.Haas, P. J., and J. van Strijp. 2007. Anaphylatoxins: their role in bacterial infection and inflammation. Immunol. Res. 37:161-175. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, L. E., and M. Perego. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 186:5629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, P., and P. Ralph. 1985. Human leukemic models of myelomonocytic development: a review of the HL-60 and U937 cell lines. J. Leukoc. Biol. 37:407-422. [DOI] [PubMed] [Google Scholar]
- 14.Hauert, A. B., S. Martinelli, C. Marone, and V. Niggli. 2002. Differentiated HL-60 cells are a valid model system for the analysis of human neutrophil migration and chemotaxis. Int. J. Biochem. Cell Biol. 34:838-854. [DOI] [PubMed] [Google Scholar]
- 15.Herzberg, M. C. 2000. Persistence of infective endocarditis. ASM Press, Washington, DC.
- 16.Huber-Lang, M., J. V. Sarma, F. S. Zetoune, D. Rittirsch, T. A. Neff, S. R. McGuire, J. D. Lambris, R. L. Warner, M. A. Flierl, L. M. Hoesel, F. Gebhard, J. G. Younger, S. M. Drouin, R. A. Wetsel, and P. A. Ward. 2006. Generation of C5a in the absence of C3: a new complement activation pathway. Nat. Med. 12:682-687. [DOI] [PubMed] [Google Scholar]
- 17.Ji, Y., L. McLandsborough, A. Kondagunta, and P. P. Cleary. 1996. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect. Immun. 64:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makinen, P. L., D. B. Clewell, F. An, and K. K. Makinen. 1989. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (“gelatinase”) from Streptococcus faecalis (strain 0G1-10). J. Biol. Chem. 264:3325-3334. [PubMed] [Google Scholar]
- 19.Makinen, P. L., and K. K. Makinen. 1994. The Enterococcus faecalis extracellular metalloendopeptidase (EC 3.4.24.30; coccolysin) inactivates human endothelin at bonds involving hydrophobic amino acid residues. Biochem. Biophys. Res. Commun. 200:981-985. [DOI] [PubMed] [Google Scholar]
- 20.McCormick, J. K., H. Hirt, G. M. Dunny, and P. M. Schlievert. 2000. Pathogenic mechanisms of enterococcal endocarditis. Curr. Infect. Dis. Rep. 2:315-321. [DOI] [PubMed] [Google Scholar]
- 21.Megran, D. W. 1992. Enterococcal endocarditis. Clin. Infect. Dis. 15:63-71. [DOI] [PubMed] [Google Scholar]
- 22.Moreillon, P., and Y. A. Que. 2004. Infective endocarditis. Lancet 363:139-149. [DOI] [PubMed] [Google Scholar]
- 23.Mylonakis, E., and S. B. Calderwood. 2001. Infective endocarditis in adults. N. Engl. J. Med. 345:1318-1330. [DOI] [PubMed] [Google Scholar]
- 24.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 25.Nesterenko, M. V., M. Tilley, and S. J. Upton. 1994. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods 28:239-242. [DOI] [PubMed] [Google Scholar]
- 26.Park, S. Y., K. M. Kim, J. H. Lee, S. J. Seo, and I. H. Lee. 2007. Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect. Immun. 75:1861-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, S. Y., Y. P. Shin, C. H. Kim, H. J. Park, Y. S. Seong, B. S. Kim, S. J. Seo, and I. H. Lee. 2008. Immune evasion of Enterococcus faecalis by an extracellular gelatinase that cleaves C3 and iC3b. J. Immunol. 181:6328-6336. [DOI] [PubMed] [Google Scholar]
- 28.Potempa, J., and R. N. Pike. 2009. Corruption of innate immunity by bacterial proteases. J. Innate Immun. 1:70-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21:510-515. [DOI] [PubMed] [Google Scholar]
- 32.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidtchen, A., I. M. Frick, E. Andersson, H. Tapper, and L. Bjorck. 2002. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol. 46:157-168. [DOI] [PubMed] [Google Scholar]
- 34.Sifri, C. D., E. Mylonakis, K. V. Singh, X. Qin, D. A. Garsin, B. E. Murray, F. M. Ausubel, and S. B. Calderwood. 2002. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect. Immun. 70:5647-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh, K. V., S. R. Nallapareddy, E. C. Nannini, and B. E. Murray. 2005. Fsr-independent production of protease(s) may explain the lack of attenuation of an Enterococcus faecalis fsr mutant versus a gelE-sprE mutant in induction of endocarditis. Infect. Immun. 73:4888-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, V. C., Y. Hiromasa, N. Harms, L. Thurlow, J. Tomich, and L. Hancock. 2009. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol. Microbiol. 72:1022-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas, V. C., L. R. Thurlow, D. Boyle, and L. E. Hancock. 2008. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 190:5690-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waters, C. M., M. H. Antiporta, B. E. Murray, and G. M. Dunny. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J. Bacteriol. 185:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woo, C. H., M. H. Yoo, H. J. You, S. H. Cho, Y. C. Mun, C. M. Seong, and J. H. Kim. 2003. Transepithelial migration of neutrophils in response to leukotriene B4 is mediated by a reactive oxygen species-extracellular signal-regulated kinase-linked cascade. J. Immunol. 170:6273-6279. [DOI] [PubMed] [Google Scholar]