Abstract
Porphyromonas gingivalis secretes a serine phosphatase enzyme, SerB, upon contact with gingival epithelial cells in vitro. The SerB protein plays a critical role in internalization and survival of the organism in epithelial cells. SerB is also responsible for the inhibition of interleukin-8 (IL-8) secretion from gingival epithelial cells infected with P. gingivalis. This study examined the ability of a P. gingivalis SerB mutant to colonize the oral cavity and induce gingival inflammation, immune responses, and alveolar bone resorption in a rat model of periodontal disease. Both P. gingivalis ATCC 33277 and an isogenic ΔSerB mutant colonized the oral cavities of rats during the 12-week experimental period. Both of the strains induced significant (P < 0.05) systemic levels of immunoglobulin G (IgG) and isotypes IgG1, IgG2a, and IgG2b, indicating the involvement of both T helper type 1 (Th1) and Th2 responses to infection. Both strains induced significantly (P < 0.05) higher levels of alveolar bone resorption in infected rats than in sham-infected control rats. However, horizontal and interproximal alveolar bone resorption induced by the SerB mutant was significantly (P < 0.05) lower than that induced by the parental strain. Rats infected with the ΔSerB mutant exhibited significantly higher levels of apical migration of the junctional epithelium (P < 0.01) and polymorphonuclear neutrophil (PMN) recruitment (P < 0.001) into the gingival tissues than rats infected with the wild type. In conclusion, in a rat model of periodontal disease, the SerB phosphatase of P. gingivalis is required for maximal alveolar bone resorption, and in the absence of SerB, more PMNs are recruited into the gingival tissues.
One of the predominant polymicrobial infections of humans is expressed clinically as periodontal disease. A bacterial etiology for periodontal diseases is well established, and a group of Gram-negative, mostly anaerobic bacteria is associated with the initiation and progression of disease. Porphyromonas gingivalis is considered one of the more pathogenic members of this group, and elevated levels of this organism are associated with an increased risk of periodontal breakdown (4, 54). P. gingivalis produces a variety of virulence factors that enable colonization of the periodontal pocket and destruction of the structural components of the periodontium. These virulence factors include proteolytic enzymes that can damage host cells, tissues, and immune response mediators; toxic metabolites; surface components with immune-modulating activity; and adherence factors that promote colonization and persistence (7, 8, 15, 18, 21, 22, 35, 38). Moreover, the role of some of these virulence factors, such as fimbriae and proteases, has been verified in animal models of periodontal disease (36, 44, 46). P. gingivalis is also an intracellular organism that can invade gingival epithelial cells in culture (37, 39, 58). In addition, P. gingivalis has been observed within epithelial cells in ex vivo samples such as gingival biopsy samples (45, 49), and high numbers of P. gingivalis have been observed within gingival and buccal epithelial cells obtained from healthy and disease subjects (5, 50, 51). Mechanistically, P. gingivalis invasion is mediated through fimbria-mediated attachment to gingival epithelial cell integrin receptors and cytoskeletal rearrangements that allow the bacteria to enter the cell (64, 65). A key effector of the invasive process is SerB, a haloacid dehydrogenase family phosphoserine phosphatase enzyme. SerB is present in the outer membrane of P. gingivalis, and contact with gingival epithelial cells induces secretion into the extracellular milieu (67). Cell-associated and extracellular SerB impacts host cell signal transduction pathways that control the cytoskeletal architecture, and treatment of epithelial cells with SerB induces actin microfilament and tubulin microtubule rearrangements (19, 59). Furthermore, SerB is required for intracellular persistence, as a mutant lacking SerB is compromised in intracellular survival (59). An additional role of SerB concerns involvement in the stealth-like properties of P. gingivalis (18). P. gingivalis is capable of both inhibiting secretion of interleukin-8 (IL-8) from gingival epithelial cells and antagonizing IL-8 production stimulated by other organisms (9, 23, 26, 60). SerB activity is required for this innate immune suppression, known as localized chemokine paralysis (9, 19).
While SerB is important for invasion, intracellular survival, and immune suppression in vitro, its contribution to in vivo pathogenicity has not been addressed. Study of the in vivo role of P. gingivalis virulence factors has employed a variety of animal models (16). Among these, the oral infection model in rodents has been used to study colonization, periodontal inflammation, immune responses, and induction of alveolar bone resorption by P. gingivalis (3, 29, 32). The pathological processes induced by oral infection with periodontal pathogens in rodent models, including gingival and periodontal ligament destruction and resorption of the alveolar bone, resemble those that occur in humans (34). In this study, we examine the in vivo role of SerB in colonization, inflammation, immune response, alveolar bone resorption, and induction of periodontal disease in rats.
MATERIALS AND METHODS
Bacterial strains and microbial inocula.
P. gingivalis strain ATCC 33277 and its isogenic serB-defective mutant, ΔSerB (59), were maintained anaerobically at 37°C on blood agar plates as described previously (32). For infection, bacteria were suspended at 2 × 1010 bacteria ml−1, as determined by the optical density at 600 nm (OD600). Bacteria were mixed with equal volumes of sterile 4% (wt/vol) low-viscosity carboxymethylcellulose (CMC; Sigma-Aldrich, St. Louis, MO) in phosphate-buffered saline (PBS), and 0.25 ml was used for infection (2.5 × 109 cells) by oral gavage as described previously (32, 61, 62).
P. gingivalis oral infections and sampling.
Female Sprague-Dawley rats (8 to 9 weeks old; Charles River Laboratories, Boston, MA) were maintained in groups and housed in microisolator cages. Rats were fed standard powdered chow (Teklad Global 18% protein rodent diet 2918; Harlan, Madison, WI) and water ad libitum and were kept at 25°C with alternating 12-h periods of light and dark. All animal procedures were performed in accordance with the approved protocol guidelines set forth by the Institutional Animal Care and Use Committee of the University of Florida. In addition, adequate measures were taken to minimize pain or discomfort of rats during oral infection and sampling. Rats were administered kanamycin (20 mg/rat) and ampicillin (20 mg/rat) daily for 4 days in the drinking water (32, 33, 53), and the oral cavity was swabbed with 0.12% chlorhexidine gluconate (Peridex; 3M Espe Dental Products, St. Paul, MN) mouth rinse (32, 53) to inhibit endogenous organisms and to promote subsequent colonization of P. gingivalis. After a 3-day antibiotic washout period, rats were randomized into three groups (n = 9). The microbial inocula were administered by oral gavage for 4 consecutive days per week on 6 alternate weeks for a total of 24 inoculations during 12 weeks of the experimental infection period (Fig. 1). Sham-infected control rats received the vehicle (sterile CMC) only. Oral microbial samples obtained from isoflurane-anesthetized rats were collected at pre- and postinfections, as described previously (32, 53). In order to monitor the colonization/infection with minimal disruption of the biofilms, a total of 2 postinfection microbial samples (following the fourth and sixth infections) were collected from all infected rats. Rats were sacrificed, blood specimens were collected, and sera were stored at −20°C for immunoglobulin G (IgG), IgA, IgM, and IgG isotype antibody analysis (61, 62). Rat skulls were removed, autoclaved, and mechanically defleshed with a periodontal scaler for evaluation of alveolar bone resorption by morphometric and radiographic analysis. The rat jaws were also suspended in 10% buffered formalin and decalcified for histology and histomorphometry (61, 62).
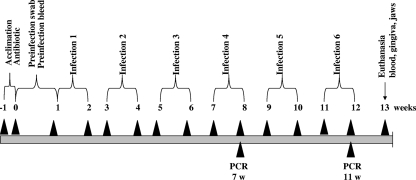
FIG. 1.
Schematic diagram illustrating the experimental design, including rat acclimation, antibiotics treatment, Peridex swabbing, preinfection, oral microbial sample collection, bacterial infection (6 infections), oral microbial sample collections (two times), PCR analysis, euthanasia, and gingival tissue and alveolar bone collection. For detailed information, see Materials and Methods.
Determination of P. gingivalis colonization.
DNA was isolated from rat oral samples using a Wizard genomic DNA purification kit (Promega, Madison, WI). Subsequently, PCR was performed with a Bio-Rad thermal cycler using the following 16S rRNA gene species-specific PCR oligonucleotide primers: 5′-TGTAGATGACTGATGGTGAAAACC-3′ (forward) and 5′-ACGTCATCCCCACCTTCCTC-3′ (reverse). Genomic DNAs extracted from P. gingivalis parental and mutant strains served as positive controls, and PCR performed with no template DNA was the negative control. Each PCR assay could detect at least 0.05 pg of DNA standard.
Serum antibody and IgG subclass analysis.
Sera obtained from infected rats (n = 9) at 12 weeks were used to determine IgG, IgM, IgA, and IgG subclass (IgG1, IgG2a, IgG2b, and IgG2c) antibody concentrations against whole cells of the strain of P. gingivalis (wild type or ΔSerB) used for infection with a standard enzyme-linked immunosorbent assay (ELISA) (61, 62). Briefly, P. gingivalis cells were treated overnight with 0.5% formalin in buffered saline (FK cells), washed, diluted to an OD600 of 0.3, and deposited in the wells of microtiter plates (32). Diluted rat sera (1:100 for IgG and 1:20 for IgM, IgA, and subclasses of IgG) were reacted with the bacteria for 2 h at room temperature. After being washed, goat anti-rat IgG, IgG1, IgG2a, IgG2b, IgG2c, IgM, or IgA conjugated to alkaline phosphatase (1:5,000) (Bethyl Laboratories, Montgomery, TX) was added (1:500) to the plates, and the assay was developed with p-nitrophenyl phosphate (Sigma). The assay reactions were terminated by the addition of 3 M NaOH and analyzed at an OD405 using a Bio-Rad microplate reader. Serum antibody concentrations were determined using a gravimetric standard curve. The standard curve consisted of 8 rat IgG concentrations (Sigma), which were coated onto wells, detected, and developed as described above (61, 62).
Alveolar bone resorption.
The pattern of horizontal or interproximal alveolar bone resorption induced by P. gingivalis was measured by morphometric or radiographic methods, respectively. For horizontal bone loss, the rat maxillae and mandibles (n = 6) were immersed in 3% (vol/vol) hydrogen peroxide overnight, washed with deionized water, air dried, and stained for 1 min in an aqueous solution of 0.1% (wt/vol) methylene blue to delineate the cementoenamel junction (CEJ) (48, 61, 62). Digital images of both buccal and lingual root surfaces of all molar teeth were captured under a 10× stereo dissecting microscope (SteReo Discovery V8; Carl Zeiss MicroImaging, Inc., Thornwood, NY), after superimposition of buccal and lingual cusps to ensure reproducibility and consistency. The line tool was used to make horizontal bone resorption measurements from the CEJ to the alveolar bone crest (ABC). The surface perimeters of the CEJ and ABC were traced using the calibrated line tool (AxioVision LE 29A software version 4.6.3.). Two blinded investigators were used, all measurements were performed twice by the same examiner at separate times, and the means of the measurements for each of the four quadrants were obtained.
For interproximal bone loss, the maxillae and mandibles were trimmed to reduce the buccolingual dimensions in order to allow close proximity of the teeth to the digital Kodak 6000 sensor (Carestream Health, Rochester, NY). Digital radiographs of the distal and mesial sides of the molars were acquired using orthogonal projection geometry and an exposure time of 0.08 s at 60 peak kV [kVp] and 15 mA. All radiographic images were exported into the TACT (tuned aperture computed tomography) Workbench, and histograms were equalized. The line tool was used to make vertical bone resorption (interproximal defect) measurements on the distal and mesial sides of each interproximal surface (2 sites [mesial and distal] per tooth) for each of the molars in each quadrant from the CEJ to the ABC (i.e., resorption). Measurements were made by investigators blinded to the group designation. The summation of alveolar bone loss in millimeters was tabulated and analyzed for intra- and intergroup differences (32, 53, 61, 62).
Histomorphometric analysis of periodontal tissue.
Infected and control rat jaws (n = 3) were removed randomly and fixed in 10% buffered formalin. Bone was decalcified in Immunocal (Decal Chemical, Tallman, NY) for 28 days at 4°C (61, 62). The decalcified tissue was embedded in paraffin blocks, 4-μm sections were prepared and stained with hematoxylin and eosin, and whole slides were digitally scanned with a ScanScope CS system (Aperio, Vista, CA). The scanned slides were viewed with ImageScope viewing software (Aperio). The inflammation of the supracrestal gingival connective tissue between the molars in each specimen at consecutive sections or levels 10 and 20 was examined based on multiple parameters, including apical migration of the junctional epithelium (JE), rete ridge elongation, interdental crestal alveolar bone resorption, lymphocyte numbers, and blood vessel density (14, 57). The number of lymphocytes per unit area (0.05 mm by 0.05 mm) was counted in the area of the JE and adjacent connective tissue (14, 57, 61, 62). The apical migration of the JE, elongation of rete ridges, and resorption of alveolar bone was measured using ImageScope software with a microgrid at a magnification of ×200. The distances from the CEJ to the coronal portion of the connective tissue attachment (apical migration), from the CEJ to the apical portion of the rete ridge, and from the CEJ to the level of the alveolar bone crest were measured (14, 57, 61, 62).
Quantitation of neutrophils.
Immunohistochemical analysis for polymorphonuclear neutrophils (PMNs) was performed on paraffin-embedded rat tissue sections as described previously (17). Tissue sections were deparaffinized in xylene and rehydrated in decreasing concentrations of alcohol. Unmasking was performed using sodium citrate, and endogenous peroxidase activity was blocked with 3% hydrogen peroxide. Slides were reacted with rabbit anti-rat PMN antibody (Accurate Chemical and Scientific Corp., Westbury, NY) followed by secondary alkaline phosphatase anti-rabbit IgG. Visualization was performed with Vector red alkaline phosphatase (Vector Laboratories, Burlingame, CA), followed by hematoxylin QS counterstaining (Vector Laboratories). PMNs were counted in 10 randomly chosen high-powered fields per slide (3 slides/group).
Osteoclast analysis of alveolar bone.
Following 12 weeks of infection, rat right maxillae (n = 3) were decalcified and embedded in paraffin, and sections at 4 μm were prepared and stained for tartrate-resistant acid phosphatase (TRAP; Sigma). The TRAP-stained whole slides were digitally scanned immediately with a ScanScope CS system (Aperio) to minimize color fading. The scanned slides were viewed with ImageScope viewing software (Aperio). The right maxillary interdental areas of the crestal alveolar bone from the first molar to third molar were used to quantify osteoclasts (31, 42, 68). Activated osteoclasts were identified as multinucleated dark red cells.
Statistical analysis.
Antibody and alveolar bone resorption data were analyzed using Kruskal-Wallis analysis of variance (ANOVA) with Dunn's correction for multiple comparisons. Inflammatory marker and osteoclast data were analyzed by the Mann-Whitney test with Welch's correction. PMN counts were compared using the Tukey-Kramer multiple-comparison test.
RESULTS
Oral bacterial infections.
Prior to infection, we examined all rats by PCR of oral samples using P. gingivalis-specific primers, and all rats were negative for P. gingivalis genomic DNA. PCR evaluations of the oral samples collected at 2 time points postinfection demonstrated that 100% of rats were infected with P. gingivalis during the experimental periodontal disease period. PCR products were examined by electrophoresis and scanning densitometry, and no differences were observed between samples obtained from parent strain- or mutant-infected rats. None of the sham-infected control rats were positive for P. gingivalis during the study period.
Antibody responses to oral infections.
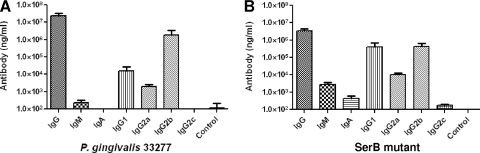
To investigate the antibody responses to P. gingivalis wild-type and ΔSerB mutant infection, we measured the levels of species-specific IgG, IgM, IgA, and IgG subclass (IgG1, IgG2a, IgG2b, and IgG2c) antibodies in rat sera after 12 weeks of oral infection. Sera obtained from rats in the P. gingivalis wild-type-infected group displayed robust levels of anti-whole-cell-specific IgG antibody following oral infection compared with those levels in sera obtained from sham-infected control rats (Fig. 2A). In addition, rats infected with the SerB mutant also produced significant levels of anti-whole-cell-specific IgG antibody (Fig. 2B). The P. gingivalis wild type induced low levels of IgM and undetectable levels of IgA antibodies. In comparison to the parental strain, the SerB mutant induced approximately 10-fold-lower IgG levels, along with >10-fold-higher IgM and IgA levels (P < 0.001).
FIG. 2.
IgG, IgG subclass (IgG1, IgG2a, IgG2b, and IgG2c), IgM, and IgA antibody levels in sera collected from rats at the end of 12 weeks of infection. (A) P. gingivalis wild-type-infected rats (n = 9); (B) P. gingivalis ΔSerB-infected rats (n = 9). The control used was sera obtained from sham-infected rats. Whole cells of the P. gingivalis wild type or SerB mutant (according to the infection condition) were used as the antigen in an ELISA. Error bars indicate standard deviations.
In order to more fully evaluate the characteristics of the IgG antibody responses, we determined the IgG antibody subclass responses. In P. gingivalis wild-type infection, the IgG2b subclass levels were significantly (P < 0.001) higher than the IgG1, IgG2a, and IgG2c antibody levels (Fig. 2A). In ΔSerB mutant infection, the IgG1 and IgG2b subclass levels were equal and were higher (P < 0.001) than the IgG2a and IgG2c antibody levels (Fig. 2B). Additionally, SerB mutant infection induced a higher (P < 0.001) concentration of IgG1, IgG2a, and IgG2c subclass antibodies than that induced by wild-type P. gingivalis.
Horizontal alveolar bone resorption.
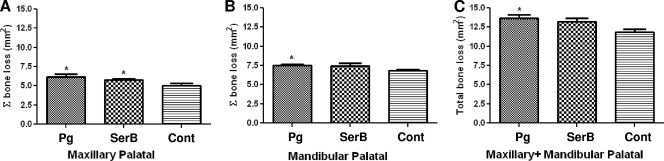
Infection with either wild-type or SerB mutant strains induced maxillary palatal areas of alveolar bone resorption, which were measured morphometrically and found to be significantly greater (P < 0.05) than those of the control uninfected rats (Fig. 3A). However, infection with the SerB mutant induced less (P < 0.05) mandibular alveolar bone loss than wild-type P. gingivalis (Fig. 3B), and combined total palatal (mandibular and maxillary) bone loss was not significantly higher in the SerB mutant-infected animals than in control animals (Fig. 3C).
FIG. 3.
Maxillary (A), mandibular (B), and combined (C) morphometric horizontal area alveolar bone resorption in rats (n = 6) following infection with the P. gingivalis wild type or SerB mutant strain. Each bar indicates the mean level of horizontal area alveolar bone loss for the distance measured between the cementoenamel junction and alveolar bone crest of three molar teeth. Error bars indicate standard deviations. Asterisks denote significant differences from values obtained with sham-infected control rats (P < 0.05). Pg, P. gingivalis; SerB, ΔSerB; Cont, sham-infected control.
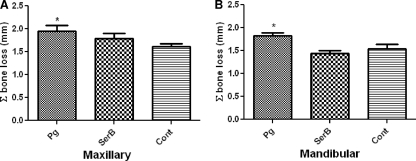
Interproximal alveolar bone resorption.
To corroborate our observations of horizontal area alveolar bone loss, digital radiographic analyses of vertical alveolar bone loss were also performed. All interproximal bone loss measurements were taken from the CEJ to the most coronal level of the crestal alveolar bone to the mesial and distal sides of the three molars. P. gingivalis wild-type infection resulted in significantly increased maxillary and mandibular interproximal alveolar bone loss at 12 weeks of infection compared with that of sham-infected rats (P < 0.05) (Fig. 4). In contrast, infection with the SerB mutant demonstrated decreased vertical bone loss compared to that resulting from wild-type P. gingivalis infection (P < 0.05) and showed no significant increase over that of sham-infected control rats (Fig. 4). These data confirm the in vivo-altered virulence potential of the SerB-deficient mutant.
FIG. 4.
Maxillary (A) and mandibular (B) radiographic interproximal alveolar bone resorption in rats (n = 6) following infection with the P. gingivalis wild type or SerB mutant strain. Each bar indicates the mean level of interproximal alveolar bone loss for the distance measured between the cementoenamel junction and alveolar bone crest at mesial and distal sites of three molar teeth (6 sites). Error bars indicate standard deviations. Asterisks denote significant differences from values obtained with sham-infected control rats (P < 0.05). Pg, P. gingivalis; SerB, ΔSerB; Cont, sham-infected control.
Histological evaluation of inflammation and bone loss.
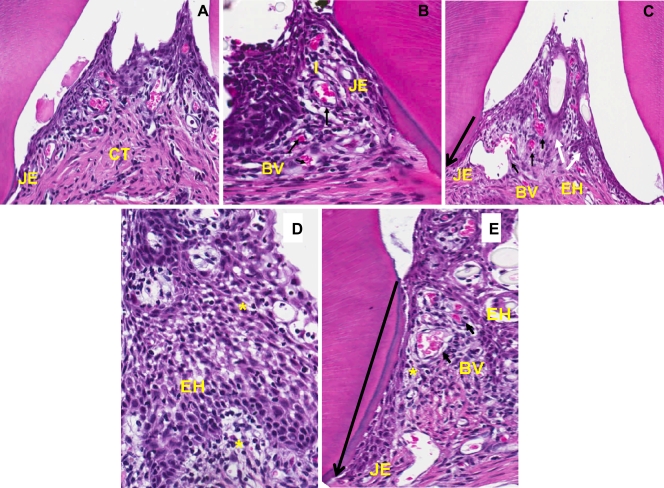
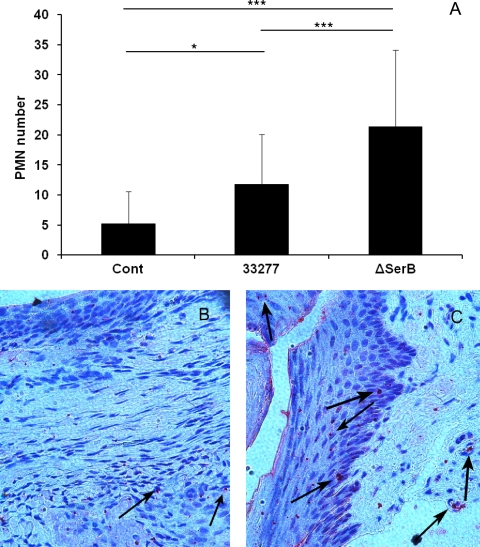
In order to examine the levels of inflammation associated with wild-type and SerB mutant infection, sections of maxillae at consecutive levels 1, 10, and 20 were examined histologically for inflammatory markers. The sham-infected control rat maxilla exhibited minimal hyperplasia of the crevicular epithelium with scattered chronic inflammatory cells (PMNs and lymphocytes), lack of migration of the junctional epithelium (JE), and minimal inflammation in the connective tissue (Fig. 5 A). In contrast, the rats infected with both parental and mutant P. gingivalis strains showed prominent epithelial hyperplasia, apical migration of the JE, an increase in the number of blood vessels, and dense inflammatory infiltrates in the connective tissue (Fig. 5B and C). In addition, there was extensive intra- and subepithelial edema in response to both parent and mutant strains (Fig. 5D and E). Quantitation of inflammatory markers (Table 1) confirmed a significant increase in apical migration of the JE, lymphocyte infiltration, and blood vessel density induced by the wild-type and ΔSerB strains in comparison to those in sham-infected control tissues. Furthermore, both strains induced significant elongation of rete ridges. Thus, while the SerB mutant caused significantly less alveolar bone resorption, the inflammatory response to the mutant was not abrogated; indeed, apical migration of the JE was significantly higher in rats infected with the mutant than in parent strain-infected rats. These results suggest that inflammation is not directly related to alveolar bone resorption and, further, that the SerB enzyme can limit, to some extent, inflammatory responses to P. gingivalis. As SerB, suppresses production of the neutrophil chemokine IL-8 in vitro (9), we counted the numbers of PMNs in the tissues of infected and control rats. PMN recruitment into the gingival tissues was significantly (P < 0.001) increased in rats infected with the SerB mutant compared to that in rats infected with the parental strain (Fig. 6). Thus, the ability of SerB to antagonize IL-8 production in vitro is reflected in vivo through decreased neutrophil chemotaxis.
FIG. 5.
Comparative histology (hematoxylin and eosin staining) of alveolar bone sections from the maxillae of rats after 12 weeks of infection with the P. gingivalis wild type or ΔSerB strain. (A) Section from a control rat displaying minimal inflammation, lack of migration of the junctional epithelium (JE), and minimal inflammation in the connective tissue (CT). (B) Section from a P. gingivalis wild-type-infected rat showing migration of the JE, an increase in the number of blood vessels (BV) (short black arrows), and dense inflammatory infiltrates (I). (C) Section from a SerB mutant-infected rat also exhibiting migration of the JE (long arrow shows direction of apical migration), prominent epithelial hyperplasia (EH) (white arrows), dilated blood vessels, and dense inflammation. Images are shown at 200× magnification. (D) P. gingivalis wild-type-infected tissue demonstrating extensive intra- and subepithelial edema (*), with prominent epithelial hyperplasia. (E) SerB mutant-infected tissue exhibiting extensive intra- and subepithelial edema (*), proliferation of blood vessels, epithelial hyperplasia, and migration of the JE. Images are shown at 600× magnification and are representative of over 5 rats from each group.
TABLE 1.
Histometrical analysis of rat periodontal tissue after infection with P. gingivalis ATCC 33277 or the SerB mutant
| Histological parameter | Value forf: |
||
|---|---|---|---|
| Control | ATCC 33277 | ΔSerB | |
| Apical migration (μm)g | 24.4 ± 14.4 | 80.5 ± 27.8c | 96.5 ± 10.3b,d |
| Rete ridge elongation (μm)h | 36.1 ± 12.9 | 73.7 ± 26.3a | 84.1 ± 21.2b |
| Alveolar bone resorption (μm)i | 234.0 ± 31.9 | 330.8 ± 44.6c | 266.7 ± 33.7e |
| No. of lymphocytes per 0.05- by 0.05-mm area | 4.0 ± 1.4 | 8.0 ± 3.8a | 6.2 ± 2.4a |
| Blood vessel density (no. of blood vessels per 0.05- by 0.05-mm area) | 2.7 ± 1.4 | 5.2 ± 1.9b | 7.0 ± 2.1c |
Significantly higher than values obtained with the sham-infected control group (P < 0.05).
Significantly higher than values obtained with the sham-infected control group (P < 0.01).
Significantly higher than values obtained with the sham-infected control group (P < 0.001).
Significantly higher than values obtained with the P. gingivalis group (P < 0.05).
Significantly different from values obtained with the P. gingivalis group (P < 0.01).
Values are presented as means ± standard deviations (n = 5 to 11).
Distance from the CEJ to the coronal portion of the connective tissue attachment (apical migration of the JE).
Distance from the CEJ to the apical portion of the rete ridge.
Distance from the CEJ to the level of the ABC.
FIG. 6.
PMN recruitment in rat tissue following infection with the P. gingivalis wild type (33277) or ΔSerB. Cont, sham-infected control. (A) Mean PMN counts in a microscope field. Error bars indicate standard deviations (n = 30). *, P < 0.05; ***, P < 0.001. (B, C) P. gingivalis wild-type-infected (B) and SerB mutant-infected (C) rat tissue stained with anti-PMN antibody (200× magnification). Images are representative of 30 examined in each group.
Osteoclast analysis of alveolar bone.
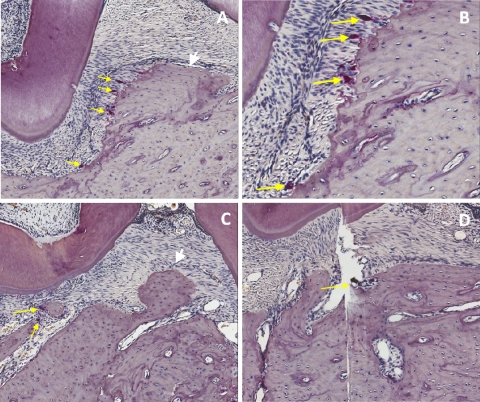
To determine if the P. gingivalis strains induced differing levels of osteoclasts, which could be responsible for the alveolar bone resorption pattern, osteoclasts in rat maxillae were quantitated. In the interdental area from molars 1 to 3, wild-type P. gingivalis infection induced a mean of 10.0 ± 1.18 activated osteoclasts (Fig. 7A and B), which was significantly (P < 0.005; n = 8) higher than the mean of 5.1 ± 0.81 osteoclasts found in the same area in sham-infected control rats (Fig. 7D). Infection with the SerB mutant strain (Fig. 7C) did not show a statistically significant increase in osteoclast number (5.1 ± 0.81) compared to those of the controls (P > 0.05). These results indicate that the reduced crestal alveolar bone loss in the SerB mutant-infected animals is due to fewer activated osteoclasts.
FIG. 7.
Representative histological sections with TRAP staining showing osteoclastic activity from the right maxillae of rats. (A) P. gingivalis wild-type infection demonstrating an alveolar bone crest and tooth with numerous osteoclasts (yellow arrows) in the area of the alveolar bone crest (white arrow). Magnification of ×100. (B) Higher magnification (×200) of the same area as shown in panel A, demonstrating Howship's lacunae and osteoclasts. (C) Infection with the SerB mutant demonstrating the alveolar bone crest (white arrow), interdental area, and few osteoclasts (yellow arrow). Magnification of ×100. (D) Sham-infected control with minimal osteoclastic activity (yellow arrow). Magnification of ×100.
DISCUSSION
Tightly controlled patterns of protein phosphorylation/dephosphorylation control a multitude of signal transduction processes in both eukaryotes and prokaryotes. In bacteria, signal transduction frequently involves histidine phosphorylation/dephosphorylation-based two-component systems. However, serine/threonine kinases and phosphatases are emerging as major contributors to bacterial control mechanisms, and these enzymes have been described in Yersinia, Pseudomonas aeruginosa, Listeria monocytogenes, Streptococcus agalactiae, Staphylococcus aureus, Bacillus anthracis, and Streptococcus mutans, where they contribute to virulence (11, 24, 47). Serine/threonine kinases and phosphatases have been shown to play a role in the molecular dialog between bacteria and host cells. For example, Shigella flexneri produces a dually specific phosphatase, OspF, that dephosphorylates mitogen-activated protein kinase (MAPK), which in turn prevents histone H3 phosphorylation (1). A reduction in the level of H3 phosphorylation impedes access of the transcription factor NF-κB to the chromosome, and thus, transcription of NF-κB-responsive genes such as IL-8 is reduced. The P. gingivalis serine phosphatase SerB is involved in internalization and survival in gingival epithelial cells (59). SerB is functional within epithelial cells, interacting with cytoplasmic GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and HSP90 and modulating microtubule dynamics. SerB also regulates the dynamics of the actin cytoskeleton and inhibits the production of IL-8 (19). SerB thus plays numerous roles in P. gingivalis-host cell interaction, and in this study we established a direct role of SerB in P. gingivalis virulence in a rat model of periodontitis.
The SerB mutant induced less alveolar bone loss than the parental strain. Both horizontal alveolar bone loss and interproximal alveolar bone loss was lower in rats infected with the ΔSerB strain than in those infected with the wild type. These results indicate that loss of SerB function in P. gingivalis reduces virulence and establish a direct role for the SerB serine phosphatase in alveolar bone loss. As both the parental and mutant strains colonized the rat oral cavity to the same degree, these results also provide indirect evidence for the importance of intracellular invasion in P. gingivalis pathogenicity. Moreover, reduced virulence of the invasion-deficient SerB mutant corroborates reports that P. gingivalis strains isolated from diseased sites possess greater in vitro invasion efficiencies than strains isolated from healthy sites (25). Together with studies that P. gingivalis mutants lacking FimA fimbriae, a major invasion effector, are less virulent in animal models (44), a picture emerges of intracellular invasion as an important contributor to alveolar bone loss, and disruption of the invasive process renders P. gingivalis less virulent. Interestingly, despite the reduced bone loss resulting from infection with the SerB mutant, the maxillary gingival inflammatory response to both organisms was similar and, indeed, in some cases elevated in response to the SerB mutant. Rete ridge elongation, blood vessel density, and lymphocyte infiltration were elevated to similar extents by both the wild-type and mutant strains. However, apical migration of the junctional epithelium (JE) and neutrophil infiltration were significantly increased in response to ΔSerB. The adverse effects on the JE may result from the inability of the SerB mutant to penetrate the tissue and thus remain extracellular. Extracellular P. gingivalis organisms are more intensely proteolytic than their intracellular counterparts (63), and protease activity may contribute to apical migration of the JE (20, 30). Enhanced PMN infiltration in response to the mutant is likely the direct result of the loss of the SerB enzyme. In vitro, SerB inhibits production of IL-8 from gingival epithelial cells (9). Reduced amounts of IL-8 in the gingival tissue will impede recruitment of PMNs, and hence, in the absence of SerB, PMN recruitment will increase. These results show that localized chemokine paralysis induced by P. gingivalis also occurs in vivo through the action of SerB. In addition, the proinflammatory properties of the SerB mutant also support the concept that acute inflammation is not a proximate cause of bone resorption (43, 55). Furthermore, neutrophil responses in this model system are associated with protection against bone loss. This notion is supported by the finding of increased periodontitis severity in congenital diseases such as leukocyte adhesion deficiency, Chediak-Higashi syndrome, Papillon-Lefèvre syndrome, and chronic/cyclic neutropenia, in which neutrophil recruitment is compromised (12). Moreover, IL-17 receptor-deficient mice show greater P. gingivalis-induced bone loss as a result of a defective ability to stimulate PMN migration (66). Impaired PMN activity may fail to adequately control the bacterial challenge or may disrupt cytokine networks that control bone formation and resorption.
An ELISA of rat sera showed that infection with either parent or mutant P. gingivalis induced elevated IgG antibody responses. However, the level of IgG induced by the SerB mutant was approximately 10-fold less than that induced by the parent strain. These results indicate that antibody responses to infection by P. gingivalis are not protective against bone loss induced by the infecting organisms. Similarly, previous studies in mice and rats have found that serum IgG to P. gingivalis does not protect against bone loss (2, 46, 61, 62). Furthermore, disease severity in humans can show a positive correlation with levels of IgG against P. gingivalis (6, 13, 52). In contrast, IgM and IgA responses to ΔSerB were higher than those to the parental strain. Elevated levels of IgA may be a reflection of the impaired ability of the SerB mutant to enter epithelial cells, and thus, these bacteria remain extracellular and capable of stimulating mucosa-associated lymphoid tissue. Higher IgM and lower IgG levels may also reflect an inability of the ΔSerB strain to penetrate the gingival tissues. The predominant IgG subclass response following infection with either the parental or mutant strain of P. gingivalis was IgG2b (T helper type 1 [Th1]) followed by IgG1 (Th2) and IgG2a (Th1), indicating stimulation of both Th1 and Th2 responses. Both Th1- and Th2-type responses have been reported in experimental animals infected with P. gingivalis and in humans with periodontal disease (10, 27, 28, 40, 41, 56), indicating that complex inflammatory and immune responses are involved in the progression of periodontal disease, and the balance between Th1 and Th2 activities may be important in determining whether the responses induced are protective or destructive.
Acknowledgments
We thank Sarah Whitmore for assistance with the manuscript.
This work was supported by grants DE11111, DE016509, and T32 DE007200 and a University of Florida Student Summer Research Fellowship.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 30 August 2010.
REFERENCES
- 1.Arbibe, L., D. W. Kim, E. Batsche, T. Pedron, B. Mateescu, C. Muchardt, C. Parsot, and P. J. Sansonetti. 2007. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat. Immunol. 8:47-56. [DOI] [PubMed] [Google Scholar]
- 2.Baker, P. J., S. Carter, M. Dixon, R. T. Evans, and D. C. Roopenian. 1999. Serum antibody response to oral infection precedes but does not prevent Porphyromonas gingivalis-induced alveolar bone loss in mice. Oral Microbiol. Immunol. 14:194-196. [DOI] [PubMed] [Google Scholar]
- 3.Baker, P. J., R. T. Evans, and D. C. Roopenian. 1994. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch. Oral Biol. 39:1035-1040. [DOI] [PubMed] [Google Scholar]
- 4.Byrne, S. J., S. G. Dashper, I. B. Darby, G. G. Adams, B. Hoffmann, and E. C. Reynolds. 2009. Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol. Immunol. 24:469-477. [DOI] [PubMed] [Google Scholar]
- 5.Colombo, A. V., C. M. da Silva, A. Haffajee, and A. P. Colombo. 2007. Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J. Periodontal Res. 42:236-243. [DOI] [PubMed] [Google Scholar]
- 6.Craig, R. G., R. Boylan, J. Yip, D. Mijares, M. Imam, S. S. Socransky, M. A. Taubman, and A. D. Haffajee. 2002. Serum IgG antibody response to periodontal pathogens in minority populations: relationship to periodontal disease status and progression. J. Periodontal Res. 37:132-146. [DOI] [PubMed] [Google Scholar]
- 7.Curtis, M. A., J. Aduse-Opoku, and M. Rangarajan. 2001. Cysteine proteases of Porphyromonas gingivalis. Crit. Rev. Oral Biol. Med. 12:192-216. [DOI] [PubMed] [Google Scholar]
- 8.Cutler, C. W., J. R. Kalmar, and C. A. Genco. 1995. Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis. Trends Microbiol. 3:45-51. [DOI] [PubMed] [Google Scholar]
- 9.Darveau, R. P., C. M. Belton, R. A. Reife, and R. J. Lamont. 1998. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 66:1660-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeCarlo, A. A., Y. Huang, C. A. Collyer, D. B. Langley, and J. Katz. 2003. Feasibility of an HA2 domain-based periodontitis vaccine. Infect. Immun. 71:562-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeVinney, R., O. Steele-Mortimer, and B. B. Finlay. 2000. Phosphatases and kinases delivered to the host cell by bacterial pathogens. Trends Microbiol. 8:29-33. [DOI] [PubMed] [Google Scholar]
- 12.Dixon, D. R., B. W. Bainbridge, and R. P. Darveau. 2004. Modulation of the innate immune response within the periodontium. Periodontol. 2000 35:53-74. [DOI] [PubMed] [Google Scholar]
- 13.Ebersole, J. L., and M. A. Taubman. 1994. The protective nature of host responses in periodontal diseases. Periodontol. 2000 5:112-141. [DOI] [PubMed] [Google Scholar]
- 14.Ekuni, D., T. Yamamoto, R. Yamanaka, K. Tachibana, and T. Watanabe. 2003. Proteases augment the effects of lipopolysaccharide in rat gingiva. J. Periodontal Res. 38:591-596. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, F. C. III, and C. A. Genco. 2007. Porphyromonas gingivalis mediated periodontal disease and atherosclerosis: disparate diseases with commonalities in pathogenesis through TLRs. Curr. Pharm. Des. 13:3665-3675. [DOI] [PubMed] [Google Scholar]
- 16.Graves, D. T., D. Fine, Y. T. Teng, T. E. Van Dyke, and G. Hajishengallis. 2008. The use of rodent models to investigate host-bacteria interactions related to periodontal diseases. J. Clin. Periodontol. 35:89-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grigoryants, V., K. K. Hannawa, C. G. Pearce, I. Sinha, K. J. Roelofs, G. Ailawadi, K. B. Deatrick, D. T. Woodrum, B. S. Cho, P. K. Henke, J. C. Stanley, M. J. Eagleton, and G. R. Upchurch. 2005. Tamoxifen up-regulates catalase production, inhibits vessel wall neutrophil infiltration, and attenuates development of experimental abdominal aortic aneurysms. J. Vasc. Surg. 41:108-114. [DOI] [PubMed] [Google Scholar]
- 18.Hajishengallis, G. 2009. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 11:637-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasegawa, Y., G. D. Tribble, H. V. Baker, J. J. Mans, M. Handfield, and R. J. Lamont. 2008. Role of Porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infect. Immun. 76:2420-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hintermann, E., S. K. Haake, U. Christen, A. Sharabi, and V. Quaranta. 2002. Discrete proteolysis of focal contact and adherens junction components in Porphyromonas gingivalis-infected oral keratinocytes: a strategy for cell adhesion and migration disabling. Infect. Immun. 70:5846-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holt, S. C., and J. L. Ebersole. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38:72-122. [DOI] [PubMed] [Google Scholar]
- 22.Holt, S. C., L. Kesavalu, S. Walker, and C. A. Genco. 1999. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20:168-238. [DOI] [PubMed] [Google Scholar]
- 23.Huang, G. T., H. B. Zhang, H. N. Dang, and S. K. Haake. 2004. Differential regulation of cytokine genes in gingival epithelial cells challenged by Fusobacterium nucleatum and Porphyromonas gingivalis. Microb. Pathog. 37:303-312. [DOI] [PubMed] [Google Scholar]
- 24.Hussain, H., P. Branny, and E. Allan. 2006. A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans. J. Bacteriol. 188:1628-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jandik, K. A., M. Belanger, S. L. Low, B. R. Dorn, M. C. Yang, and A. Progulske-Fox. 2008. Invasive differences among Porphyromonas gingivalis strains from healthy and diseased periodontal sites. J. Periodontal. Res. 43:524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji, S., Y. Kim, B. M. Min, S. H. Han, and Y. Choi. 2007. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria. J. Periodontal. Res. 42:503-510. [DOI] [PubMed] [Google Scholar]
- 27.Katz, J., K. P. Black, and S. M. Michalek. 1999. Host responses to recombinant hemagglutinin B of Porphyromonas gingivalis in an experimental rat model. Infect. Immun. 67:4352-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katz, J., and S. M. Michalek. 1998. Effect of immune T cells derived from mucosal or systemic tissue on host responses to Porphyromonas gingivalis. Oral Microbiol. Immunol. 13:73-80. [DOI] [PubMed] [Google Scholar]
- 29.Katz, J., D. C. Ward, and S. M. Michalek. 1996. Effect of host responses on the pathogenicity of strains of Porphyromonas gingivalis. Oral Microbiol. Immunol. 11:309-318. [DOI] [PubMed] [Google Scholar]
- 30.Katz, J., Q. B. Yang, P. Zhang, J. Potempa, J. Travis, S. M. Michalek, and D. F. Balkovetz. 2002. Hydrolysis of epithelial junctional proteins by Porphyromonas gingivalis gingipains. Infect. Immun. 70:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawai, T., R. Eisen-Lev, M. Seki, J. W. Eastcott, M. E. Wilson, and M. A. Taubman. 2000. Requirement of B7 costimulation for Th1-mediated inflammatory bone resorption in experimental periodontal disease. J. Immunol. 164:2102-2109. [DOI] [PubMed] [Google Scholar]
- 32.Kesavalu, L., S. Sathishkumar, V. Bakthavatchalu, C. Matthews, D. Dawson, M. Steffen, and J. L. Ebersole. 2007. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect. Immun. 75:1704-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kesavalu, L., B. Vasudevan, B. Raghu, E. Browning, D. Dawson, J. M. Novak, M. C. Correll, M. J. Steffen, A. Bhattacharya, G. Fernandes, and J. L. Ebersole. 2006. Omega-3 fatty acid effect on alveolar bone loss in rats. J. Dent. Res. 85:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klausen, B. 1991. Microbiological and immunological aspects of experimental periodontal disease in rats: a review article. J. Periodontol. 62:59-73. [DOI] [PubMed] [Google Scholar]
- 35.Krauss, J. L., J. Potempa, J. D. Lambris, and G. Hajishengallis. 2010. Complementary Tolls in the periodontium: how periodontal bacteria modify complement and Toll-like receptor responses to prevail in the host. Periodontol. 2000 52:141-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuboniwa, M., A. Amano, S. Shizukuishi, I. Nakagawa, and S. Hamada. 2001. Specific antibodies to Porphyromonas gingivalis Lys-gingipain by DNA vaccination inhibit bacterial binding to hemoglobin and protect mice from infection. Infect. Immun. 69:2972-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamont, R. J., and O. Yilmaz. 2002. In or out: the invasiveness of oral bacteria. Periodontol. 2000 30:61-69. [DOI] [PubMed] [Google Scholar]
- 40.Lappin, D. F., C. P. MacLeod, A. Kerr, T. Mitchell, and D. F. Kinane. 2001. Anti-inflammatory cytokine IL-10 and T cell cytokine profile in periodontitis granulation tissue. Clin. Exp. Immunol. 123:294-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leshem, O., S. S. Kashino, R. B. Goncalves, N. Suzuki, M. Onodera, A. Fujimura, H. Sasaki, P. Stashenko, and A. Campos-Neto. 2008. Th1 biased response to a novel Porphyromonas gingivalis protein aggravates bone resorption caused by this oral pathogen. Microbes Infect. 10:664-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, R., H. S. Bal, T. Desta, N. Krothapalli, M. Alyassi, Q. Luan, and D. T. Graves. 2006. Diabetes enhances periodontal bone loss through enhanced resorption and diminished bone formation. J. Dent. Res. 85:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, Y. C., U. H. Lerner, and Y. T. Teng. 2010. Cytokine responses against periodontal infection: protective and destructive roles. Periodontol. 2000 52:163-206. [DOI] [PubMed] [Google Scholar]
- 44.Malek, R., J. G. Fisher, A. Caleca, M. Stinson, C. J. van Oss, J. Y. Lee, M. I. Cho, R. J. Genco, R. T. Evans, and D. W. Dyer. 1994. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J. Bacteriol. 176:1052-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noiri, Y., K. Ozaki, H. Nakae, T. Matsuo, and S. Ebisu. 1997. An immunohistochemical study on the localization of Porphyromonas gingivalis, Campylobacter rectus and Actinomyces viscosus in human periodontal pockets. J. Periodontal Res. 32:598-607. [DOI] [PubMed] [Google Scholar]
- 46.Pathirana, R. D., N. M. O'Brien-Simpson, G. C. Brammar, N. Slakeski, and E. C. Reynolds. 2007. Kgp and RgpB, but not RgpA, are important for Porphyromonas gingivalis virulence in the murine periodontitis model. Infect. Immun. 75:1436-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajagopal, L., A. Clancy, and C. E. Rubens. 2003. A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J. Biol. Chem. 278:14429-14441. [DOI] [PubMed] [Google Scholar]
- 48.Rajapakse, P. S., N. M. O'Brien-Simpson, N. Slakeski, B. Hoffmann, and E. C. Reynolds. 2002. Immunization with the RgpA-Kgp proteinase-adhesin complexes of Porphyromonas gingivalis protects against periodontal bone loss in the rat periodontitis model. Infect. Immun. 70:2480-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rautemaa, R., A. Jarvensivu, K. Kari, J. Wahlgren, A. DeCarlo, M. Richardson, and T. Sorsa. 2004. Intracellular localization of Porphyromonas gingivalis thiol proteinase in periodontal tissues of chronic periodontitis patients. Oral Dis. 10:298-305. [DOI] [PubMed] [Google Scholar]
- 50.Rudney, J. D., R. Chen, and G. J. Sedgewick. 2005. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J. Dent. Res. 84:59-63. [DOI] [PubMed] [Google Scholar]
- 51.Rudney, J. D., R. Chen, and G. J. Sedgewick. 2001. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect. Immun. 69:2700-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakai, Y., H. Shimauchi, H. O. Ito, M. Kitamura, and H. Okada. 2001. Porphyromonas gingivalis-specific IgG subclass antibody levels as immunological risk indicators of periodontal bone loss. J. Clin. Periodontol. 28:853-859. [DOI] [PubMed] [Google Scholar]
- 53.Sathishkumar, S., A. Meka, D. Dawson, N. House, W. Schaden, M. J. Novak, J. L. Ebersole, and L. Kesavalu. 2008. Extracorporeal shock wave therapy induces alveolar bone regeneration. J. Dent. Res. 87:687-691. [DOI] [PubMed] [Google Scholar]
- 54.Socransky, S. S., and A. D. Haffajee. 1992. The bacterial etiology of destructive periodontal disease: current concepts. J. Periodontol. 63:322-331. [DOI] [PubMed] [Google Scholar]
- 55.Teng, Y. T. 2006. Protective and destructive immunity in the periodontium: part 1—innate and humoral immunity and the periodontium. J. Dent. Res. 85:198-208. [DOI] [PubMed] [Google Scholar]
- 56.Teng, Y. T. 2006. Protective and destructive immunity in the periodontium: part 2—T-cell-mediated immunity in the periodontium. J. Dent. Res. 85:209-219. [DOI] [PubMed] [Google Scholar]
- 57.Tomofuji, T., T. Yamamoto, N. Tamaki, D. Ekuni, T. Azuma, T. Sanbe, K. Irie, K. Kasuyama, M. Umakoshi, J. Murakami, S. Kokeguchi, and M. Morita. 2009. Effects of obesity on gingival oxidative stress in a rat model. J. Periodontol. 80:1324-1329. [DOI] [PubMed] [Google Scholar]
- 58.Tribble, G. D., and R. J. Lamont. 2010. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol. 2000 52:68-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tribble, G. D., S. Mao, C. E. James, and R. J. Lamont. 2006. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc. Natl. Acad. Sci. U. S. A. 103:11027-11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vankeerberghen, A., H. Nuytten, K. Dierickx, M. Quirynen, J. J. Cassiman, and H. Cuppens. 2005. Differential induction of human beta-defensin expression by periodontal commensals and pathogens in periodontal pocket epithelial cells. J. Periodontol. 76:1293-1303. [DOI] [PubMed] [Google Scholar]
- 61.Verma, R. K., I. Bhattacharyya, A. Sevilla, I. Lieberman, S. Pola, M. Nair, S. M. Wallet, I. Aukhil, and L. Kesavalu. 30 May 2010. Virulence of major periodontal pathogens and lack of humoral immune protection in a rat model of periodontal disease. Oral Dis. doi: 10.1111/j.1601-0825.2010.01678.X. [Epub ahead of print.] [DOI] [PubMed]
- 62.Verma, R. K., S. Rajapakse, A. Meka, C. Hamrick, S. Pola, I. Bhattacharyya, M. Nair, S. M. Wallet, I. Aukhil, and L. Kesavalu. 2010. Lack of humoral immune protection during Porphyromonas gingivalis and Treponema denticola infection in a rat model of periodontal disease. Interdiscip. Perspect. Infect. Dis. 2010:605125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia, Q., T. Wang, F. Taub, Y. Park, C. A. Capestany, R. J. Lamont, and M. Hackett. 2007. Quantitative proteomics of intracellular Porphyromonas gingivalis. Proteomics 7:4323-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yilmaz, O., K. Watanabe, and R. J. Lamont. 2002. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell. Microbiol. 4:305-314. [DOI] [PubMed] [Google Scholar]
- 65.Yilmaz, O., P. A. Young, R. J. Lamont, and G. E. Kenny. 2003. Gingival epithelial cell signalling and cytoskeletal responses to Porphyromonas gingivalis invasion. Microbiology 149:2417-2426. [DOI] [PubMed] [Google Scholar]
- 66.Yu, J. J., M. J. Ruddy, G. C. Wong, C. Sfintescu, P. J. Baker, J. B. Smith, R. T. Evans, and S. L. Gaffen. 2007. An essential role for IL-17 in preventing pathogen-initiated bone destruction: recruitment of neutrophils to inflamed bone requires IL-17 receptor-dependent signals. Blood 109:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, Y., T. Wang, W. Chen, O. Yilmaz, Y. Park, I. Y. Jung, M. Hackett, and R. J. Lamont. 2005. Differential protein expression by Porphyromonas gingivalis in response to secreted epithelial cell components. Proteomics 5:198-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zubery, Y., C. R. Dunstan, B. M. Story, L. Kesavalu, J. L. Ebersole, S. C. Holt, and B. F. Boyce. 1998. Bone resorption caused by three periodontal pathogens in vivo in mice is mediated in part by prostaglandin. Infect. Immun. 66:4158-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]